Abstract
Diversification processes acting across geographically continuous populations have been rarely documented in Amazonia, because of the lack of fine-scale sampling over extensive areas. We aimed to determine the geographic effects of an environmental transition zone and large rivers on the intraspecific population structure of the Manaus slender-legged tree frog (Osteocephalus taurinus) along a ~ 900 km transect of tropical rain forest. Using one mitochondrial (16S), two nuclear genes (TYR, POMC) and three microsatellites, we estimated the population structure, phylogenetic relationships and geographic variation of 262 O. taurinus and 5 O. oophagus (a close relative) along the Purus–Madeira interfluve (PMI) and opposite banks of the central Amazon and upper Madeira rivers, at central-southern Amazonia, Brazil. Six genetic clusters were identified: two corresponding to sympatric populations of O. taurinus and O. oophagus from their type locality, north of Amazon river. Within PMI, there were three distinct O. taurinus genetic clusters distributed along the geographic gradient with one main phylogeographic break found (concordant between 16S and TYR), that corresponds to a transition zone (ecotone) between dense and open rain forest ecotypes. The sixth cluster was an O. taurinus population isolated at the east bank of the upper Madeira river. In addition, restricted haplotype sharing was identified from the west to east banks at upper Madeira river. Within PMI, parapatric genetic structure is explained by a potential association of the genetic clusters to the different forest ecotypes they inhabit coupled with isolation by distance, thus supporting the gradient hypothesis for diversification. Differentiation of populations that are external to the PMI is most likely explained by the barrier effect of the Madeira and Amazon rivers. Our findings provide new evidence on diversification processes across continuous Amazonian landscapes; however, the specific mechanisms underlying the origin and maintenance of the identified phylogeographic break need to be further studied.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The origin and geographic distribution of the high biodiversity in Amazonia result from long and complex interactions between geologic, climatic and ecological processes (Hoorn and Wesselingh 2010; Smith et al. 2014). Among proposed explanations for the distribution patterns of Amazonian biota, the riverine barrier hypothesis (Wallace 1852) has acquired notorious importance for groups like birds, primates, anurans, squamates, butterflies and plants (Hall and Harvey 2002; Ribas et al. 2012, 2018; Boubli et al. 2015; Dias-Terceiro et al. 2015; Moraes et al. 2016; Nazareno et al. 2017; Godinho and Da Silva 2018; but see Santorelli et al. 2018). This hypothesis predicts that major Amazonian rivers act as barriers to gene flow between populations inhabiting opposite river banks, thus promoting speciation (Wallace 1852; Leite and Rogers 2013). Therefore, the intervening land regions (interfluves) between the largest Amazonian rivers have been proposed to represent biogeographic units known as Areas of Endemism (AoE; Cracraft 1985).
On the other hand, the gradient hypothesis (Endler 1977) has received considerably less attention in Amazonian phylogeography (see reviews in Moritz et al. 2000; Leite and Rogers 2013). This hypothesis predicts parapatric structuration of the populations along a continuous geographical or environmental cline (Endler 1977); therefore, involving potential gene flow between diverging populations (Moritz et al. 2000). Under this scenario, populations differentiate following a model of isolation by distance, often facilitated by ecological adaptation of the populations to adjacent but different environments (Endler 1977; Nosil 2012).
Recent studies have added complexity to the understanding of Amazonian AoE, showing that the distribution of taxa is not completely homogeneous within them, as some species and intraspecific lineages have distribution limits concordant with transition zones and smaller tributaries (Simões et al. 2008; Naka et al. 2012; Fernandes et al. 2012; Simões et al. 2014). Identifying transition zones (regions containing phylogeographic breaks and zones of contact, suture, tension and hybridization) across relatively continuous landscapes provides unique opportunities to study the combination of geography, environment and the organisms’ ecology on the speciation processes (Naka et al. 2012).
Transition zones can result from primary contact, when parapatric populations differentiate in situ; and from secondary contact, when populations initially isolated encounter after they already started to differentiate (Thorpe 1984). In continuous populations, contact zones tend to match with transitions between different environments; however, defining the nature of contact zones (primary or secondary) is still a major challenge for the gradient hypothesis (Endler 1977; Thorpe 1984; Grahame et al. 2006). Therefore, it is often difficult to evaluate the mechanisms interacting between environmental heterogeneity and ecologically-based selection on the differentiation patterns of the populations (Nosil 2012; Stein et al. 2014).
Amphibians are a model group to detect ecological and biogeographical signatures associated with population divergence. Because of their strong association with local environmental conditions, high phylopatry and low dispersal abilities, in general amphibians show high genetic structure at several geographic scales (Zeisset and Beebee 2008). Previous studies reporting associations between environmental factors with processes of population connectivity (gene flow), population structure and speciation in amphibians (without physical barriers) have focused mostly on terrestrial and understory species (e.g., Graham et al. 2004; Richardson 2012; Wang 2012; Richter-Boix et al. 2013; Buskirk 2014). Therefore, studies investigating spatial genetic structure in tropical arboreal anurans, which use several vertical forest strata as home range and to disperse, could give novel insights into population differentiation processes associated to variation on physic structure, floristic composition and climatic conditions of rain forests.
Osteocephalus taurinus (Steindachner 1862) is a large hylid frog considered a specialist dweller of primary and secondary rain forests, and widely distributed through the Amazonia (Trueb and Duellman 1971; Jungfer et al. 2013). Osteocephalus taurinus taxonomy has been historically problematic, because of the huge morphological variation across its distribution (Trueb and Duellman 1971; Estupiñán-Tristancho 2001) and suggested high levels of cryptic diversity (De la Riva et al. 1995; Jungfer et al. 2013). The last taxonomic review (Jungfer et al. 2013) considered O. taurinus sensu stricto (our study taxon and hereafter referred only as O. taurinus) as a widely distributed species through southern Amazonia and allocated it to the O. taurinus species group, which also includes O. oophagus (a close relative).
Standardized and fine-scale studies within a given Amazonian AoE are scarce due to large distances and difficult access to most areas. In recent years, the Program for Planned Biodiversity and Ecosystem Research (PPBio) installed research sites along the BR-319 highway covering most of the middle and lower sections of the Purus–Madeira interfluve (PMI), and on both margins of the upper Madeira river (Magnusson et al. 2013). Combined, all research sites represent the largest geographic gradient sampled within a single Amazonian interfluve. This study system has shown that physical soil and hydrologic characteristics combined with weather conditions drive the forest structure and its dynamics within the PMI (Cintra et al. 2013; Martins et al. 2015; Schietti et al. 2016). Therefore, based on the association of O. taurinus with rain forests, we were interested in determining the effects of the most conspicuous landscape features on its intraspecific population structure and geographic variation. We asked if the spatial genetic structure of the populations could be associated with the main vegetation formations within the PMI, and if large rivers could reduce the potential dispersal of individuals inhabiting different interfluves.
Materials and methods
Study area
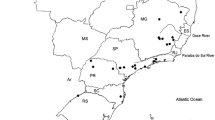
Purus–Madeira interfluve (PMI) is a mostly flat area (30–100 m a.s.l.) located in central-southern Amazonia (Fig. 1). The geologic sedimentary origin of the PMI is relatively recent (late Pleistocene–early Holocene; Sombroek 2000; Rossetti et al. 2005). Vegetation cover corresponds to lowland tropical rain forest, composed of two main forest formations (ecotypes): Ombrophilous dense forest in the northeastern half of PMI and Ombrophilous open forest in the southwestern half (Fig. 1). In general, open forests at the southern border of Amazonia present more gaps and, depending on the region, a higher presence of palms, lianas, bamboo (Guadua spp.) and “sororoca” (Phenakospermum guyannense) than the dense forests, which are more abundant at the Amazonian core. The distinction between these two rain forest ecotypes was defined using Landsat images, in situ photographs and botanical surveys (IBGE 2012). There is variation in the number of trees and their biomass within the PMI dense forest ecotype (Schietti et al. 2016). In addition, considerable areas of savanna and transition between open forest-savanna occur in the extreme south section of the PMI (Fig. 1). Mean annual precipitation varies from 2200 in the southwestern PMI (open forest) to 2800 mm in the northeastern PMI (dense forest) (Alvares et al. 2013). Biogeographically, the PMI constitutes the eastern limit of the Inambari AoE (Cracraft 1985). We also sampled adjacent areas outside PMI: Reserva Ducke (RD) on the north bank of the central Amazon river; and four localities on the east bank of the upper Madeira river (Fig. 1).
Study area of Osteocephalus taurinus in central-southern Amazonia, depicting vegetation cover. The ecotone arrow represents the sharp transition between dense and open rain forest ecotypes. Results from population structure analyses conducted in BAPS are presented as colored circles representing the membership proportion of individuals in each locality to the identified genetic clusters for 16S mtDNA (inner circles) and TYR nDNA (outer circles); see Fig. 2 for expanded results. For sample sizes see Table 1, except for Lábrea (n = 2). Samples from RD (type locality) are represented as ss (sensu stricto)
Sampling design
Sampling was conducted in 22 localities, with a maximum transect-line distance of 883 km between RD and M15 (Fig. 1; Table 1). Collected specimens and tissues were deposited in the Collection of Amphibians and Reptiles (INPA-H) and Collection of Tissues (INPA-HT) of Instituto Nacional de Pesquisas da Amazônia. Sympatric O. taurinus and O. oophagus samples of RD are from their type locality (Jungfer and Schiesari 1995; Jungfer et al. 2013). Excluding J1–J3, the remaining localities correspond to PPBio research sites, which follow the RAPELD method (Magnusson et al. 2013). Research sites consist of modules (M1–M18) and a grid (RD) distributed at mean distances of ~ 50 km apart (Fig. 1). Given the large body size and arboreal life-style of O. taurinus, we assumed that dispersal of individuals could occur within the 5 km2 area encompassing each module, but it is expected to be considerably lower among modules within PMI, so the landscape resolution matched the biology of our study organism.
DNA extraction, amplification and sequencing/genotyping
We extracted genomic DNA from 267 individuals (262 O. taurinus; 5 O. oophagus) using the Wizard® extraction kit (Promega) from muscular/hepatic tissue preserved in 99% ethanol. Using the polymerase chain reaction (PCR), we amplified one mitochondrial (16S rRNA, 526 bp) and two nuclear genes (TYR-tyrosinase, 555 bp; POMC-proopiomelanocortin, 497 bp). For detailed laboratory protocols, see Table S1 in Appendix 1 as supplementary material. We also tested 28 polymorphic microsatellites described for other species in Hylidae; however, 25 were discarded due to low transferability for O. taurinus. Therefore, we obtained polymorphic genotypes for three microsatellites (HEB226, 3897, Ha-T53) (Tables S1 and S2 in Appendix 1). For sequencing reactions, we used BigDye® Terminator (ThermoFisher Scientific). For microsatellites, genotypes were determined using GeneScan-500 ROX® (ThermoFisher Scientific). Sample readings for sequences and genotypes were conducted on an ABI Prism® 3130 Genetic Analyzer (ThermoFisher Scientific). Sample sizes are provided on Table 1, and voucher information and GenBank accessions on Table S3 in Appendix 1.
Population structure analyses
Consensus mitochondrial (mtDNA) and nuclear (nDNA) sequences were assembled in GENEIOUS 5.4 (Kearse et al. 2012), aligned in MAFFT 7 (Katoh and Standley 2013) and manually adjusted in ALIVIEW 1.18 (Larsson 2014). Presence of pseudogenes in nDNA was discarded by translation using the EMBL-EBI framework (Li et al. 2015). We estimated the most probable number of genetic clusters (k) using a Bayesian approach implemented in BAPS 6 (Corander and Tang 2007; Corander et al. 2008), without imposing a geographic constraint on individuals. We analyzed each gene independently and combined in three different data sets: total (16S + TYR + POMC), mtDNA and nDNA genes with the strongest signal (16S + TYR), and only nDNA genes (TYR + POMC). For combined gene analyses, we did not include individuals with missing genes. The three microsatellites were analyzed together. As BAPS requires resolved phases of diploid nuclear genes (TYR and POMC), the two most probable haplotypes for each individual were reconstructed in PHASE 2.1 (Stephens et al. 2001; Stephens and Donnelly 2003), using files generated in SEQPHASE (Flot 2010).
As the analysis of combined haploid (mtDNA) and diploid (nDNA) sequence markers is technically no feasible in BAPS, we conducted two approaches in order to assess the overall genetic information of the combined data sets. First, we conducted a haploid analysis using the two phased haplotypes of nDNA genes as representing an individual each, combined with its correspondent mtDNA sequence (J. Corander pers. comm.). Second, we conducted a diploid analysis considering both phased haplotypes of nDNA for each individual combined with its correspondent mtDNA sequence, which was duplicated in order to not allow a missing allele. As both approaches did not differ to each other, we presented the results of the second approach (individuals based). For each gene and combination of genes or microsatellites, we first ran the mixture model using a k range of 1–22 (upper limit equal to n localities), and analyses were run four times. Then we used this result to run the admixture model (Corander and Marttinen 2006) using 100 interactions, 200 reference individuals, and 10 interactions per individual. Admixed individuals were considered as significant at P < 0.05.
Phylogenetic analyses
Phylogenetic relationships were estimated using Maximum Likelihood (ML) and Bayesian Inference (BI) approaches for each gene and for a concatenated data set (1578 bp). We analyzed two data sets for nuclear genes (phased and unphased sequences). Substitution models and partition schemes were defined in PARTITIONFINDER 1.1 (Lanfear et al. 2012). ML analyses were conducted in RAXML 8 (Stamatakis 2014) using 1000 replicates and the GTRCAT approximation. BI analyses were conducted in MRBAYES 3.2 (Ronquist et al. 2012). Effective sample sizes (ESS) of the parameters and the stability and convergence between independent runs were evaluated in TRACER 1.6 (Rambaut et al. 2014). In the ingroup, we included available GenBank sequences of the O. taurinus species group; and used as outgroups Osteocephalus spp., Dryaderces, Tepuihyla and Trachycephalus (Table S3 in Appendix 1).
Mitochondrial and microsatellites variation
For 16S mtDNA, we estimated the number of haplotypes and segregating sites in DNASP 5.1 (Librado and Rozas 2009) and elaborated a haplotype network in HAPLOVIEW 4.2 (Barrett et al. 2005). Haplotype and nucleotide diversity indexes and two neutrality tests [Tajima’s D (Tajima 1989) and Fu’s Fs (Fu 1997)] were calculated in ARLEQUIN 3.5 (Excoffier and Lischer, 2010) with 1000 interactions. We assessed the relative partitioning of genetic variation between the two main vegetation formations (forest ecotypes) within the PMI using an analysis of molecular variation (AMOVA). We implemented this analysis in ARLEQUIN using the FST (fixation statistic) as differentiation metric. We did not conducted an AMOVA among interfluves because the comparatively lower sampling outside the PMI. Finally, we calculated FST in ARLEQUIN and mean uncorrected p-distance in MEGA 6 (Tamura et al. 2013) among the genetic clusters recovered by BAPS, and among all localities without defining genetic clusters.
For microsatellites, alleles were scored in GENEMARKER® 2.6 (SoftGenetics). Scoring issues and presence of null alleles were evaluated in MICRO-CHECKER 2.2.3 (Van Oosterhout et al. 2004). Summary statistics of observed and expected heterozygosity and among locality FST were calculated in GENETIX 4.05 (Belkhir et al. 2004). Conformance to expectations of Hardy–Weinberg equilibrium (HWE) and Linkage Disequilibrium were calculated in GENEPOP 4.2 (Raymond and Rousset 1995; Rousset 2008). Calculation of FST and mean uncorrected p-distance for 16S mtDNA and FST for microsatellites used 1000 interactions, and the Benjamini–Hochberg procedure was applied to detect type-I errors.
Isolation by distance (association between geographic and genetic distances) was tested among localities within PMI using a simple Mantel test based on 16S mtDNA and microsatellites (by means of FST). We analyzed data for the entire PMI gradient (839 km), and for each forest ecotype: dense forest (544 km) and open forest (295 km). Geographic distances were calculated with ‘geosphere’ and Mantel test with ‘vegan’ packages within R environment (R Core Team 2017).
Results
Population structure
The most likely number of genetic clusters (k) was: 16S = 6, TYR = 5, POMC = 4, and microsatellites = 1 (no structure). The 16S clusters (A–F) were: O. taurinus (A) and O. oophagus (B) from Reserva Ducke (RD), north of the Amazon river; a population confined to the east bank of upper Madeira river (C); and three other clusters with a parapatric distribution along the PMI (Figs. 1, 2a). Cluster D had the largest geographic distribution (M1–M11) and was dominant in the dense forest ecotype (M1–M10). Cluster F was dominant in M11, which correspond to the open forest ecotype, and was present but less frequent in M12 and M8. The geographical region between M9 and M11 is coincident with the ecotone between dense and open rain forests (Fig. 1). Cluster E included localities from J2–M15, distributed in a region of transition between open forest-savanna (Fig. 1). Very few individuals from this cluster were also present on the opposite bank of upper Madeira river (in M16 and M18), but no individuals of cluster C were detected on the west bank.
For 16S, we also conducted an additional and expanded population structure analysis (results not shown) to test if the existence of the six recovered groups from our study area was an artifact of BAPS sensitivity to geographic scale. For this analysis, we used all available sequences of the O. taurinus species group (Table S3 in Appendix 1) encompassing a broader geographic distribution in Amazonia. In general, this analysis reflected the clades present in our current 16S phylogeny (Fig. S1 in Appendix 2), and recovered the same six genetic groups for our study area. We show herein results restricted only to our study area as we had dense sampled localities and additional markers to 16S (nDNA and microsatellites), and because discussion of the expanded results is beyond the scope of this paper.
Among 16S GenBank sequences, however, we identified one locality of interest within PMI (Lábrea), located on the east bank of Purus river, approximately between M11 and J2 (Figs. 1, 2a). This locality was included only in BAPS analyses and is located in an area of ecological tension where up to three types of environments contact (dense forest, open forest, and the transition between open forest-savanna) in combination with the presence of the Purus river and one of its tributaries (Fig. 1). The two individuals from this locality clustered each to a different group (clusters D and E); therefore, Lábrea seems to be the southern and northern distribution limits for these clusters, respectively (Figs. 1, 2a). Overall, for 16S there was a notable dominance of cluster D in dense forest; while clusters E and F were dominant in open forest, indicating that the main phylogeographic break occurred between M10–M11. Although there was no clear distribution limit between the two open forest clusters, a deeper subdivision might suggest that cluster F was dominant exclusively in open forest, while cluster E was dominant in the transition between open forest-savanna (Fig. 1). In addition, four individuals within PMI (in M2, Lábrea and J2) had a considerable portion of admixture from cluster C (Fig. 2a).
Barplots of population structure analyses conducted in BAPS for Osteocephalus taurinus in central-southern Amazonia, resulting from mtDNA (a 16S) and nDNA (b TYR and c POMC). Sample size (n individuals) and the most probable number of genetic clusters (k) is provided for each marker. Each genetic cluster is assigned to a different letter and color, and each bar represent an individual with the proportion of its admixed genome (P < 0.05). Two physical (central Amazon and upper Madeira rivers) and one environmental barrier (dense-open forest ecotone) divide our study area in four biogeographical regions (see Fig. 1). For sample sizes in each locality see Table 1, except for L (Lábrea; n = 2). Oo = O. oophagus from RD
For the five recovered clusters (A–E) in TYR, one corresponded to O. oophagus (B) and another to the east bank of upper Madeira river (C). The remaining three clusters were distributed at RD and the entire PMI. Clusters A and D were most frequent at RD and M1–M10; while cluster E was most frequent from M11–M15 (Figs. 1, 2b). In concordance with 16S, but less structured, latitudinal substitution of clusters within PMI was observed and the most conspicuous phylogeographic break for TYR also occurred between M10–M11. TYR also showed evidence of restricted haplotype sharing at the upper Madeira river (Figs. 1, 2b). In addition, there were different levels of admixture for TYR; the most noticeable case was an O. taurinus from RD with a considerable proportion of O. oophagus (Fig. 2b).
For the four recovered clusters (A–D) in POMC, one was present at RD, east bank of the upper Madeira river, and within PMI from M1–M7 and J2–J1 (A). The remaining three clusters were present mainly within PMI: one from M1–M12 and M18 (B), another most frequent from M6–M13 (C), and the last one corresponding to a single individual from M12 (D; Fig. 2c).
The combined gene analyses recovered intermediate patterns from those observed independently and supported the existence of a phylogeographic break in the ecotone area, as well as different clusters occurring at both sides of the Amazon and Madeira rivers (Fig. 3). The total data set (16S + TYR + POMC; k = 5) recovered O. taurinus and O. oophagus from RD as a single cluster (A), a cluster occurring on the east bank of the upper Madeira river (C), a cluster occurring in the dense forest within PMI (D), and two clusters occurring in the open forest within PMI (E and B). The last one represented by a single individual from M12 (Fig. 3a). The 16S + TYR data set (k = 6) produced similar results than the total data set but it recovered O. oophagus as a different cluster (B), and it also identified an additional group occurring in dense forest (F). In addition, there were high levels of admixture within the dense forest ecotype (between clusters D and F; Fig. 3b). The nDNA data set (TYR + POMC; k = 4) recovered a cluster mostly dominant in RD and M1–M4 (A), a cluster mostly dominant from M5–M10 (D), other dominant in open forest at both sides of the upper Madeira river (C), and a cluster represented by the single individual from M12 (B; Fig. 3c).
Barplots of population structure analyses conducted in BAPS for Osteocephalus taurinus in central-southern Amazonia, resulting from combined mitochondrial (16S) and nuclear (TYR, POMC) data sets: a 16S + TYR + POMC, b 16S + TYR and c TYR + POMC. Sample size (n individuals) and the most probable number of genetic clusters (k) is provided for each combination of genes. Each genetic cluster is assigned to a different letter and color, and each bar represent an individual with the proportion of its admixed genome (P < 0.05). Two physical (central Amazon and upper Madeira rivers) and one environmental barrier (dense-open forest ecotone) divide our study area in four biogeographical regions (see Fig. 1). Oo = O. oophagus from RD
Phylogenetic relationships
Gene trees were mostly congruent between ML and BI approaches. The 16S mtDNA tree recovered the strongest geographic and taxonomic structure (Fig. 4; Fig. S1 in Appendix 2). For nDNA, phased and unphased data sets produced similar results. For easy of interpretation, results are presented for the unphased data sets. TYR and concatenated trees were moderately structured; while POMC was unstructured (Figs. S2–S4 in Appendix 2). We selected the 16S tree as our best phylogenetic hypothesis (Fig. 4) because its recovered clades (for our study area and surroundings) represented the six genetic clusters (A–F) identified by BAPS based on 16S, with support of TYR and combined data sets (Figs. 2, 3). In contrast, TYR tree recovered only two clades (B and C) of the five genetic clusters identified by BAPS based on TYR. These two clades corresponded to populations outside the PMI: B) O. oophagus and C) east bank of upper Madeira river. However, the support (ML/BI) for most of clades and their relationships was overall low in all gene and concatenated trees (Fig. 4).
Simplified phylogeny of the Osteocephalus taurinus species group in Amazonia, based on Maximum Likelihood (ML) topology of 16S mtDNA gene (526 bp). The six clades (A–F) recovered from our study area are depicted in different colors and show their geographical distribution (PMI = Purus–Madeira interfluve), forest ecotype and nodal values (ML bootstrap support/Bayesian Inference posterior probabilities). We also provide nodal values when a specific clade was recovered by TYR nDNA (555 bp) and concatenated (1578 bp) trees. Detailed phylogenies are available on Appendix 2
The 16S tree recovered the genus Osteocephalus (98/1) and the O. taurinus group (78/1) as monophyletic (Fig. S1 in Appendix 2). Other recognized Osteocephalus species groups also were monophyletic, except for the O. buckleyi group. In general, relationships among and within Osteocephalus species groups were not supported. Within the O. taurinus group, only clade A) O. taurinus sensu stricto (RD and surroundings at north of Amazon river), and four of the five candidate species from outside our study area (proposed by Jungfer et al. 2013) had high nodal support (Fig. S1 in Appendix 2). As noticed, clades B–F had low support (Fig. 4); however, within them some subclades presented high support for samples from localities close to each other (Fig. S1 in Appendix 2).
Genetic diversity and geographic differentiation
The 16S mtDNA had 41 haplotypes and 40 segregating sites (Table 2), and its haplotype network mirrored the six genetic clusters identified by BAPS based on 16S (Fig. 5). Osteocephalus taurinus (A) and O. oophagus (B) from RD were the most distant groups. The most conspicuous pattern from the network indicated that clusters C, D and E (with large number of individuals) were formed by one or two more common, shared haplotypes over the others (Fig. 5). Haplotype diversity (Hd) was slightly different among the six clusters, with highest values recorded in clusters E and F (Table 2).
Haplotype network based on 16S mtDNA gene (526 bp) for Osteocephalus taurinus from central-southern Amazonia. In concordance with population structure (BAPS) and phylogenetic analyses, haplotype groups are named and colored as A–F. The substitution represented as a star in the middle of the network separates haplotypes belonging to groups C–F. A single haplotype containing one individual (INPA-H37105 from M8) belongs to cluster F according BAPS analyses (see Figs. 1, 2a; Table 2), but it appears within cluster D in the haplotype network as the phylogenetic tree used to elaborate it did not recovered this haplotype within cluster F (see Fig. S1 in Appendix 2). Oo = O. oophagus from RD
Neutrality tests for O. taurinus and both grouped species (O. taurinus + O. oophagus) indicated significant neutrality deviance for Fu’s Fs but not for Tajima’s D for 16S (Table 2). When calculated for each genetic cluster, however, only cluster C (east bank of upper Madeira river) and cluster E (on the opposite west bank) showed significant deviance from neutrality. When neutrality was tested for O. taurinus and both grouped species after excluding clusters C and E, no significant deviance was found. Mean uncorrected p-distance for 16S was low and varied from 0.7–2.3% among the six clusters, with the higher values (including Fst) being those of O. oophagus versus all O. taurinus clusters (Table S4 in Appendix 3).
The AMOVA for 16S (Table S5 in Appendix 3) indicated that the greatest fraction of genetic variation was that between the two forest ecotypes (72.86%); while the variation among localities within forest ecotypes was low (7.14%) and slightly greater within localities (20%). The 16S mean p-distance among O. taurinus localities within the PMI ranged from 0–1.2% (average: 0.7). Genetic differentiation by means of Fst was low among immediate localities along PMI, but became higher rapidly when geographic distance increased (Table S6 in Appendix S3). We found significant isolation by distance in 16S for the entire PMI (r = 0.82; P < 0.001) and dense-forest localities (r = 0.71; P < 0.001), and slightly no significant for open-forest localities (r = 0.9; P = 0.083).
For microsatellites, only 2.45% of the total scored alleles (n = 1144) were out of the size range described in literature. The average number of alleles among the three loci varied from 3.3–9 across localities (see summary statistics in Table S7 in Appendix 3). Evidence of null alleles and deviance from HWE was found in one loci: Ha-T53. No significant values of Linkage Disequilibrium were observed between each pair of loci. In general, there was not a clear pattern of differentiation by means of FST among localities, except for M8 and O. oophagus versus the rest of O. taurinus localities, which showed the highest values. However, these were not considered significant after the Benjamini–Hochberg procedure (Table S7 in Appendix 3). There was no evidence of isolation by distance for microsatellites for the entire PMI (r = − 0.24; P = 0.993), dense-forest (r = − 0.17; P = 0.791) or open-forest localities (r = 0.05; P = 0.4).
Discussion
Our results over a fine-scale sampled region encompassing a ~ 900 km transect showed spatial genetic structure for O. taurinus populations in central-southern Amazonia, which was shaped by the presence of an environmental transition zone and large rivers. Therefore, these findings support both the gradient and riverine barrier hypotheses, respectively, for Neotropical diversification.
Parapatric structuration for O. taurinus populations within the Purus–Madeira interfluve (PMI) was mostly determined by a sharp phylogeographic break, concordant between 16S and TYR genes and combined data sets. Geographically, this break approximately matched the ecotone between dense and open forest ecotypes (Fig. 1), suggesting that the distribution of O. taurinus populations might be associated to these different forest formations. Isolation by distance was also detected for the entire PMI based on 16S, suggesting that low dispersion among sampling modules also could contribute to this pattern. However, isolation by distance by itself does not explain the abrupt change in gene frequencies at the ecotone area (between M10–M11). Our interpretation is supported by studies that reported adaptive divergence of populations (differences in morphology and behavior of tadpoles coupled with divergence on neutral genetic markers) to different habitat types across a continuum landscape in two frog species, thus suggesting incipient ecological specialization and assortive mating with same habitat conspecifics (Richter-Boix et al. 2013; Buskirk 2014). In addition, it has been reported that forest-specialist anurans tend to have stronger genetic structure than open-area specialists (Rodríguez et al. 2015). Therefore, we propose that differences in the macro habitat (forest structure and climate) could have produced distinct ecological responses (e.g., in physiological preferences and dispersal capabilities) in continuous O. taurinus populations distributed along the PMI environmental gradient, thus promoting their genetic differentiation.
Dense forests at northeastern PMI are more humid and less seasonal. Therefore, O. taurinus cluster D (description of clusters hereafter is based on 16S) could be adapted to more climatically stable, buffered microhabitats (Scheffers et al. 2014); and individuals probably need to cross shorter dispersal distances between trees, as supported by evidence of isolation by distance found (in 16S). In contrast, open forests at southwestern PMI are more dry and seasonal (IBGE 2012; Alvares et al. 2013). Open forests and savannas of this region (near to Humaitá and Porto Velho cities) experienced an extremely cold and dry climate during glacial periods in the late Pleistocene (Cohen et al. 2014). Therefore, O. taurinus clusters E and F on the west bank, and cluster C on the east bank at the upper Maderia river could have been historically exposed to more climatically unstable and ephemeral conditions; and therefore, need to disperse over larger distances in search of resources, such as reproductive sites (Rodríguez et al. 2015). Isolation by distance was no significant for open-forest localities (in 16S), which might support a higher dispersion of individuals inhabiting this ecotype. However, this lack of significance also may represent a type II error because the low number of localities and shorter distances among them. In addition, the presence of two genetic groups in open forest with the highest genetic variability (clusters E and F), coupled with evidence of deviance from neutrality (clusters C and E) suggest a scenario of population expansion for O. taurinus clusters within the open forest ecotype.
A recent study conducted in our same study area within the PMI (M1–M11) supports our findings. It showed that the intraspecific variation in plumage color and brightness in three passerine birds was associated to (among other geographical and environmental variables) the basal area of trees and palms from the sampling sites, thus suggesting that different phenotypes might be adapted to differences in the amount of light present in each habitat (i.e., dense vs. open forest) in order to increase their conspicuousness or crypsis (Teófilo et al. 2018). In the latter study, an abrupt cline in color (particularly the ultraviolet spectra) also was found for Gymnopithys salvini males between M10–M11 (lighter plumages in dense forest and darker ones in open forest; Teófilo et al. 2018), thus exactly matching the phylogeographic break identified for O. taurinus populations.
We acknowledge that O. taurinus individuals inhabiting different forest ecotypes can disperse to the adjacent habitats, but they seem to be filtered by the environment as the proportion of these individuals was low in localities where two groups contact (in 16S), all located in the proximities of the ecotone area (i.e., M8, M11 and M12; Fig. 1). Previous studies on rain forest frogs have suggested that steep genetic clines and contact zones are maintained by reinforcement due to morphological, acoustic and genetic character displacement as prezygotic mechanisms (Hoskin et al. 2005); and hybrid unviability as postzygotic mechanisms (Simões et al. 2012; Smith et al. 2013). The evolution of these mechanisms in transition zones are thought to reduce the likelihood of heterospecific encounters (Nosil 2012), thus maintaining the geographic differentiation of parapatric populations (Endler 1977). Although we identified a main phylogeographic break coincident with the ecotone between dense and open forests for O. taurinus populations, further research is needed to determine its relevance for other taxonomic groups and the mechanisms promoting its maintenance.
Other landscape features also could be related to the differentiation of the two main vegetation formations within the PMI, and subsequently to the O. taurinus population structure. Each forest ecotype is associated with a different drainage basin: northeastern dense forest tributaries drain into the Madeira river depression, while southwestern open forest tributaries drain in an opposite direction into the Purus river depression (Fig. 1). Probably, the location of an ancient structure, the Purus arch (approximately coincident with the dense-open forest ecotone), drove the first division of these drainage basins. The type and age of sedimentary deposits are also different in each basin, with older sediments on the west side of Purus arch and younger on the east side (Rossetti et al. 2005). Different draining dynamics coupled with differences in sedimentary deposition age in each basin might have had an effect on the vegetation establishment of major forest ecotypes within the PMI (Cohen et al. 2014).
In addition, the Purus arch itself was an important barrier separating western and eastern Amazonia during the Oligocene through the middle Miocene (34–11 Ma; Antonelli et al. 2010). Large scale landscape reconfiguration and establishment of rain forests in western Amazonia during the late Miocene (11–7 Ma) are thought to be crucial for the diversification of several Amazonian organisms (Antonelli et al. 2010). However, the lack of knowledge on the age and ancestral distribution of O. taurinus species group within Amazonia limit our inferences to determine if the Purus arch might have contributed to the differentiation of O. taurinus populations within the PMI. For instance, the role of ancient geomorphological barriers as drivers of geographic differentiation (rather than rivers) involving secondary contact has been proposed for some Amazonian frogs, particularly dendrobatids (Gascon et al. 1998; Lougheed et al. 1999; Symula et al. 2003; Simões et al. 2008). However, we could not determine the type of contact (primary or secondary) for O. taurinus populations within PMI, and this issue must be addressed in future research.
We detected a barrier effect of the upper Madeira and Amazon rivers by separating O. taurinus (clusters A and C) and O. oophagus (cluster B) outside the PMI. However, our inferences on a riverine barrier effect for the Amazon river are limited to only one locality in each bank at its middle section (RD and M1). In contrast, for the upper Madeira river area (4–6 localities in each bank) we identified different populations in each bank (clusters C and E) recovered by 16S, TYR genes and the combined data sets (Figs. 1, 2, 3). In addition, we detected restricted and unidirectional haplotype sharing from west to east banks at upper Madeira river. This last result is not necessarily evidence against a barrier effect of this river, as higher gene flow is expected near headwaters (Leite and Rogers 2013). It has been recently reported that anurans with large sizes and more generalist arboreal habits like O. taurinus are more likely to disperse between river banks than terrestrial and understory species (Fouquet et al. 2015). Therefore, the proximity to Madeira river headwaters and the ecology of O. taurinus could explain the observed pattern. However, it is not clear why the restricted haplotype sharing is unidirectional outside PMI (despite dense sampling in both river banks at the upper Madeira river). Although with less sampling effort, additional 16S GenBank sequences supported a barrier effect of the Madeira river at its middle and lower sections (Fig. S1 in Appendix 1). With the exception of a single sample (MTR 12668), no samples east of Madeira river were related to clades within PMI along its entire extent. Therefore, our findings support previous studies indicating the Madeira river as an important barrier for anurans (Ron 2000; Simões et al. 2008; Kaefer et al. 2013; Dias-Terceiro et al. 2015; Simões 2016; Godinho and Da Silva 2018).
The overall congruence between mtDNA and nDNA structure is a reliable indicator of the observed phylogeographic pattern, and also is suggestive of recent diversification since the variability is expected to be higher and coalescence time faster in mtDNA than in nDNA (Zink and Barrowclough 2008). Although phylogeographic breaks could appear in mtDNA stochastically and not to be related to any landscape structure (Irwin 2002), the break we found within PMI was consistent between independent molecular markers. Admixture levels for 16S were low and present in very few individuals, while in TYR they were higher. As the groups within PMI were highly structured with only few contact zones for 16S, we assumed that gene flow among them was low. Therefore, the admixture levels observed in 16S and TYR genes, as well as between clusters D and F in the 16S + TYR data set, were more probably related to unsorted ancestral polymorphisms. In addition, phylogenies were poorly supported and rendered O. taurinus as paraphyletic with respect to O. oophagus, as previously reported by Jungfer et al. (2013). Therefore, the relationships among the six genetic clusters from our study area remain unresolved. A future and better supported phylogeny will allow to determine whether the PMI populations occurring in each forest ecotype are sister to each other, which might provide insights onto the contact type between them.
We found no resolution for microsatellites, despite its potential to uncover population structure and gene flow patterns. We recognize that our inferences on microsatellites were limited due to the low number of loci (3) and HWE deviations in one of them. Overall, cross-amplification of heterologous microsatellites was low on O. taurinus. Therefore, our microsatellite results should be considered as rough estimates. We encourage the use of single nucleotide polymorphisms (SNPs) through next-generation sequencing approaches in future studies in order to obtain higher numbers of variable markers. SNPs are powerful tools to detect demographic process and loci involved with local adaptation (Lemmon and Lemmon 2013; McKinney et al. 2017). If O. taurinus populations are being selected to different environments, then future research must focus in loci involved with these processes, and explore their relations with gene expression and morphological fitness depending on environmental variation.
In conclusion, O. taurinus demonstrated to be a promising evolutionary system to track signs of ecologically based diversification in Amazonia. This study showed that the spatial genetic structure of the populations was associated with both the environmental variation present along the continuous landscape within PMI (different forest ecotypes), in agreement with the gradient hypothesis; and the presence of large rivers, in agreement with the riverine barrier hypothesis. One main phylogeographic break was identified among parapatric populations coincident with a transition zone (ecotone) between dense and open rain forests. Despite the potential for dispersal of O. taurinus, the ecotone area seemed to act as an environmental filter impeding the dispersion of individuals from genetic clusters inhabiting adjacent but different environments. To further test the origin and maintenance of this transition zone, future research must involve mating experiments and fitness assessments between O. taurinus populations from both sides of this transition zone; as well as multi-taxa research to determine its relevance for organisms with different ecologies.
References
Alvares CA, Stape JL, Sentelhas PC, de Moraes Gonçalves JL, Sparovek G (2013) Köppen’s climate classification map for Brazil. Meteorol Z 22:711–728
Antonelli A, Quijada-Mascareñas A, Crawford AJ, Bates JM, Velazco PM, Wolfgang W (2010) Molecular studies and phylogeography of Amazonian tetrapods and their relation to geological and climatic models. In: Hoorn C, Wesselingh FP (eds) Amazonia: landscape and species evolution. A look into the past. Wiley, Oxford, pp 421–433
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (2004) Genetix, version 4.05. Laboratoire génome, populations, interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier
Boubli JP, Ribas C, Lynch Alfaro JW, Alfaro ME, da Silva MNF, Pinho GM, Farias IP (2015) Spatial and temporal patterns of diversification on the Amazon: a test of the riverine hypothesis for all diurnal primates of Rio Negro and Rio Branco in Brazil. Mol Phylogenet Evol 82:400–412
Buskirk JV (2014) Incipient habitat race formation in an amphibian. J Evol Biol 27:585–592
Cintra BBL, Schietti J, Emillio T, Martins D, Moulatlet G, Souza P, Levis C, Quesada CA, Schöngart J (2013) Soil physical restrictions and hydrology regulate stand age and wood biomass turnover rates of Purus–Madeira interfluvial wetlands in Amazonia. Biogeosciences 10:7759–7774
Cohen MCL, Rossetti DF, Pessenda LCR, Friaes YS, Oliveira PE (2014) Late Pleistocene glacial forest of Humaitá–western Amazonia. Palaeogeogr Palaeoclimatol Palaeoecol 415:37–47
Corander J, Marttinen P (2006) Bayesian identification of admixture events using multi-locus molecular markers. Mol Ecol 15:2833–2843
Corander J, Tang J (2007) Bayesian analysis of population structure based on linked molecular information. Math Biosci 205:19–31
Corander J, Marttinen P, Sirén J, Tang J (2008) Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinform 9:539
Cracraft J (1985) Historical biogeography and patterns of differentiation within the South American avifauna: areas of Endemism. Ornithol Monogr 36:49–84
De la Riva I, Márquez R, Bosch J (1995) Advertisement calls of eight Bolivian hylids (Amphibia, Anura). J Herpetol 29:113–118
Dias-Terceiro RG, Kaefer IL, de Fraga R, de Araújo MC, Simões PI, Lima AP (2015) A matter of scale: historical and environmental factors structure anuran assemblages from the upper Madeira river, Amazonia. Biotropica 47:259–266
Endler JA (1977) Geographic variation, speciation, and clines. Princeton University Press, Princeton
Estupiñán-Tristancho RA (2001) Variação geográfica de Osteocephalus taurinus Steindachner, 1862 (Amphibia: Anura: Hylidae). MSc Dissertation, Universidade Federal do Pará—Museu Parense Emílio Goeldi
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Fernandes AM, Wink M, Aleixo A (2012) Phylogeography of the chestnut-tailed antbird (Myrmeciza hemimelaena) clarifies the role of rivers in Amazonian biogeography. J Biogeogr 39:1524–1535
Flot JF (2010) Seqphase: a web tool for interconverting phase input/output files and fasta sequence alignments. Mol Ecol Resour 10:162–166
Fouquet A, Courtois EA, Baudain D, Lima JD, Souza SM, Noonan BP, Rodrigues MT (2015) The trans-riverine genetic structure of 28 Amazonian frog species is dependent on life history. J Trop Ecol 31:361–373
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Gascon C, Lougheed SC, Bogart JP (1998) Patterns of genetic population differentiation in four species of Amazonian frogs: a test of the riverine barrier hypothesis. Biotropica 30:104–119
Godinho MBC, Da Silva FR (2018) The influence of riverine barriers, climate, and topography on the biogeographic regionalization of Amazonian anurans. Sci Rep 8:3427
Graham CH, Ron SR, Santos JC, Schneider CJ, Moritz C (2004) Integrating phylogenetics and environmental niche models to explore speciation mechanisms in dendrobatid frogs. Evolution 58:1781–1793
Grahame JW, Wilding CS, Butlin RK (2006) Adaptation to a steep environmental gradient and an associated barrier to gene exchange in Littorina saxatilis. Evolution 60:268–278
Hall JPW, Harvey DJ (2002) The phylogeography of Amazonia revisited: new evidence from riodinid butterflies. Evolution 56:1489–1497
Hoorn C, Wesselingh FP (eds) (2010) Amazonia: landscape and species evolution. A look into the past. Wiley, Oxford
Hoskin CJ, Higgie M, McDonald KR, Moritz C (2005) Reinforcement drives rapid allopatric speciation. Nature 437:1353–1356
Instituto Brasileiro de Geografia e Estatística (2012) Manual técnico de vegetação brasileira, 2nd edn. Manuais técnicos em geociências, Rio de Janeiro
Irwin DE (2002) Phylogeographic breaks without geographic barriers to gene flow. Evolution 56:2383–2394
Jungfer KH, Schiesari LC (1995) Description of a central Amazonian and Guianan treefrog, genus Osteocephalus (Anura, Hylidae), with oophagus tadpoles. Alytes 13:1–13
Jungfer KH, Faivovich J, Padial JM et al (2013) Systematics of spiny-backed treefrogs (Hylidae: Osteocephalus): an Amazonian puzzle. Zool Scr 42:351–380
Kaefer IL, Tsuji-Nishikido BM, Mota EP, Farias IP, Lima AP (2013) The early stages of speciation in Amazonian forest frogs: phenotypic conservatism despite strong genetic structure. Evol Biol 40:228–245
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Kearse M, Moir R, Wilson A et al (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Lanfear R, Calcott B, Ho SYW, Guindon S (2012) PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol 29:1695–1701
Larsson A (2014) AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278
Leite RN, Rogers DS (2013) Revisiting Amazonian phylogeography: insights into diversification hypotheses and novel perspectives. Org Divers Evol 13:639–664
Lemmon EM, Lemmon AR (2013) High-throughput genomic data in systematics and phylogenetics. Annu Rev Ecol Evol Syst 44:99–121
Li W, Cowley A, Uludag M, Gur T, McWilliam H, Squizzato S, Park YM, Buso N, Lopez R (2015) The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res 43:W580–W584
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Lougheed SC, Gascon C, Jones DA, Bogart JP, Boag PT (1999) Ridges and rivers: a test of competing hypotheses of Amazonian diversification using a dart-poison frog (Epipedobates femoralis). Proc R Soc Lond B Biol Sci 266:1829–1835
Magnusson WE, Braga-Neto R, Pezzini F et al (2013) Biodiversity and integrated environmental monitoring. Áttema Editorial, Santo André
Martins DL, Schietti J, Feldpausch TR et al (2015) Soil-induced impacts on forest structure drive coarse woody debris stocks across central Amazonia. Plant Ecol Divers 8:229–241
McKinney GJ, Larson WA, Seeb LW, Seeb JE (2017) RADseq provides unprecedented insights into molecular ecology and evolutionary genetics: comment on Breaking RAD by Lowry et al. (2016). Mol Ecol Resour 17:356–361
Moraes LJCL, Pavan D, Barros MC, Ribas CC (2016) The combined influence of riverine barriers and flooding gradients on biogeographical patterns for amphibians and squamates in south-eastern Amazonia. J Biogeogr 43:2113–2124
Moritz C, Patton JL, Schneider CJ, Smith TB (2000) Diversification of rainforest faunas: an integrated molecular approach. Annu Rev Ecol Syst 31:533–563
Naka LN, Bechtoldt CL, Henriques LMP, Brumfield RT, Heard AESB, McPeek EMA (2012) The role of physical barriers in the location of avian suture zones in the Guiana Shield, northern Amazonia. Am Nat 179:E115–E132
Nazareno AG, Dick CW, Lohmann LG (2017) Wide but not impermeable: testing the riverine barrier hypothesis for an Amazonian plant species. Mol Ecol 26:3636–3648
Nosil P (2012) Ecological speciation. Oxford University Press, Oxford
R Core Team (2017) R: a language and environment for statistical computing. R foundation for statistical computing, Vienna
Rambaut A, Suchard MA, Xie D, Drummond AJ (2014) Tracer version 1.6. Molecular evolution, phylogenetics and epidemiology, Edinburgh
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Heredity 86:248–249
Ribas CC, Aleixo A, Nogueira ACR, Miyaki CY, Cracraft J (2012) A palaeobiogeographic model for biotic diversification within Amazonia over the past three million years. Proc R Soc Lond B Biol Sci 279:681–689
Ribas CC, Aleixo A, Gubili C, d’Horta F, Brumfield RT, Cracraft J (2018) Biogeography and diversification of Rhegmatorhina (Aves: Thamnophilidae): implications for the evolution of Amazonian landscapes during the Quaternary. J Biogeogr 45:917–928
Richardson JL (2012) Divergent landscape effects on population connectivity in two co-occurring amphibian species. Mol Ecol 21:4437–4451
Richter-Boix A, Quintela M, Kierczak M, Franch M, Laurila A (2013) Fine-grained adaptive divergence in an amphibian: genetic basis of phenotypic divergence and the role of nonrandom gene flow in restricting effective migration among wetlands. Mol Ecol 22:1322–1340
Rodríguez A, Börner M, Pabijan M, Gehara M, Haddad CFB, Vences M (2015) Genetic divergence in tropical anurans: deeper phylogeographic structure in forest specialists and in topographically complex regions. Evol Ecol 29:765–785
Ron SR (2000) Biogeographic area relationships of lowland Neotropical rainforest based on raw distributions of vertebrate groups. Biol J Linn Soc Lond 71:379–402
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Rossetti DF, de Toledo PM, Góes AM (2005) New geological framework for western Amazonia (Brazil) and implications for biogeography and evolution. Quat Res 63:78–89
Rousset F (2008) Genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Santorelli S, Magnusson WE, Deus CP (2018) Most species are not limited by an Amazonian river postulated to be a border between endemism areas. Sci Rep 8:2294
Scheffers BR, Edwards DP, Diesmos A, Williams SE, Evans TA (2014) Microhabitats reduce animal’s exposure to climate extremes. Glob Change Biol 20:495–503
Schietti J, Martins D, Emilio T et al (2016) Forest structure along a 600 km transect of natural disturbances and seasonality gradients in central-southern Amazonia. J Ecol 104:1335–1346
Simões PI (2016) A new species of nurse-frog (Aromobatidae, Allobates) from the Madeira river basin with a small geographic range. Zootaxa 4083:501–525
Simões PI, Lima AP, Magnusson WE, Hödl W, Amézquita A (2008) Acoustic and morphological differentiation in the frog Allobates femoralis: relationships with the upper Madeira river and other potential geological barriers. Biotropica 40:607–614
Simões PI, Lima AP, Farias IP (2012) Restricted natural hybridization between two species of litter frogs on a threatened landscape in southwestern Brazilian Amazonia. Conserv Genet 13:1145–1159
Simões PI, Stow A, Hödl W, Amézquita A, Farias IP, Lima AP (2014) The value of including intraspecific measures of biodiversity in environmental impact surveys is highlighted by the Amazonian brilliant-thighed frog (Allobates femoralis). Trop Conserv Sci 7:811–828
Smith KL, Hale JM, Kearney MR, Austin JJ, Melville J (2013) Molecular patterns of introgression in a classic hybrid zone between the Australian tree frogs, Litoria ewingii and L. paraewingi: evidence of a tension zone. Mol Ecol 22:1869–1883
Smith BT, McCormack JE, Cuervo AM et al (2014) The drivers of tropical speciation. Nature 515:406–409
Sombroek W (2000) Amazon landforms and soils in relation to biological diversity. Acta Amazon 30:81–100
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Stein A, Gerstner K, Kreft H (2014) Environmental heterogeneity as a universal driver of species richness across taxa, biomes and spatial scales. Ecol Lett 17:866–880
Stephens M, Donnelly P (2003) A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73:1162–1169
Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989
Symula R, Schulte R, Summers K (2003) Molecular systematics and phylogeography of Amazonian poison frogs of the genus Dendrobates. Mol Phylogenet Evol 26:452–475
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Teófilo FHA, Schietti J, Anciães M (2018) Spatial and environmental correlates of intraspecific morphological variation in three species of passerine birds from the Purus–Madeira interfluvium, Central Amazonia. Evol Ecol 32:191–214
Thorpe RS (1984) Primary and secondary transition zones in speciation and population differentiation: a phylogenetic analysis of range expansion. Evolution 38:233–243
Trueb L, Duellman WE (1971) A synopsis of neotropical hylid frogs, genus Osteocephalus. Occ Pap Mus Nat Hist Univ Kansas 1:1–47
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Wallace AR (1852) On the monkeys of the Amazon. Proc Zool Soc Lond 20:107–110
Wang IJ (2012) Environmental and topographic variables shape genetic structure and effective population sizes in the endangered Yosemite toad. Divers Distrib 18:1033–1041
Zeisset I, Beebee TJC (2008) Amphibian phylogeography: a model for understanding historical aspects of species distributions. Heredity 101:109–119
Zink RM, Barrowclough GF (2008) Mitochondrial DNA under siege in avian phylogeography. Mol Ecol 17:2107–2121
Acknowledgements
This study was funded by the Brazilian Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Amparo à Pesquisa do Estado do Amazonas (FAPEAM) under grants conceded to A.P.L. (CNPq: Programa Ciência sem Fronteiras process 401327/2012-4; FAPEAM/CNPq: PRONEX process 445653/2009) and to F.P.W. (CNPq: process 475559/2013-4; FAPEAM: process 062.00665/2015). D.A.O. received a MSc scholarship by the Programa Estudantes-Convênio de Pós-Graduação—PEC-PG, CAPES/CNPq—Brazil. F.P.W. thanks grants from Partnerships for Enhanced Engagement in Research from the U.S. National Academy of Sciences and U.S. Agency of International Development PEER NAS/USAID AID-OAA-A-11-00012) and L’Oréal-UNESCO For Women In Science awards. Specimens were collected under permit numbers 13777-1 (A.P.L.) and 44832-1 (F.P.W.) issued by Instituto Chico Mendes de Conservação da Biodiversidade-ICMBio of the Ministry of Environment, Government of Brazil. These permits were subject to approval of all procedures for collecting and euthanizing frogs. R. de Fraga and M. Ferrão participated in the fieldwork. Laboratory work was conducted at the Thematic Laboratory of Molecular Biology—LTBM/INPA. C. Gubili provided pivotal advice on laboratory/statistical analyses for microsatellites. D. Pazmiño, O. Colatrelli and E. Choueri provided general advice on laboratory and statistical analyses. D. Pazmiño helped to elaborate figures of phylogenies. I. Kaefer, R. Leite, J. Alves-Gomes, C. Ribas, S. Borges, I. Farias and W. Magnusson provided comments on the study design and early versions of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest
The specimens involved in this study were euthanized using approximately 1 g of ointment (containing 0.2 g of Benzocaine) applied in the buccal mucous membrane. After the death of the animal, muscle and liver tissue were collected in 99% ethanol. The whole specimen was first fixed in 10% formalin, and then transferred to 70% ethanol for long-term storage.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ortiz, D.A., Lima, A.P. & Werneck, F.P. Environmental transition zone and rivers shape intraspecific population structure and genetic diversity of an Amazonian rain forest tree frog. Evol Ecol 32, 359–378 (2018). https://doi.org/10.1007/s10682-018-9939-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10682-018-9939-2