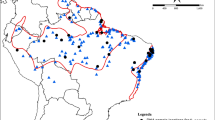
Abstract
The Rufous Gnateater, Conopophaga lineata, is a small insectivorous understory bird which is endemic to and widely distributed in the tropical and subtropical Atlantic forest of South America. Its distribution makes it ideally suited for testing two major hypotheses for the origin of biodiversity, namely, the riverine barrier and the forest refuge hypotheses. In this study, we sequenced mitochondrial (control region) and nuclear markers (intron 5 of the β-fibrinogen gene) for individuals distributed in the southern Atlantic forest and obtained a strong genetic structure with one clear discontinuity in northern Brazilian state of São Paulo. We consistently detected signals of demographic expansion for both markers, with estimates indicating that expansion started in the Late Pleistocene (250,000 years ago), suggesting that the forest refuge hypothesis potentially explains Rufous Gnateater’s diversification. We also found evidence of gene flow between populations from each side of this discontinuity, with a possible secondary contact zone occurring in the states of Minas Gerais, São Paulo, and Rio de Janeiro.
Zusammenfassung
Die populationsgenetische Struktur des in der Mata Atlântica endemischen Rotkehl-Mückenfressers ( Conopophaga lineata , Passeriformes: conopophagidae) lässt eine Kontaktzone in der Mata Atlântica erkennen
Der Rotkehl-Mückenfresser, Conopophaga lineata, ist ein kleiner insektivorer Vogel, der im Unterholz der tropischen und subtropischen Mata Atlântica in Südamerika endemisch und weit verbreitet ist. Die Verbreitung des Rotkehl-Mückenfressers macht ihn besonders dafür geeignet, zwei Haupthypothesen zum Ursprung von Biodiversität zu testen, die „Flüsse als Barrieren“-Hypothese (“riverine barrier hypothesis”) und die „Wälder als Rückzugsgebiete“-Hypothese (“forest refuges hypothesis”). Wir haben mitochondriale Marker (Kontrollregion) und Zellkernmarker (Intron 5 von β-Fibrinogen) für in der südlichen Mata Atlântica verbreitete Individuen sequenziert und eine ausgeprägte genetische Struktur mit einer klaren Diskontinuität im Norden des Bundesstaates São Paulo gefunden. Für beide Marker haben wir durchweg Signale demographischer Ausbreitung entdeckt, die schätzungsweise im späten Pleistozän (vor 250000 Jahren) begann, was darauf hindeutet, dass die „Wälder als Rückzugsgebiete“-Hypothese einen möglichen Einflussfaktor bei der Diversifikation des Rotkehl-Mückenfressers darstellt. Wir haben auch Hinweise auf Genfluss zwischen Populationen auf beiden Seiten der Diskontinuität gefunden, mit einer möglichen sekundären Kontaktzone in den Bundesstaaten Minas Gerais, São Paulo und Rio de Janeiro.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Rufous Gnateater (Conopophaga lineata; Weir and Schluter 2008) (Conopophagidae: Passeriformes) is a small insectivorous understory bird, endemic to the Atlantic forest (Sick 1997; Whitney 2003). This species is distributed in tropical and subtropical forests of South America (Sigrislt 2005), from Paraguay and northeast Argentina to northeast Brazil (Sick 1997; Whitney 2003). This biome is considered to be one of the most important areas for conservation in the world, due to its high biodiversity, high levels of endemism, and degradation (Ribeiro et al. 2009). Although approximately 11 % of the primary forest still remains—in a highly fragmented state (Ribeiro et al. 2009)—new species are still being discovered in the area (Weber et al. 2005; Miranda et al. 2006). Its biota is the result of a complex evolutionary history, but the processes shaping it are poorly understood (Mustrangi and Patton 1997; Costa et al. 2000; Geise et al. 2001; Pellegrino et al. 2005). The evolutionary processes involved in the formation of the Atlantic forest can be indirectly studied based on phylogeographic analyses of endemic species. In addition, knowledge of the range limits of lineages and species is fundamental for the effective conservation of biodiversity (Moritz 2002).
There are several hypotheses for the origin of extreme biodiversity in tropical forests. The riverine barrier hypothesis (Wallace 1852) predicts that some rivers limit regional faunas and therefore may represent gene flow barriers, reshaping much of the biota’s distribution. In the Atlantic forest, the Doce river (Costa et al. 2000; Silva et al. 2004; Brown 2005), Jequitinhonha river (Lacerda et al. 2007), São Francisco river, and Paraíba do Sul river (Silva and Straube 1996; Pellegrino et al. 2005) may be barriers to dispersal for many animals. Additionally, the tectonic activity associated with the formation of such geographical landmarks could also have influenced the distribution of biodiversity, mainly in the southern Atlantic forest where a complex relief exists, with many mountain ranges and valleys, as also in the region of Serra do Mar (Petri and Fulfaro 1983; Riccomini et al. 1989; Cabanne et al. 2008).
Isolation in forest refugia during the cycles of climate change that occurred during the Pleistocene may also have contributed to diversification, mainly in the Amazon basin (Haffer 1969; Vanzolini and Williams 1970; Brown and Ab’Saber 1979; Haffer and Prance 2001). The Pleistocene refuge hypothesis proposes that during glacial maxima, rainforests were reduced to patches (refugia) isolated by open areas and that organisms isolated in these refugia diverged into new species by drift and/or divergent selection. In the following interglacial period, the forest expanded, resulting in secondary contact among the now reproductively isolated new clades. Brown and Ab’Saber (1979) proposed that open areas dominated the current Atlantic forest’s landscape during the maximum of the Late Pleistocene glaciations, suggesting that the refuge hypothesis could be very useful for gaining a better understanding of the biological diversification of the biome.
A paleoclimate model for the distribution of the Atlantic forest predicted that there were two large and stable forest refugia at the last glacial maxima during the Pleistocene—one in the state of Pernambuco, north of the Atlantic forest, and another one in the state of Bahia (Carnaval and Moritz 2008). According to this model, throughout the Pleistocene, the forest was more stable in the north region than in the southern region, i.e., south of the Doce river, where several smaller forest refugia occur. This model is partially compatible with phylogeographic or population genetic data on Passeriformes (Cabanne et al. 2007, 2008; d’Horta et al. 2011; Maldonado-Coelho 2012), bats (Pavan et al. 2011), lizards (Pellegrino et al. 2005), and frogs (Carnaval et al. 2009; Amaro et al. 2012).
Few studies have investigated the evolutionary history of the Rufous Gnateater, despite its large distribution, and remarkable geographical variation in song and plumage (Willis et al. 1983; Whitney 2003; Dantas et al. 2007; Dantas et al. 2009). Three subspecies are recognized: Conopophaga lineata (C. l.) cearae is found in northeastern Brazil, from Ceará to Pernambuco; C. l. lineata occurs from Pernambuco to southern Bahia, Central Brazil, and in southern Goiás and northern Mato Grosso do Sul; C. l. vulgaris is found in southeastern Brazil, from southern Espírito Santo to Rio Grande do Sul, and in eastern Paraguay, Misiones, and eastern Uruguay (Whitney 2003). A phylogeny of the genus Conopophaga based on mitochondrial data indicates that C. l. cearae does not form a monophyletic clade with the other two subspecies (C. l. lineata and C. l. vulgaris; Pessoa 2007). C. lineata is a species which is endemic to the Atlantic forest, and as such it represents a good model for evolutionary studies in this biome. The Atlantic forest is distributed along eastern Brazil, eastern Paraguay, and north-eastern Argentina (Gusmão Câmara 2003).
In this context, the aim of our study is to evaluate the phylogeography of the Rufous Gnateater in the Atlantic forest, based on mitochondrial (the “Control” region) and nuclear [intron 5 of the β-fibrinogen [FIB5)] DNA sequences, in order to evaluate if the previous gene flow barriers described for other vertebrates in the Atlantic forest could also have influenced this species. First, if rivers were important barriers to gene flow for the Rufous Gnaeater, we would expect to find breaks concordant with river positions, i.e., a presence of low or no gene flow between the two sides of the river. Alternatively, if forest refuges did act as gene flow barriers for the Rufous Gnaeater, we would expect to find more than one break; this would not necessarily be concordant with river positions, but would be associated to a clear signal of population expansion due to the expansion of the forest after the glacial period.
Methodology
Study area and samples
Samples (blood or muscle, N = 146; see "Appendix") were collected between 1999 and 2009 in the Brazilian states of Minas Gerais, Rio de Janeiro, São Paulo, Paraná, Santa Catarina, and Rio Grande do Sul (Fig. 1). Blood was collected (approximately 0.1 mL) from the largest vein in the right cervical region using insulin syringes, and muscle was obtained from specimens, which were deposited at the Museu de Zoologia da Universidade de São Paulo and the Taxonomic Collection Center at the Universidade Federal de Minas Gerais. Tissue samples have been deposited at the Laboratório de Biodiversidade e Evolução Molecular (Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Brazil) or at the Laboratório de Genética e Evolução Molecular de Aves (Instituto de Biociências, Universidade de São Paulo, Brazil). Total DNA was obtained from blood or muscle samples by a conventional proteinase K–sodium dodecyl sulfate digestion, organic extraction with phenol–chloroform, and ethanol precipitation (Bruford et al. 1992).
Sequences of the control region
The mitochrondrial (mt)DNA of the Control region was amplified with primers L1 (5′-TCCACACTCGACATCTCATT-3′; designed by E.H. Sari for this work) and H16137 (5′-AAAATRYCAGCTTTGGGAGTTG-3′; Lacerda et al. 2007). The PCR reaction volume (10 μL) contained 20 ng of DNA, 1× Taq buffer, 200 μM of each dNTP, 1.0 μM of each primer, and 0.5 U of Taq polymerase. Amplifications were performed with an initial step at 94 °C for 2 min and 35 cycles of 30 s at 94 °C, 40 s at 57 °C, and 90 s at 72° C, followed by final extension of 10 min at 72 °C. The PCR products were cleaned up by precipitation using 20 % polyethyleneglycol with 2.5 M NaCl. Sequences were obtained in a MegaBACE 1000 sequencer (Amersham-Biosciences, GE Healthcare, Little Chalfont, UK) or in ABI Prima 377 and ABI 3700 DNA sequencers (Applied Biosystems, Foster City, CA). The sequencing was conducted using intern primers LEC2 and H4 (Lacerda et al. 2007).
Nuclear sequences
The nuclear intron 5 from the β-fibrinogen gene was amplified with the primers FIB5 and FIB6 (Marini and Hackett 2002). The PCR reaction volume (10 μL) contained 20–40 ng of total DNA, 1× Taq buffer, 200 μM of each dNTP, 0.5 μM of each primer, and 0.5 U of Taq polymerase. Amplifications were performed with an initial step at 95 °C for 4 min and 37 cycles of 45 s at 94 °C, 45 s at 53.5 °C, and 1 min at 72 °C, followed by a final extension of 10 min at 72 °C.
PCR products were cleaned by precipitation using 20 % polyethyleneglicol with 2.5 M NaCl. The sequences were obtained in a MegaBACE 1000 sequencer (Amersham-Biosciences) using the same primers. The alignments were made by eye using Bioedit version 5.06 (Hall 2001). The heterozygous nucleotide positions were identified by their high-quality double peaks in the electropherograms. All resolved positions received a posterior probability of 1 in the phasing analyses. To identify both haplotypes of heterozygous individuals with multiple variable sites, we used a Bayesian approach in the program PHASE 2.0 (Stephens et al. 2001; Stephens and Donnelly 2003), implemented in DNAsp 5.1 software (Librado and Rozas 2009).
Analytical methods
Descriptive analyses, including estimates of haplotype diversity (h) and nucleotide diversity (π), were performed in DNAsp v.5.10.01 (Librado and Rozas 2009). We applied the Network software version 4.6 (www.fluxus-technology.com) to obtain separate networks for intron 5 from the β-fibrinogen gene (410 bp) and Control region (472 bp) using the median joining method (Bandelt et al. 1999; Polzin and Daneschmand 2003). We also calculated Tajima’s (1989), Fu and Li’s (1993), and Ramos-Onsins and Rozas’ neutral deviation parameters (D, D*, F*, F) of the R2 statistic with DNAsp v.5.10.01 (Librado and Rozas 2009). We selected these test statistics due to their power to detect population expansion scenarios in specific sampling conditions and with a specified population expansion rate, time since the expansion, sample size, and number of segregation sites (Ramos-Onsins and Rozas 2002). We used the Akaike information criterion (AIC) as implemented in jModel test software (Posada and Cradall 1998) to select the evolutionary model that best fit the Control region. The evolutionary model selected was TPM3 + I, with a proportion of invariable site (I) of 0.74, and FIB5 (Jukes and Cantor model), with a proportion of invariable sites (I) of 0.352 and a discrete gamma distribution (α = 0.3860).
In addition, to analyze population size dynamics through time, we reconstructed the phylogenies using the Extended Bayesian Skyline Plot method (EBSP; Heled and Drummond 2008) implemented in BEAST 1.6.1 (Drummond and Rambaut 2007). We performed four independent runs of 30,000,000 steps, logged every 1,000 steps, and a burn-in of 3,000,000 runs. For the BEAST analysis we considered the mutation rate to be 2.1 % per mya (Weir and Schluter 2008) compared to the Control region and 0.53 % compared to FIB5 (Axelsson et al. 2004). To evaluate the convergence of parameters between runs and the performance of analysis (ESS values >200), we used TRACER version 1.5 (http://beast.bio.ed.ac.uk/Tracer) (Drummond and Rambaut 2007). To check the level of population genetic structure among localities, we performed an analysis of molecular variance (AMOVA) with two hierarchical levels using ARLEQUIN 3.5 (Excoffier and Lischer 2010). Additionally, we performed a Bayesian analysis of population structure using sequences of FIB5 in BAPS version 5.4. (The Bayesian Analysis of Population Structure; Corander and Tang 2007.) BAPS estimates hidden population substructure by clustering sampled populations into panmictic groups. This analysis takes into account each individual and the probability at which it fits into each cluster, uninformed by the population origin of the individual. We tested the mixture analysis with two through to 20 populations, with the population allele frequencies and structure parameters automatically set to be uninformative. This approach produced a joint posterior distribution of partitions of the sampled populations into panmictic groups, along with their respective allele frequencies. We ran BAPS ten times for 105 interactions after a burn-in period of 20,000 steps. The resulting partitions were averaged based on their plotted posterior probabilities.
We performed divergence time estimates between pairs of C. lineata lineages with Isolation–Migration (IM) software (Nielsen and Wakeley 2001; Hey and Nielsen 2004). We then estimated the effective population sizes for current (θ 1, θ 2) and ancestral (θ a) populations, the divergence time between lineages, and migration between populations (m 1, m 2) using the IM model. To define the priors, we carried out initial runs followed by five posterior runs for each analysis, applied a burn-in of 2,000,000 steps, and finished with at least 20,000,000 steps in the chain after the burn-in stage and a minimum effective sample size value of 50. To obtain the divergence times in years, we considered t = t/u (Hey and Nielsen 2004). To calculate the dates, we assumed a divergence of 0.53 % per million years for FIB5 (Axelsson et al. 2004; Cabanne et al. 2008).
Results
We sequenced 472 bp of the Control region from 208 individuals of C. lineata, revealing 310 invariable sites, 133 variable sites, and 29 indels, corresponding to 71 haplotypes. The Control region of C. lineata showed high genetic diversity (haplotype diversity h = 0.937, nucleotide diversity π = 0.04051, θ S = 22.46) (Table 1; Fig. 2a). We obtained FIB5 sequences (410 bp) from 120 individuals (240 haploid sequences) of C. lineata (GenBank acession number FIB5 KJ835862-KJ836073, Dloop KJ836074-KJ836281). There were no indels and no evidence of recombination. We identified 80 haplotypes and 70 polymorphic sites, showing high genetic diversity [haplotype diversity (h) = 0.929; nucleotide diversity (π) = 0.00942, θ S = 10.47)] (Table 1; Fig. 2b).
Median joining network based on 472 bp of the Control region (a) and 410 bp of intron 5 of the β-fibrinogen gene (b) of C. lineata individuals from the Brazilian states of Bahia (horizontal lines), Minas Gerais (gray coloration), São Paulo (black coloration), Rio de Janeiro (white), Paraná (dark gray), Santa Catarina and Rio Grande do Sul (Vertical lines) and the Argentinian state of Missiones (red) (color figure online)
Demographic history
The neutrality tests for the Control region did not show significant bias to C. lineata. Although the mismatch distribution showed five distinct peaks, this is an expected result due to the existence of four mtDNA clades (see below) (Fig. 3a). The haplotype network showed four clades: one corresponding to Minas Gerais and Bahia states (MG clade); a second one including Rio de Janeiro, south of Minas Gerais and north of São Paulo (RJ clade); a third clade corresponding to a region near Serra do Mar (São Paulo state) and Paraná state (SP clade); the fourth clade including samples from west of São Paulo state (Paranapanema region), and the states of Santa Catarina, Rio Grande do Sul, and Misiones, Argentina (SUL clade) (Fig. 2a). In addition, the skyline plot analyses that were generated to reconstruct the effective population size through time also indicated population expansion starting at 250,000 years ago for the mtDNA Control region (Fig. 4a).
Bayesian skyline plot based on the analysis of the Control region (a) and intron 5 of β-fibrinogen (b) from C. lineata with a log-normal restricted clock. y-Axis Effective number of individuals, thick solid line median estimate, shaded area 95 % highest posterior density limit. Time on the x-axis is scaled as millions of years ago (mya)
In contrast, the neutrality test for intron 5 of the β-fibrinogen gene showed a significant bias for all tests, with Fs = −75.52 (p = 0.00), Tajima’s D = −1.91 (p = 0.001), R2 = 0.0301 (p = 0.010), and the raggedness statistic r = 0.0141 (p = 0.033), suggesting a recent demographic expansion scenario. This outcome is congruent with the mismatch distribution that showed a wave signal consistent with a population expansion [estimated initial theta (θ I) of 1.45 and tau (τ) of 2.36] (Fig. 3b). The haplotype network showed two clades, one corresponding to the Minas Gerais and Bahia states (MG clade) and another to São Paulo state (SP clade). The two clades have star shapes, indicating population expansion, with one abundant haplotype and several haplotypes at low frequencies (Fig. 2b). This star-like network configuration corroborated the neutrality test results (Tajima’s D, Fs, R2). The skyline plot analyses also indicated population expansion starting at 250,000 years ago for FIB5 (Fig. 4b).
Phylogeography, date estimates, and population structure
A phylogeographic study of C. l. lineata based on mitochondrial data resulted in four groups, with a break between Minas Gerais and São Paulo state [fixation index (F ST) = 0.66, p < 0.001) (Fig. 6), followed by another break in Serra do Mar among São Paulo and Rio de Janeiro (F ST = 0.88, p < 0.001), and a third in the Paranapanema region. The haplotype network based on FIB5 sequences corroborated the phylogeographic break in the north of São Paulo state (Fig. 2b). The F ST between these regions was 0.422 (p < 0.001) (Table 2), and the genetic diversity and expansion signal were maintained when the two geographic regions (São Paulo and Minas Gerais) were analyzed separately (Table 1). The same pattern was observed when the four clades with separate segegration of mtDNA were analyzed separately (Tables 3, 4).
The estimate of effective population sizes based on the Control region was 848,326 for the SP clade and 482,589 for the MG clade; that for an ancestral population was estimated to be 30,835. The estimated migration rate from SP to MG (N m = 0.115) was higher than that from MG to SP (N m = 0.005). The effective population size based on FIB5 of the SP and the MG clade was 315,320 and 140,253, respectively; that of an ancestral population was estimated to be 1,69,665. The N m from SP to MG (0.165) was higher than that from MG to SP (0.055) (Fig. 5).
The outcome of the Bayesian analysis of genetic population structure outcome for FIB5 showed two clear groups that correspond to the SP and MG clades, with some individuals collected in one place, but genetically related to another. For example, three individuals sampled in Minas Gerais state were related to the SP clade, and nine individuals sampled in São Paulo state were related to the MG clade (Fig. 6).
Discussion
Nucleotide diversity found for C. lineata in the FIB5 (π = 0.009) and Control regions (π = 0.016) were similar to those obtained for other species of Passeriformes from the Atlantic forest. Cabanne et al. (2011, 2012) obtained a value of 0.0168 for the nucleotide diversity of the Dendrocolaptes platyrostris Control region. For the same locus (FIB5) in Xinphorhynchus fuscus, Cabanne et al. (2008) found π = 0.0063, while Batalha-Filho et al. (2012) found π = 0.00403 in Basileuterus leucoblepharus, and d’Horta et al. (2011) found π = 0.0043 for FIB7 in Sclerurus scansor. Thus, the genetic diversity observed in C. lineata falls within the expected range for Passeriformes in the Atlantic forest.
Our analysis of C. lineata revealed a clear signal of population expansion based on all neutrality tests for the FIB5 (D’Tajima’s D = −1.92, Fu and Li’s statistic = −75.52, R2 = 0.03) and Control regions (Fu and Li’s statistic = −11.82). This outcome was corroborated by the Skyline plot analyses that showed a population expansion starting 250,000 years ago, possibly associated with climate changes during the Late Pleistocene. Recent population expansions have also been observed for other vertebrates in the Atlantic forest, such as frogs (Amaro et al. 2012), lizards (Carnaval et al. 2009), snakes (Grazziotin et al. 2006), birds (Cabanne et al. 2007, 2008; d’Horta et al. 2011; Batalha-Filho et al. 2012; Maldonado-Coelho 2012), and mammals (Martins 2009). Thus, the consensus is that the Atlantic forest underwent geological or climatological events that marked the genetics of several groups. However, studies vary in their explanations of such events that allowed these lineages to separate and consequently expand as genetically distinct subpopulations. Alternatively, expansion after a vicariant event will generate contacts between historically divergent populations and—likely—current gene flow (secondary contact).
Our study of C. lineata based on analysis of the Control region and FB5 revealed a significant population genetic structure. The FIB5 showed two main lineages—one associated with specimens collected from northern Minas Gerais and one associated with those collected from São Paulo and Paraná (F ST = 0.422) (Fig. 2b). These results provide evidence for a possible phylogeographic break in northern São Paulo. Other species of forest vertebrates, such as frogs [Proceratophrys boie (Amaro et al. 2012); Thoropa miliaris complex (Fitzpatrick et al. 2009)], snakes [Bothrops jararaca (Grazziotin et al. 2006)], and birds [Xinphorhynchus fuscus (Cabanne et al. 2008)] also show well-defined phylogeographic breaks. In comparison, analysis of the mtDNA Control region revealed a major structure distributed into four clades: (1) a northern clade that included south Bahia and area northeast of Minas Gerais (MG clade); (2) a clade including Minas Gerais, Rio de Janeiro, and the area northeast of São Paulo (RJ clade); (3) a clade including the areas from São Paulo and Paraná (SP clade); (4) a clade including individuals from the area west of São Paulo state (Pontal do Paranapanema) and Santa Catarina, Rio Grande do Sul to Misiones, Argentina (RS clade). The differences in the observed population structure could be related to the differences between markers, as theoretically the mtDNA genome is one-quarter the effective size of the nuclear DNA one. Thus, the recent events of population reduction could have marked the mitochondrial genome more intensively than the nuclear genome. In this context, we believe that the lineages first split around 2,50,000 years ago to form the two major clusters (denoted here as the SP and MG clades) and that during the Last Glacial Maximum a second split occurred, form two additional clades.
Recent studies using palaeoclimatic dating have revealed that the Atlantic forest passed through many temperature and humidity changes during the Pleistocene (Behling 2002; Behling and Pillar 2007), and phylogeography studies in Passeriformes have confirmed that these climatic changes during the Pleistocene affected the distribution of lineages in the region (Cabanne et al. 2007; Maldonado-Coelho 2012). It is to be expected that species living in isolated forest habitats (refuges) will have passed through a more recent population expansion after a bottleneck during the Pleistocene period. These demographic oscillations should mark the genome of the species, and we found such markers in our study. Evidence of such oscillations include the results of mismatch distribution analyses, Tajima’s D, Fu and Li’s statistic, and the R2 tests, as well as Bayesian skyline plots, all of which show that a relatively recent demographic event, with population growth, was associated to most of the coalescent events that occurred before the expansion. Consequently, samples of these populations have gene genealogies stretched near the external nodes and compressed near the root (i.e., star-like genealogies) (Ramos-Onsins and Rozas 2002).
Cabanne et al. (2008) proposed that populations in the southern Atlantic forest (which present low forest stability) that were affected by Pleistocene refugia (vegetational change) would exhibit low genetic diversity and strong signatures of demographic expansion. Fitzpatrick et al. (2009) affirmed that demographic expansion in the southern Atlantic forest, south of the Doce river, should be attributed to demographic and geographical expansions of populations that occupied very small refugia. Conopophaga lineata lineages were found to show a strong signature of demographic expansion and genetic diversity, comparable to that of other Passeriformes of the Atlantic forest, such as X. fuscus (Cabanne et al. 2007, 2008), B. leucoblepharus (Batalha-Filho et al. 2012), and S. scansor (d’Horta et al. 2011).
Alternatively, other studies have corroborated the hypothesis of rivers as barriers (e.g., Pellegrino et al. 2005; Lacerda et al. 2007). Pellegrino et al. (2005) found two lineages in geckos (Gymnodactylus darwinii complex) that diverged between southern Minas Gerais and São Paulo, suggesting that this divergence resulted from the formation of the Paraíba do Sul valley. However, if the Paraíba do Sul river were to be responsible for the splitting of the C. lineata lineages, we would expect that these lineages would be older than the Pleistocene period and have accumulated mutations for the last 15 million years (the date of the formation of the Paraiba valley), thereby showing a remarkable differentiation and low gene flow.
In contradiction to this latter possibility, our results demonstrate recent demographic expansion for both lineages (the MG and SP clades), associated with date estimates of expansion occurring in the Pleistocene period (2,50,000 years ago), and seem to indicate that forest refugia during glaciations affected the evolutionary history of C. lineata in the southern Atlantic forest. We believe that the lineages from Minas Gerais were most likely derived from the Bahia refugia and that the lineages from São Paulo were derived from São Paulo refugia. In addition, our results for FIB5 using BAPS confirmed the structure in two major clades, but indicated gene flow between them, mainly observed in 12 individuals from the regions of Buri, Teodoro Sampaio, and Euclides da Cunha in São Paulo state and the regions of Nova Lima, Santa Barbara and Caratinga in Minas Gerais state. These results corroborate the IM inference that indicates a major gene flow from São Paulo to Minas Gerais (south to north). However, the recent migration of individuals indicated by BAPS suggests that this could also be a merging of lineages due to secondary contact. Similar patterns of divergence and secondary contact were suggested by Cabanne et al. (2007) for Xinphorhynchus fuscus in the valley of the Paraíba do Sul river, who estimated the divergence between mtDNA lineages to have occurred approximately 70,000 years ago.
Our study with the mtDNA Control region and FIB5 corroborate the structure observed for other vertebrates in the Atlantic forest, and even though it was not possible to estimate the divergence between lineages, our estimate for population expansion is similar to that of both markers (2,50,000 years ago; Fig. 4a, b). These outcomes were corroborated by neutrality tests (Tables 1, 2, 3, 4). The difference observed in the level of genetic structure of the Control region and the nuclear intron (FIB5) were expected due these genetic markers having different effective population sizes and modes of inheritance. Consequently, it is expected that demographic events will shape the variation in the markers in distinct ways (Zink and Barrowclough 2008). However, the high congruence between the nuclear and mitochondrial marker revealed in our study leads us to suggest that the evolutionary history of C. lineata consists of major population splits during the Pleistocene followed by population expansions.
Cabanne et al. (2008) proposed that minor geographic features that are traditionally considered to be inefficient barriers could be important for explaining a number of observed biogeographic patterns in the Atlantic forest, where climatic changes appear as a central factor modulating the effect of barriers to gene flow. In this context, the Paraíba do Sul valley and the mountain ranges of Serra da Mantiqueira and Serra do Mar could be highly susceptible to climatic alterations, such as changes in water flow and vegetation. This partial barrier effect may be considered together with more widespread changes in the forest distribution during glaciations to explain the divergence and transitions of lineages in the southern Atlantic forest. Consequently, the pattern of C. lineata genetic structure we documented using the mtDNA Control region could be generated by vicariance events occurring in the Pleistocene followed by secondary contact in the Holocene. Batalha-Filho et al. (2012) studied Basileuterus leucoblepharus in the southern Atlantic forest in Brazil and did not find any significant structure, suggesting that this species with different habitat requirements could show a more homogeneous phylogeographic distribution. Thus, while some endemic taxa from the Atlantic forest may be highly influenced by selective ecological filters working as barriers, other species with higher a tolerance to habitat fragmentation would be able to maintain gene flow or disperse across these barriers. Contrary to expectations, C. lineata is a species relatively tolerant of habitat fragmentation (Dantas et al. 2007) that showed a clear signal of population structure. On the other hand, Salisbury et al. (2012) suggested that canopy birds tend to be less genetically differentiated than understory birds, which is a likely explanation for the genetic structure we observed in C. lineata.
Evolution in isolation and secondary contact provides a possible explanation for the current phylogeographic structure of C. lineata. Costa (2003) suggested that speciation in the Neotropics could not be explained by any single model of vicariance or climatic change. Thus, it is possible that no general pattern for the diversification of organisms in the Atlantic forest will be determined—rather, a complex range of scenarios shall be described. The refuge hypothesis, the influence of geography, and rivers as barriers are among the most discussed models in the study of Neotropical diversification. Our data do not support a primary influence of rivers on the divergence between the two main mitochondrial lineages of C. lineata in the south-central Atlantic forest. Our data do support the influence of isolation by distance in separated forest relicts as a main vicariant event, followed by subsequent demographic expansions, in shaping the current distribution and phylogeographic structure of C. lineata. Thus, vicariance and demographic expansions leading to secondary contact appear to be related to recent natural forest landscape dynamics affecting the population of birds, and perhaps other organisms endemic to this region.
References
Amaro RC, Carnaval AC, Yonenaga-Yassuda Y, Trefaut MR (2012) Demographic processes in the montane Atlantic rainforest: molecular and cytogenetic evidence from the endemic frog Proceratophrys boiei. Mol Phylogenet Evol 63:880–888
Axelsson E, Smith NGC, Sundstrom H, Berlin S, Ellegren H (2004) Male-biased mutation rate and divergence in autosomal, Z-linked and W-linked introns of chicken and turkey. Mol Biol Evol 21:1538–1547
Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Batalha-Filho H, Cabanne GS, Miyaki CY (2012) Phylogeography of an Atlantic Forest passerine reveals demographic stability through the last glacial maximum. Mol Phylogenet Evol 65:892–902
Behling H (2002) South and southeast Brazilian grasslands during late quaternary times: a synthesis. Palaeogeogr Paleoclimatol Palaeoecol 177:19–27
Behling H, Pillar VDP (2007) Late quaternary vegetation, biodiversity and fire dynamics on the southern Brazilian highland and their implication for conservation and management of modern Araucaria forest and grassland ecosystems. Phil Trans R Soc B 362:243–251
Brown KS (2005) Geological, evolutionary, and ecological bases of the diversification of neotropical butterflies: implications for conservation. In: Berminhgam E, Dick CW, Moritz C (eds) Tropical Rainforest: past present and future. University of Chicago Press, Chicago, pp 166–200
Brown KS, Ab’Saber AN (1979) Ice-age forest refuges and evolution in Neotropics: correlation of paleoclimatological, geomorphological and pedological data with biological endemism. Paleoclimas 5:1–30
Bruford MW, Hanotte O, Brookfield JFY, Burke T (1992) Single-locus and multilocus DNA fingerprinting. In: Hoelzel AR (ed) Molecular genetic analysis of populations—a practical approach. IRL Press, New York, pp 287–336
Cabanne GS, Santos FR, Miyaki CY (2007) Phylogeography of Xiphorhynchus fuscus (Passeriformes, Dendrocolaptidae): vicariance and recent demographic expansion in southern Atlantic Forest. Biol J Linn Soc 91:73–84
Cabanne GS, d’Horta FM, Sari EH, Santos FR, Miyaki CY (2008) Nuclear and mitochondrial phylogeography of the Atlantic Forest endemic Xiphorhynchus fuscus (Aves: dendrocolaptidae): biogeography and systematics implications. Mol Phylogenet Evol 49:760–773
Cabanne GS, D´Horta FM, Meyer D, Silva JMC, Myiaki CY (2011) Evolution of dendrocolaptes platyrostris (Aves:Furnariidae) between the south American open vegetation corridor and the Atlantic Forest. Biol J Linn Soc 103:801–820
Cabanne GS, Sari HER, Meyer D, Santos FR, Miyaki CY (2012) Matrilineal evidence for demographic expansion, low diversity and lack of phylogeographic structure in the Atlantic forest endemic Grenish Schiffornis Schiffornis virescens (Aves: tityridae). J Ornithol 154:371–384
Carnaval AC, Moritz C (2008) Historical climate modeling predicts patterns of current biodiversity in the Brazilian Atlantic Forest. J Biogeogr 35:1187–1201
Carnaval AC, Hickerson MJ, Haddad CFB, Rodrigues MT, Moritz C (2009) Stability predicts genetic diversity in the Brazilian Atlantic forest hotspot. Science 323:785–789
Corander J, Tang J (2007) Bayesian analysis of population structure based on linked molecular information. Math Biosci 205:19–31
Costa LP, Leite YLR, da Fonseca GAB, da Fonseca MT (2000) Biogeography of south American forest mammals: endemism and diversity in the Atlantic forest. Biotropica 32:872–881
Costa LP (2003) The historical bridge between the Amazon and the Atlantic Forest of Brazil: a study of molecular phylogeography with small mammals. J Biogeogr 30:71–86
d’Horta F, Cabanne GS, Meyer D, Miyaki CY (2011) The genetic effects of late quaternary climatic changes over a tropical latitudinal gradient: diversification of an Atlantic Forest passerine. Mol Ecol 20:1932–1935
Dantas GPM, Santos FR, Marini MÂ (2007) Genetic variability of Conopophaga lineata (Conopophagidae) in Atlantic Forest fragments. Braz J Biol 67:631–637
Dantas GPM, Santos FR, Marini MÂ (2009) Efeitos de fragmentação na razão sexual de Conopophaga lineata em fragmentos de Mata Atlântica no Estado de Minas Gerais. Iheringia Série Zool 99:115–119
Drummond AJ, Rambaut A (2007) BEAST: bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214
Excoffier L, Lischer HLE (2010) Arlequim ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res 10:564–567
Fitzpatrick SW, Brasileiro CA, Haddad CF, Zamudio KR (2009) Geographical variation in genetic structure of an Atlantic coastal forest frog reveals regional differences in habitat stability. Mol Ecol 18:2877–2896
Fu Y-X, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709
Geise L, Smith MF, Patton JL (2001) Diversification in the genus Akodon (Rodentia: sigmodontinae) in southeastern South America: mitochondrial DNA sequence analysis. J Mamm 82:92–101
Grazziotin FG, Monzel M, Echeverrigarauy S, Bonato SL (2006) Phylogeography of the Bothrops jararaca complex (Serpentes: viperidae): past fragmentation and island colonization in the Brazilian Atlantic Forest. Mol Ecol 15:3969–3982
Gusmão Câmara I (2003) Brief history of conservation in the Atlantic forest. In: Galindo-Leal C, Gusmão Câmara I(eds). The state of the hotspots: The Atlantic Forest. Island Press, Washington DC, pp 31–42
Haffer J (1969) Speciation in Amazonian forest birds. Science 165:131–137
Haffer J, Prance GT (2001) Climatic forcing of evolution in Amazonia during the Cenozoic: on the refuge theory of biotic differentiation. Amazoniana 16:579–605
Hall TA (2001) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Heled J, Drummond AD (2008) Bayesian inference of population size history from multiple loci. BMC Evol Biol 8:289
Hey J, Nielsen R (2004) Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167:747–760
Lacerda DR, Marini MÂ, Santos FR (2007) Mitochondrial DNA corroborates the species distinctiveness of the Planalto (Thamnophilus pelzelni Hellmayr, 1924), the Sooretama (T. ambiguus Swainson, 1825) Slaty-antshrikes (Passeriformes: thamnophilidae). Braz J Biol 67:873–882
Librado P, Rozas J (2009) DnaSP v. 5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Maldonado-Coelho M (2012) Climatic oscillations shape the phylogeographical structure of Atlantic forest fire-eye antbirds (Aves: thamnophilidae). Biol J Linn Soc 105:900–924
Marini MÂ, Hackett SJ (2002) A multifaceted approach to the characterization of an intergeneric hybrid manakin (Pipridae) from Brazil. Auk 119:1114–1120
Martins FM (2009) The Brazilian Atlantic Forest historical biogeography and the Carnaval-Moritz model of Pleistocene refugia: what do the phylogeographical studies tell us? Biol J Linn Soc 104:499–509
Miranda JM, Bernardi IP, Passos FC (2006) A new species of Eptesicus (Mammalia: chiroptera) from Atlantic Forest Brazil. Zootaxa 1383:57–68
Moritz C (2002) Strategies to protect biological diversity and the evolutionary process that sustain it. Syst Biol 51:238–254
Mustrangi MA, Patton JL (1997) Phylogeography and systematics of the slender mouse opossum Marmosops (Marsupialia: didelphidae). Univ Calif Publ Zool 130:1–86
Nielsen R, Wakeley J (2001) Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics 158:885–896
Pavan AC, Martins FM, Santos FR, Ditchfield A, Redondo RAF (2011) Patterns of diversification in two species of short-tailed bats (Carollia Gray, 1838): the effects of historical fragmentation of Brazilian rainforests. Biol J Linn Soc 102:527–539
Pellegrino KCM, Rodrigues MI, Waite AN, Morando M, Yassuda YY, Sites JW (2005) Phylogeography and species limits in the Gymnodactylus darwinii complex (Gekkonidae, Squamata): genetic structure coincides with river systems in the Brazilian Atlantic Forest. Biol J Linn Soc 85:13–26
Pessoa RO (2007) Sistemática e Biogeografia Histórica da Família Conopophagidae (Aves: Passeriformes): Especiação nas Florestas da América do Sul. PhD thesis. Instituto de Biociências da Universidade de São Paulo, São Paulo
Petri S, Fulfaro VJ (1983) Geologia do Brasil—Fanerozoico. Editora da Universidade de São Paulo, São Paulo
Polzin T, Daneschmand SV (2003) On Steiner trees and minimum spanning trees in hypergraphs. Oper Res Lett 31:12–20
Posada D, Cradall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818
Ramos-Onsins SE, Rozas J (2002) Statistical properties of new neutrality tests against population growth. Mol Biol Evol 19:2092–2100
Ribeiro MC, Metzger JP, Martensen AC, Ponzoni FJ, Hirota MM (2009) The Brazilian Atlantic Forest: how much is left, and how is the remaining forest distributed? Implications for conservation. Biol Conserv 142:1141–1153
Riccomini C, Peloggia AUG, Salón JCL, Kohner MW, Figueira RM (1989) Neotectonic activity in the Serra do Mar rift system (southeastern Brazil). J South Am Earth Sci 2:191–197
Salisbury CL, Seddon N, Cooney CR, Tobias JÁ (2012) The latitudinal grandient in dispersal constraints: ecological specialization drivers diversification in tropical birds. Ecol Lett 15:847–855
Sick H (1997) Ornitologia Brasileira. Nova Fronteira, Rio de Janeiro 912p
Sigrislt T (2005) Aves do Brasil: uma visão artística. Editora Avis Brasilis, São Paulo
Silva JMC, Straube FC (1996) Systematics and biogeography of scaled woodcreepers (Aves: dendrocolpatidae). Stud Neotrop Fauna Environ 31:3–10
Silva JMC, De Sousa MC, Castelleti CHM (2004) Areas of endemism for passerine birds in Atlantic Forest, South America. Glob Ecol Biog 13:85–93
Stephens M, Donnelly P (2003) A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73:1162–1169
Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Vanzolini PE, Williams EE (1970) South American anoles: the geographic differentiation and evolution of the Anolis chrysolepis species group (Sauria: iguanidae). Arq Zool 19:1–298
Wallace AR (1852) On the monkeys of the Amazon. Proc Zool Soc London 20:107–110
Weber LN, Gonzaga LP, Carvalho-e-Silva SP (2005) A New species of Physalaemus from the lowland Atlantic Forest of Rio de Janeiro State Brazil (Amphibia, Anura, Leptodactylidaea). Arq Mus Nac 63:677–684
Weir JT, Schluter D (2008) Calibrating the avian molecular clock. Mol Ecol 17:2321–2328
Whitney BM (2003) Family Conopophagidae. In: del Hoyo J, Elliot A, Christie DA (eds) Handbook of the birds of the world: Broadbills to Tapaculos, vol 8. Lynx Edicións, Barcelona, pp 732–747
Willis EO, Oniki Y, Silva WR (1983) On the behaviour of Rufous Gnateaters (Conopophaga lineata, Formicariidae). Naturalia 8:67–93
Zink RM, Barrowclough GF (2008) Mitochondrial DNA under siege in avian phylogeography. Mol Ecol 17:2107–2121
Acknowledgments
This work was supported by FAPEMIG (Fundação de Amparo a Pesquisa do Estado de Minas Gerais 17228), CNPq, FAPESP (BIOTA 2013/50297-0), NSF (DOB 1343578), NASA, CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), and the Research Center on Biodiversity and Computing (BioComp) of the Universidade de São Paulo (USP), supported by the USP Provost’s Office for Research. Gisele Dantas worked under a Post-doctoral grant CAPES/PNPD (2010/52590-8) and CNPq (503145/2009-2). We also thank the Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis (Brazil), Instituto Florestal de São Paulo (Brazil), and Instituto Estadual de Florestas de Minas Gerais (Brazil) for the permits to collect samples (IBAMA/MMA no 03/2004 IBAMA/MMA: 011/2000, processes 1835/2000; 053/2001, 1835/00-07; 070/2002, 02015.001835/00-07; 207/2003, 02015.023482/98-38). We gratefully acknowledge the improvements in English usage made by Caitlin Stern through the Association of Field Ornithologists’ program of editorial assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Fjeldså.
Appendix
Appendix
See Table 5.
Rights and permissions
About this article
Cite this article
Dantas, G.P.M., Sari, E.H.R., Cabanne, G.S. et al. Population genetic structure of the Atlantic Forest endemic Conopophaga lineata (Passeriformes: Conopophagidae) reveals a contact zone in the Atlantic Forest. J Ornithol 156, 85–99 (2015). https://doi.org/10.1007/s10336-014-1106-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10336-014-1106-0