Abstract
Genetic structure and evolutionary patterns of the wild olive tree (Olea europaea L.) were investigated with AFLP fingerprinting data at three geographic levels: (a) phylogenetic relationships of the six currently recognized subspecies in Eurasia and Africa; (b) lineage identification in subsp. europaea of the Mediterranean basin; and (c) phylogeography in the western Mediterranean. Two statistical approaches (Bayesian inference and analysis of molecular variance) were used to analyse the AFLP fingerprints. To determine the congruency and transferability of results across studies previous RAPD and ISSR data were analysed in a similar manner. Comparisons proved that qualitative results were mostly congruent but quantitative values differed, depending on the method of analysis. Neighbour-Joining analysis of AFLP phenotypes supported current classification of subspecies. At a Mediterranean scale no clear cut phylogeographic pattern was recovered, likely due to extensive gene flow between populations of subsp. europaea. Gene flow estimates calculated with conventional F-statistics showed that reproductive barriers separated neither populations nor lineages of O. europaea. Genetic divergence between eastern and western parts of the Mediterranean basin was observed only when geographical and population information were incorporated into the analyses through hierarchical analysis of molecular variance (AMOVA). Within the western Mediterranean, the highest genetic diversity was found in two regions: on both sides of the Strait of Gibraltar and in the Balearic archipelago. Additionally, long-lasting isolation of the northern-most populations of the Iberian Peninsula appeared to be responsible for a significant divergence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
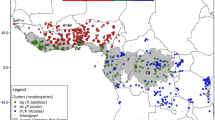
The genus Olea (Oleaceae) includes ca. 40 extant taxa distributed throughout Africa, Europe, Asia and Oceania, classified into three subgenera: Paniculatae, Tetrapilus and Olea. The latter subgenus consists of two sections, Olea and Ligustroides (Green 2002). The olive (Olea europaea L.) is the only representative of section Olea and is widely distributed in the Old World (Green and Wickens 1989). Olea europaea subsp. europaea is a distinctive element of the Mediterranean flora and serves to characterise both natural ecosystems (var. sylvestris Mill.) and agricultural landscapes (var. europaea). Five more subspecies are currently recognized (Green 2002): subsp. guanchica P. Vargas et al. (Canary Islands), subsp. cerasiformis G. Kunkel and Sundig (Madeira archipelago), subsp. maroccana (Greut. and Burd.) P. Vargas et al. (SW Morocco), subsp. laperrinei (Batt. and Trab.) Cif. (Sahara mountains) and subsp. cuspidata (Wall. ex G. Don) Cif. (S to NE Africa and SW Asia to China; Fig. 1). In recent years, attempts to clarify the taxonomic limits among the different forms of O. europaea have been undertaken, using both morphological (Green and Wickens 1989; Médail et al. 2001; Vargas et al. 2001; Green 2002) and molecular data (Angiolillo et al. 1999; Hess et al. 2000; Lumaret et al. 2000; Baldoni et al. 2002; Besnard et al. 2002a), but limited resolution was obtained. Molecular tools have also been exhaustively used to investigate the genetic diversity of cultivated and wild forms of subsp. europaea (Baldoni et al. 2002; Besnard et al. 2002b; Lumaret et al. 2000). As a result, there have been several attempts to pinpoint hotspots of present-day diversity and Pleistocene refugia around the Mediterranean basin (Besnard et al. 2002b; Lumaret and Ouazzani 2001; Lumaret et al. 2004). It was argued that phylogeographic patterns are elusive because of limited geographic barriers, the long history of olive cultivation (over 8,000 years; Zohary and Spiegel-Roy 1975) and the extensive hybridisation between wild and crop trees. In spite of these difficulties, two well-differentiated regions (eastern and western parts of the Mediterranean basin) and several cpDNA lineages were described (Besnard et al. 2002b; Lumaret et al. 2004). The most genetically diverse region corresponds to the western Mediterranean, notably the Iberian Peninsula and the Mahgreb (Besnard et al. 2002b; Lumaret and Ouazzani 2001; Lumaret et al. 2004). In these areas, some wild olive populations likely survived in situ throughout glaciations in protected riparian microenvironments (Terral et al. 2004), but precise locations of glacial refugia remain unknown.
To gain a better resolution, a fingerprint study was undertaken with amplified fragment length polymorphism (AFLP) markers. This technique has recently been found to provide the most reliable results when compared to other fingerprinting techniques (RAPD, ISSR, SSR; Nybom 2004). For the cultivated olive, a comparative study of three fingerprinting markers (RAPD, SSR and AFLP) has already been conducted (Belaj et al. 2003) to evaluate the capacity of the various techniques to discriminate cultivated genotypes. The two dominant marker systems RAPDs and AFLPs were found to be easily comparable, the latter being the most efficient (see also Owen et al. 2005).
In the present paper, we analysed AFLP data to address the following issues: (a) inference of phylogenetic relationships of the six subspecies currently recognized within the Olea europaea complex; (b) identification of major lineages in the wild olive (Olea europaea subsp. europaea); and (c) description of phylogeography and population structure of the wild olive in the W Mediterranean (Iberian Peninsula, Balearic islands and north-western Maghreb). In addition, we compared our AFLP data with data obtained previously from a similar sample with two other widely used fingerprinting techniques, RAPD (Besnard et al. 2001) and ISSR (Vargas and Kadereit 2001).
Materials and methods
Plant material, DNA extraction and AFLP fingerprinting
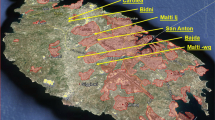
Plant material was collected in the field and total genomic DNA extracted from c. 0.5 g of silica-dried leaves using the Plant-DNeasy Minikit (QIAGEN Inc.) and the protocols provided by the manufacturer. A total of 182 individuals of the wild olive tree was chosen, based on previous studies (Besnard et al. 2001; Vargas and Kadereit 2001) to encompass high levels of taxonomic (subspecies) and biogeographic (population) diversity (see Fig. 1 and Electronic Supplementary Material Table S1).The AFLP procedure followed that of Vos et al. (1995) with the modifications of Schönswetter et al. (2004) and the primer combinations of Angiolillo et al. (1999).
Geographical distribution of the six currently recognised subspecies of Olea europaea L. and locations of the plant material examined. Numbers correspond to samples described in Table S1. a Distribution of subspp. europaea, laperrinei and cuspidata; b Distribution of subspp. cerasiformis, guanchica, maroccana and europaea (partial); c Accessions sampled in the Iberian Peninsula, the Balearic Islands and northern Morocco
Data analysis
Pairwise genetic distances among AFLP phenotypes were computed with Dice’s coefficient, using the expression of Nei and Li (1979) as implemented in PAUP 4b10* (Swofford 2000). The same software was used to construct Neighbour-Joining (NJ) trees, whose branch reliability was assessed by bootstrapping (10,000 replicates). Bayesian Analysis of Population Structure (BAPS v.3.1; Corander et al. 2003) was used to estimate the hidden structure by clustering individuals into panmictic groups. The partition with the highest log marginal likelihood was plotted onto the NJ tree.
Analyses of molecular variance AMOVA (Excoffier et al. 1992) were performed with ARLEQUIN, v 2.00 (Schneider et al. 2000) to hierarchically examine genetic structure. For a complementary approach, the AFLP data were analysed using Bayesian Inference with HICKORY v. 1.00 (Holsinger and Lewis 2003). HICKORY was run with the default parameters except for the burn-in period (50,000), number of generations (500,000) and thinning (100). Gene flow between populations and subspecies was assessed using Wright’s (1951) equation for the calculation of the number of migrants exchanged per generation: Nm = 0.25 [(1/F ST ) − 1].
Phylogenetic relationships in the Olea europaea complex
Naturalness of the different taxa and phylogenetic relationships in the O. europaea complex were investigated using 47 accessions: 21 of subsp. europaea, five of subsp. cerasiformis, five of subsp. guanchica, three of subsp. laperrinei, two of subsp. maroccana, six of subsp. cuspidata and five samples of taxa from sect. Ligustroides. Relationships among all these accessions were determined using NJ dendrograms and BAPS clusters. Genetic relationships among the different subspecies were additionally investigated using AMOVA and gene-flow estimates, the latter computed for every pair of subspecies.
Major lineages within O. europaea subsp. europaea
To determine major wild lineages of subsp. europaea with AFLPs a sample of 38 plants was used, including a subset of 16 accessions taken from the phylogeographic analysis of W Mediterranean (one from each population, see below) plus 22 accessions of representative individuals across the Mediterranean. Congruence between different fingerprinting markers was assessed by comparing the AFLP data with RAPD (Besnard et al. 2001) and ISSR data (Vargas and Kadereit 2001). NJ and BAPS reconstructions, AMOVA computed with ARLEQUIN and a Bayesian Inference analysis performed with HICKORY were used to explore genetic variation at a Mediterranean scale.
Phylogeography of the W Mediterranean
Phylogeographic analysis of the W Mediterranean was undertaken with AFLP profiles of samples from sixteen different populations. A total of 140 individuals were included in this analysis using three to ten individuals from each locality (Table S1, Electronic Supplementary Material). The resulting dataset was compared with RAPD data from Besnard et al. (2001). Population pairwise F ST obtained with ARLEQUIN were used to construct trees of populations with the NJ algorithm and to compute gene-flow estimates for both AFLPs and RAPDs. Distances between pairs of populations analysed with AFLPs were estimated with Nei’s (1978) unbiased genetic distance using POPGEN 1.32 (Yeh et al. 1998). Correlation between geographic and genetic structures within the W Mediterranean was explored by comparing the Cartesian geographic coordinates of the 16 populations analysed with AFLPs with the three-dimensional coordinate vectors obtained for each population by multidimensional scaling (PROXSCAL) of population pairwise F ST. This approach provides a rich source of information by enabling not only the detection of concordance between datasets but also the detection of any deviation from the overall trend. Large residuals indicate the points that do not match well in comparison to the overall trend. In our case, a close match between geographic and genetic differentiation (i.e. isolation by distance) would be an indication of neutral drift. Procrustean randomization analysis (10,000,000 randomizations) with PROTEST software was used for this comparison (Peres-Neto 2000; Peres-Neto and Jackson 2001).
Results
Phylogenetic relationships in the Olea europaea complex
A total of 788 AFLP fragments was scored. The data subset of 47 samples included representatives of all O. europaea subspecies. The phylogenetic reconstruction obtained with the resulting matrix of 47 × 788 (Fig. 2) revealed O. europaea as monophyletic (100% bootstrap support, bs). All subspecies within the complex were resolved as monophyletic groups. Three of them (cerasiformis, laperrinei and maroccana) had high support (> 90% bs), whereas subspecies cuspidata, europaea and guanchica displayed low support. The partition with the highest log marginal likelihood (−7909.6) produced by BAPS consisted of three clusters: one for the samples belonging to sect. Ligustroides (outgroup), a second one for all the accessions but one of subsp. cuspidata plus one sample of subsp. guanchica and a third one including all other accessions (Fig. 2). AMOVA results also supported genetic differentiation between subspecies, although most of the genetic variance was maintained within each of the subspecies (F ST = 20%, P < 0.001; Table 1). An estimation of gene flow among subspecies computed using conventional F-statistics (Table S2, Electronic Supplementary Material) revealed that the different taxa seem to have exchanged a considerable number of migrants (mean Nm ≅ 0.92). However, these results have to be interpreted cautiously, as sampling was unbalanced among subspecies and sample sizes were small in every case.
Neighbour Joining analysis of 47 individuals based on Nei and Li’s (1979) genetic distances. Numbers next to nodes indicate bootstrap support after 10,000 replicates. Every subspecies is represented by at least two individuals. The 21 individuals of subsp. europaea are a subset of samples displayed in Fig. S1 of Electronic Supplementary Material. Shaded rectangles to the right of the tree indicate the clustering with the highest log marginal likelihood according to the Bayesian analysis of population structure performed with BAPS v.3.1 (Corander et al. 2003) to estimate hidden structure by clustering individuals into panmictic groups (K = 3; log marginal likelihood = −17,909.6). The six samples marked in boldface are common to the three analyses. (a) Samples common only to AFLP and RAPD analysis
Major lineages within O. europaea subsp. europaea
The 38 samples used to determine lineage relationships in the Mediterranean basin rendered a matrix of 38 × 660. Results of the analysis of this matrix are given in Table 1 and Fig. S1 (Electronic Supplementary Material). Apart from the ingroup, which always appeared as monophyletic with low to moderate support, no major AFLP lineages could be established with confidence since significant support values were obtained only for a few tip branches in the AFLP reconstruction (S1a), while no support > 50% is observed neither for RAPDs (S1b) nor for ISSRs (S1c). BAPS analyses also failed to reveal any pattern of geographic relatedness among the analysed samples, partitions with highest log marginal likelihood were k = 2 (−2033.74), k = 4 (−933.5) and k = 4 (−2815.9) for AFLPs, RAPDs and ISSRs, respectively. Moreover, if BAPS was run with K (maximum number of groups) = 2, the grouping did not correspond with a split between the W and E Mediterranean or any other geographical regions (results not shown). Therefore, no unequivocal differentiation at a Mediterranean scale was found with any of the three fingerprinting data sets. However, when RAPD data were analysed with AMOVA, differences between the W and E Mediterranean accounted for 18.2% of the total variation (P < 0.001). Most RAPD variation was found within populations (67.3%) while there was less variation among populations (14.51%). The results of this analysis differed to some extent from those obtained previously on the same data set. While Besnard et al. (2001) estimated the degree of inter-population differentiation in the Mediterranean to be 24% with the OPEP software (Baradat and Labbé 1995), we obtained values of 24.6% with HICKORY (θ B) and 32.7% with ARLEQUIN (F ST). Thus, the two algorithms that do not consider fingerprinting patterns as haplotypes (OPEP and HICKORY) yield almost identical results, whilst ARLEQUIN (AMOVA) gives a higher F ST value.
Phylogeography in the W Mediterranean
The AFLP data matrix used for the phylogeographic analysis of the western Mediterranean consisted of 140 samples × 670 fragments. The genetic structure inferred from the analysis of this matrix is described in Fig. S2 (Electronic Supplementary Material) and Table 1. Major results of AFLP data were similar to those obtained with RAPDs. In both cases population differentiation appeared to be significant (P < 0.001; Deviance Information Criterion [DIC; Spiegelhalter et al. 2002] values of the full model more than six units smaller than that of the θ B = 0). Differences in DIC values in the case of AFLPs were due to the full model’s better average fit, but in the case of RAPD data they were mainly due to the smaller number of estimated parameters (Holsinger and Wallace 2004). Differences due to among-population variation explained between 15% (RAPD) and 20% (AFLP) of the total variation (Table 1). Two NJ dendrograms were constructed based on population pairwise F ST’s computed with ARLEQUIN, one for AFLP (S2a) and one for RAPD phenotypes (S2b). Both showed the singularity of the Cantabrian population from northern Spain, 47 San Antolín SP (Asturias) in the AFLP analysis and Asturias SP in the RAPD analysis. According to the AFLP tree, these Cantabrian plants formed the genetically most distant population from any other included in the data set (total Nei’s unbiased genetic distance = 0.096) and were related to populations located in the vicinity of the Strait of Gibraltar (20 Alcalá de los Gazules SP). Two other populations from both sides of the Strait of Gibraltar (29 Chef Chaouen MOR, 21 Almoraima SP) were genetically different from each other and from the cluster described above. High genetic differentiation is observed between any two pairs of populations situated in the vicinity of the Strait of Gibraltar except for 38 Heracles Cave MOR and 48 Tetouan MOR. The three populations from the Balearic Islands (CAB, MLL and MN) were also genetically distant to each other, in spite of their geographic proximity.
Procrustean randomization tests revealed a strong correlation between geographic and genetic structures within W Mediterranean (m 2 = 0.923; P < 0.001). The residuals for the three dimensions of the analysis (Table S3 included in the Electronic supplementary material) revealed that the populations that show a higher deviation from geographic patterns and thus from neutral drift are 29 Chef Chaouen MOR (0.484), 44 Cala Grande MLL (0.437) and 38 Heracles Cave MOR (0.419). Nei’s unbiased genetic distances and population pairwise Nm computed with conventional F-statistics are also displayed in Table S3. The estimated number of migrants (Nm) exchanged between pairs of populations indicated a high degree of gene flow among populations, with values ranging from 0.3 (47 San Antolín SP and 29 Chef Chaouen) to 8.5 (38 Heracles Cave MOR and 48 Tetouan MOR), with a mean Nm of 1.372.
Discussion
Congruency of major AFLP lineages and taxonomy
Phylogenetic reconstruction based on AFLP data supported current taxonomic treatment of the Olea europaea complex and its monophyly (Green 2002; Fig. 2). Genetic uniqueness, geographic remoteness and morphological differentiation of subsp. cerasiformis suggest a single, ancient colonization of the Madeiran archipelago. Phylogeographic relationships between the populations of the two islands of the Madeiran archipelago (Fig. 2) indicate young-to-old inter-island dispersal from Madeira to Porto Santo in spite of the remarkable time differences between island formation (Madeira, 5 Ma; Porto Santo 18 Ma). The low support in the NJ reconstruction coupled with distinct BAPS clusters and heterogeneity in plastid haplotypes (Besnard, Rubio de Casas and Vargas, submitted) seems to reflect a more complex pattern within the Canarian subsp. guanchica, as it had been previously hypothesized (Hess et al. 2000).
The two north-African endemics (subsp. laperrinei and maroccana) were resolved as sister taxa in the NJ tree (75% bs) in accordance with previous fingerprinting data (Angiolillo et al. 1999; Hess et al. 2000; Besnard et al. 2001) and ITS sequences (Besnard, Rubio de Casas and Vargas, submitted). Conversely, mitochondria and plastid haplotypes place the two Saharan taxa in independent lineages (Besnard et al. 2002b): subsp. maroccana with subsp. guanchica, and subsp. laperrinei with E Mediterranean populations of subsp. europaea. The incongruence between nuclear and organelle DNA might be due to hybridisation during waning and waxing of African lineages following climatic fluctuations. This hypothesis is supported by present-day geographic distribution in an east-to-west sequence (europaea – laperrinei - maroccana - guanchica) and by palaeobotanical records (Médail et al. 2001). We hypothesize that hybridisation was more likely to occur in the past within the taxa pairs europaea – laperrinei and guanchica – maroccana than across them. Morphological characters are also inconclusive, although certain differences are observed. While leaf morphology of subspp. laperrinei and maroccana overlaps considerably (Green 2002), width of endocarp walls discriminates both taxa: individuals of subsp. laperrinei have thinner endocarp walls (0.24–0.78 mm) than those of subsp. maroccana (0.70–2.13 mm), europaea (0.69–1.83 mm), cuspidata (0.52–2.64 mm), cerasiformis (1.20–2.31 mm), and guanchica (0.82–2.31 mm) (authors’ unpublished data).
Topological resolution of the NJ dendrogram and BAPS clustering of AFLP data revealed that the most widespread subspecies of O. europaea (subsp. cuspidata) is distinct from the other five subspecies and contains divergent lineages (Fig. 2). It had already been suggested that Asian and African populations may in fact represent different taxa (Green and Wickens 1989, but see Green 2002). However, lineages shown by our reconstructions do not clearly correspond to geographic regions, and African plants are intermingled with Asian ones. This may be due to the small sample size but it can also be related to gene-flow blurring the limits between different areas (Besnard et al. 2002b).
When the number of migrants exchanged per generation (Nm) between the subspecies was estimated using F-statistics, results proved that gene flow is taking place (Electronic Supplementary Material Table S2) as anticipated by several authors (Besnard et al. 2002b; Lumaret et al. 2004). The method employed for Nm computations has drawbacks (Whitlock and McCauley 1999) but we nevertheless consider that our estimates provide reliable support to the hypothesis that there is little genetic isolation between O. europaea taxa. Genetic barriers do not appear to be significant, neither between cultivated (var. europaea) and wild forms (var. sylvestris) (Contento et al. 2002) nor between subspecies, since fertile hybrids are observed in the wild (authors’ personal. observation.) and obtained in cultivation (Besnard et al. 2001). Geographic isolation rather than fertility barriers appears to be responsible for the differentiation patterns deduced in O. europaea.
Weak phylogeographic structure in the olive tree across the Mediterranean
The different data sources (ISSR, RAPD and AFLP) used to investigate the existence of independent lineages in the wild olive across the Mediterranean basin failed to reveal any clear-cut pattern. Although it has been suggested that E and W areas of the Mediterranean harbour disparate olive tree lineages (Besnard et al. 2001; 2002b; Lumaret et al. 2004), NJ and BAPS clustering of AFLP, RAPD and ISSR phenotypes rendered limited resolution and support (Fig. S1). This is not a surprising result since the cultivation of the olive has been widespread across the Mediterranean for several thousands of years (Zohary and Spiegel-Roy 1975) and, additionally, any gap between populations may be bridged by long distance gene flow. The olive tree has been proved to be predominantly self-incompatible, sometimes as a consequence of male sterility (Besnard et al. 2000), which promotes outcrossing between wild and cultivated individuals. Olive fruits are very widely eaten by birds and other animals (Rey and Alcántara 2000) enabling long distance-dispersal and making exchange of migrants common even between distant regions. Pollen circulation in O. europaea can also occur over long distances (Damialis et al. 2005) enhancing lineage admixture. Indeed, our gene flow estimations (Table S3) demonstrated that reproductive isolation is highly improbable and that genetic material seems to be exchanged frequently among populations.
Analysis of genetic structure of W Mediterranean populations
In order to pinpoint wild olive populations from the Iberian Peninsula, northern Morocco and the Balearic Islands harbouring high genetic diversity, we used AFLP data and re-analyzed the RAPD data from Besnard et al. (2001). Analysis of molecular variance and F ST dendrograms (Table 1, Tables S2 and Fig. S2) showed significant genetic differentiation between populations of O. europaea. We tested the transferability of results between studies based on different dominant markers by comparing AFLP and RAPD data. F ST estimates provided by HICKORY and ARLEQUIN are similar for each technique, albeit somewhat lower for RAPD analysis. Population differentiation revealed by AFLPs and RAPDs is stronger than that indicated by allozyme analysis (Lumaret et al. 2004), which appears to be a common pattern (Chung et al. 2004).
The AFLP results revealed two areas of high genetic diversity. One is located nearby the Strait of Gibraltar and the second one within the Balearic archipelago. In these two areas, O. europaea populations show a degree of genetic differentiation that is not proportional to geographic distances among them (Table S3, Figs. 1 and S2). Genetic differences among populations from the three Balearic islands included in our analysis may reflect the existence of a centre of genetic diversity for O. europaea in this archipelago, as already reported for Quercus (López de Heredia et al. 2005).
One population from the Cantabrian coast in northern Spain (48 San Antolín, Asturias) is particularly distinct from all other investigated populations according to both AFLP and RAPD data. The northern third of the Iberian Peninsula does not form part of the Mediterranean floristic region but belongs to the Eurosiberian region, where the occurrence of the olive tree is exceptional (Vargas and Kadereit 2001; Rubio de Casas et al. 2002). The NJ dendrogram of population pairwise F ST (Fig. S2) and Nei’s unbiased genetic distances (Table S3) for both AFLP and RAPD data show no close phylogeographic relationships between the Cantabrian plants and any other Iberian population. Ecological and phylogeographic singularity make Cantabrian populations of O. europaea a stock worthy of further genetic investigation.
Conclusions
Reproductive characteristics of O. europaea account for extensive gene flow among lineages. Recurrent genetic exchange, however, has not blurred a phylogenetic pattern resulting in a highly diverse group with multiple monophyletic taxa. Our results indicate monophyly of the O. europaea complex and moderate to high congruence between AFLP lineages and taxonomic circumscription of the six subspecies currently recognized: cuspidata, cerasiformis, europaea, guanchica, laperrinei and maroccana. However, the phylogenetic relationships between these taxa are not completely clear. Moreover, subsp. cuspidata appears to comprise divergent lineages displaying limited morphological and geographical differentiation.
In contrast to the phylogenetic signal obtained, extensive gene flow makes reconstruction of phylogeographic patterns difficult, although significant differences between E and W areas of the Mediterranean were retrieved by hierarchical AMOVA of RAPD data (Table 1). Within W Mediterranean, the highest diversity is found in two regions, the Strait of Gibraltar and the Balearic archipelago. Additionally, we confirmed the occurrence of singular olive genotypes in the Eurosiberian region of the Iberian Peninsula along the Cantabrian coast.
Comparison of results obtained with different fingerprinting techniques proved that, even if major conclusions are qualitatively similar, quantitative values might differ depending on genetic assumptions and the mathematical methods used to compute population structure parameters. We consider that this should be a call for cautiousness when contrasting results across studies.
References
Angiolillo A, Mencuccini M, Baldoni L (1999) Olive genetic diversity assessed using amplified fragment length polymorphisms. Theor Appl Genet 98:411–421
Baldoni L, Guerrero C, Sossey-Aloui K, Abbott AG, Angiolillo A, Lumaret R (2002) Phylogenetic relationships among Olea species based on nucleotide variation at non-coding chloroplast DNA region. Plant Biol 4:346–351
Baradat P, Labbé T (1995) OPEP: un logiciel intégré pour l’amélioration des plantes pérennes. In: Traitements statistiques des essais de sélection. Stratégies de sélection des plantes pérennes, CIRAD-CP, Montpellier, pp 303–330
Belaj A, Satovic Z, Cipriani G, Baldoni L, Testolin R, Rallo L, Trujillo I (2003) Comparative study of the discriminating capacity of RAPD, AFLP and SSR markers and of their effectiveness in establishing genetic relationships in olive. Theor Appl Genet 107:736–744
Besnard G, Baradat P, Chevalier D, Tagmount A, Bervillé A (2001) Genetic differentiation in the olive complex (Olea europaea) revealed by RAPDs and RFLPs in the rDNA genes. Genet Resour Crop Evol 48:165–182
Besnard G, Green PS, Bervillé A (2002a) The genus Olea: molecular approaches of its structure and relationships to other Oleaceae. Acta Bot Gallica 149:49–66
Besnard G, Khadari B, Baradat P, Bervillé A (2002b) Olea europaea (Oleaceae) phylogeography based on chloroplast DNA polymorphism. Theor Appl Genet 104:1353–1361
Besnard G, Khadari B, Villemur P, Bervillé A (2000) Cytoplasmic male sterility in the olive (Olea europaea L.). Theor Appl Genet 100:1018–1024
Contento A, Ceccarelli M, Gelati MT, Maggini F, Baldoni L, Cionini PG (2002) Diversity of Olea genotypes and the origin of cultivated olives. Theor Appl Genet 104:1229–1238
Corander J, Waldmann P, Sillanpaa MJ (2003) Bayesian analysis of genetic differentiation between populations. Genetics 163:367–374
Chung JD, Lin TP, Tan YC, Lin MY, Hwang SY (2004) Genetic diversity and biogeography of Cunninghamia konishii (Cupressaceae), an island species in Taiwan: a comparison with Cunninghamia lanceolata, a mainland species in China. Mol Phylogenet Evol 33:791–801
Damialis A, Gioulekas D, Lazopoulou C, Balafoutis C, Vokou D (2005) Transport of airborne pollen into the city of Thessaloniki: the effects of wind direction, speed and persistence. Int J Biometeorol 49:139–145
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Green PS (2002) A revision of Olea L. (Oleaceae). Kew Bull 57:91–140
Green PS, Wickens GE (1989) The Olea europaea complex. In: Tan K (ed) The Davis and Hedge Festschrift. Edinburgh University Press, Edinburgh, pp 287–299
Hess J, Kadereit JW, Vargas P (2000) The colonization history of Olea europaea L. in Macaronesia based on internal transcribed spacer 1 (ITS-1) sequences, randomly amplified polymorphic DNAs (RAPD), and intersimple sequence repeats (ISSR). Mol Ecol 9:857–868
Holsinger KE, Lewis PO (2003) HICKORY: a package for analysis of population genetic data V. 1.0, University of Connecticut
Holsinger KE, Wallace LE (2004) Bayesian approaches for the analysis of population genetic structure: an example from Platanthera leucophaea (Orchidaceae). Mol Ecol 13:887–894
López de Heredia U, Jiménez P, Díaz Fernández P, Gil L (2005) The Balearic Islands: a reservoir of cpDNA genetic variation for evergreen oaks. J Biogeogr 32:939–949
Lumaret R, Amane M, Ouazzani N, Baldoni L, Debain C (2000) Chloroplast DNA variation in the cultivated and wild olive taxa of the genus Olea L. Theor Appl Genet 101:547–553
Lumaret R, Ouazzani N (2001) Ancient wild olives in Mediterranean forests. Nature 413:700
Lumaret R, Ouazzani N, Michaud H, Vivier G, Deguilloux M-F, Di Giusto F (2004) Allozyme variation of oleaster populations (wild olive tree) (Olea europaea L.) in the Mediterranean Basin. Heredity 92:343–352
Médail F, Quézel P, Besnard G, Khadari B (2001) Systematics, ecology and phylogeographic significance of Olea europaea L. ssp. maroccana (Greuter and Burdet) P. Vargas et al, a relictual olive tree in south-west Morocco. Bot J Linn Soc 137:249–266
Nei M (1978) Estimation of average heterozygosity and genetic distances from a small number of individuals. Genetics 89:583–590
Nei M, Li W-H (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155
Owen CA, Bita EC, Banilas G, Hajjar SE, Sellianakis V, Aksoy U, Hepaksoy S, Chamoun R, Talhook SN, Metzidakis I, Hatzopoulos P, Kalaitzis P (2005) AFLP reveals structural details of genetic diversity within cultivated olive germplasm from the Eastern Mediterranean. Theor Appl Genet 110:1169–1176
Peres-Neto PR (2000) PROTEST. Department of Zoology, University of Toronto
Peres-Neto PR, Jackson DA (2001) How well do multivariate data sets match? The advantages of a Procrustean superimposition approach over the Mantel test. Oecologia 129:169–178
Rey PJ, Alcántara JM (2000) Recruitment dynamics of a fleshy fruited plant (Olea europaea): connecting patterns of seed dispersal to seedling establishment. J Ecol 88:622–633
Rubio de Casas R, Balaguer L, Manrique E, Pérez-Corona ME, Vargas P (2002) On the historical presence of the wild olive Olea europaea L. var. sylvestris (Miller) Lehr. in the Eurosiberian North of the Iberian Peninsula. An Jard Bot Madr 59:342–344
Schneider S, Roessli D, Excoffier L (2000) ARLEQUIN ver. 2.00: A software for population genetics data analysis, University of Geneva, Geneva, Switzerland
Schönswetter P, Paun O, Tribsch A, Niklfeld H (2003) Out of the Alps: colonization of Northern Europe by East Alpine populations of the Glacier Buttercup Ranunculus glacialis L. (Ranunculaceae). Mol Ecol 12:3373–3381
Spiegelhalter DJ, Best NG, Carlin BP, van der Linde A (2002) Bayesian measures of model complexity and fit. J R Stat Soc B 64:483–689
Swofford DL (2000) PAUP*, Phylogenetic analysis using Parsimony*and other methods, Sinauer, Sunderland, MA
Terral JF, Badal E, Heinz C, Roiron S, Thiebault S, Figueiral I (2004) A hydraulic conductivity model points to post-neogene survival of the mediterranean olive in riparian habitat. Ecology 85:3158–3165
Vargas P, Kadereit JW (2001) Molecular fingerprinting evidence (ISSR, Inter-simple sequence repeats) for a wild status of Olea europaea L. (Oleaceae) in the Eurosiberian North of the Iberian Peninsula. Flora 196:142–152
Vargas P, Muñoz Garmendia F, Hess J, Kadereit JW (2001) Olea europaea ssp guanchica and ssp maroccana (Oleaceae), two new names for olive tree relatives. An Jard Bot Madr 58:360–361
Vos P, Hogers R, Bleeker M, Reijans M, Van de Lee T, Hornes M, Fritjers A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:7213–7218
Whitlock MC, McCauley DE (1999) Indirect measures of gene flow and migration: Fst not 1/(4 Nm+1). Heredity 82:117–125
Wright S (1951) The genetical structure of populations. Ann Eugenet 15:323–324
Yeh F, Rong-Cai C, Boyle T (1998) POPGENE V. 1.31: Microsoft-based freeware for population genetic analysis. University of Alberta, Edmonton, Canada
Zohary D, Spiegel-Roy P (1975) Beginnings of fruit growing in the Old World. Science 187:319–327
Acknowledgments
We sincerely thank Esther Pérez Corona, Gerald Schneeweiss and Carlos García-Verdugo for their valuable comments. Esteban Manrique, Virginia Valcárcel, Omar Fiz and Jorge Martínez helped the authors in the field and Gudrun Kohl provided technical assistance. The research was financed by the Spanish Ministerio de Ciencia y Tecnología (REN2001-1502-C03-03) and Fundación Biodiversidad (Fundación Parques Nacionales, UCAM2005030026, Ministerio de Medio Ambiente) and Junta de Andalucía (2004X963).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by O. Savolainen.
Rights and permissions
About this article
Cite this article
Rubio de Casas, R., Besnard, G., Schönswetter, P. et al. Extensive gene flow blurs phylogeographic but not phylogenetic signal in Olea europaea L.. Theor Appl Genet 113, 575–583 (2006). https://doi.org/10.1007/s00122-006-0306-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-006-0306-2