Abstract
l-Serine is a nonessential amino acid, but plays a crucial role as a building block for cell growth. Currently, l-serine production is mainly dependent on enzymatic or cellular conversion. In this study, we constructed a recombinant Escherichia coli that can fermentatively produce l-serine from glucose. To accumulate l-serine, sdaA encoding the l-serine dehydratase, iclR encoding the isocitrate lyase regulator, and arcA encoding the aerobic respiration control protein were deleted in turn. In batch fermentation, the engineered E. coli strain YF-5 exhibited obvious l-serine accumulation but poor cell growth. To restore cell growth, aceB encoding the malate synthase was knocked out, and the engineered strain was then transformed with plasmid that overexpressed serA FR, serB, and serC genes. The resulting strain YF-7 produced 4.5 g/L l-serine in batch cultivation and 8.34 g/L l-serine in fed-batch cultivation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
l-Serine is a nonessential amino acid, but plays an important role as a building block for cell growth and is widely used in the pharmaceutical and cosmetic industries [1–3]. In Escherichia coli, about 15 % of the assimilated carbon from glucose is transformed into l-serine and its metabolites [4]. In addition, l-serine is also a predominant source of one-carbon units used for a number of anabolic processes [5].
The current l-serine production relies mainly on enzymatic or cellular conversion method. Using a hydroxymethyltransferase, l-serine could be produced enzymatically from glycine and formaldehyde [6]. Employing the resting cells of methanol-utilizing bacteria Hyphomicrobium methylovorum, l-serine production was obtained using glycine and methanol as substrate with a yield of 24 g/L [7]. Although high l-serine production was achieved in these systems, the dependence on expensive substrates and low productivity make them less attractive. Meanwhile, fermentative production of l-serine from glucose received less attention due to its complicated regulation network in vivo as a central intermediate for a number of cellular reactions. In 2005, Peters-Wendisch et al. [8] constructed an l-serine producing Corynebacterium glutamicum strain by deleting l-serine dehydratase gene, reducing expression of the serine hydroxymethyltransferase, and overexpressing the l-serine biosynthetic genes serA, serC, and serB. This strain produced 9.04 g/L l-serine directly from glucose. Later, to reduce availability of 5,6,7,8-tetrahydrofolate and control the activity of hydroxymethyltransferase, Stolz et al. [5] knocked out pabABC genes in C. glutamicum, and further increased l-serine production to 36.26 g/L in 20-L fed-batch fermentation. In addition, by blocking or attenuating the degradation of l-serine, releasing the feedback inhibition of 3-phosphoglycerate dehydrogenase, and co-expression of 3-phosphoglycerate kinase and feedback-resistant 3-phosphoglycerate dehydrogenase in C. glutamicum ATCC 13032, Lai et al. [2] also obtained a recombinant strain SER-8.
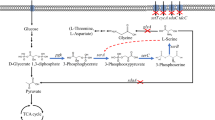
As a host, E. coli has also been widely used in the synthesis of amino acids due to its clear genetic background and amenable genetic manipulation [9–12]. However, up to now, few studies were focused on fermentative l-serine production in E. coli. By deleting three l-serine deaminase genes sdaA, sdaB, and tdcG, Li et al. [13] constructed a recombinant E. coli NW-7, which could produce 3.8 mg/L l-serine after 21 h batch fermentation. In E. coli, three enzymes, 3-phosphoglycerate dehydrogenase, phosphoserine aminotransferase and phosphoserine phosphatase, encoded by serA, serC, and serB, respectively, were responsible for catalyzing the biosynthesis of l-serine from 3-phosphoglycerate, which is an intermediate of glycolysis pathway (Fig. 1). As an important intermediate metabolite, l-serine is also the precursor of many other metabolites, therefore only a small percentage of l-serine could be accumulated and secreted into the medium from wild-type E. coli.
In this study, we constructed an l-serine producing strain from E. coli DH5α by overexpressing l-serine synthetic genes, blocking l-serine degradation pathway, and regulating the expression of glyoxylate cycle. The resulting strain YF-7 could produce 8.45 g/L l-serine in fed-batch fermentation, validating the effectiveness of our metabolic engineering strategy.
Materials and methods
Bacterial strains and plasmids construction
All strains, plasmids and oligonucleotides used in this study are listed in Tables 1 and 2. Escherichia coli DH5α was selected as a base strain for the construction of l-serine producing strain. serA FR gene, encoding the 3-phosphoglycerate dehydrogenase, has been deregulated the feedback inhibition of l-serine by deleting the last four C-terminal residues of the wild SerA [14]. Plasmids pBBR1MCS-2 and pTrc99a were applied to overexpress serA FR, serB, and serC. Plasmid pKJ-1 containing synthesized serA FR was constructed by BGI (Shanghai, China), and then this plasmid was used as a template for amplifying serA FR by oligonucleotides serA-F/serA-R. Genes serB and serC were amplified from chromosomal DNA of wild E. coli DH5α employing primers serB-F/serB-R and serC-F/serC-R, respectively. The three PCR products were then single-digested with XhoI, HindIII, and EcoRV (Fermentas) separately, and ligated into cloning vector pBBR1MCS-2 by T4 ligase (New England Biolabs, USA) sequentially to obtain pYF-2. To construct plasmid pYF-1, DNA fragment containing serA FR-serB-serC genes in pYF-2 was amplified by using serABC-F/serABC-R as the primers. The PCR products were then single digested with SacI, and ligated into the vector pTrc99a.
Gene deletion
Four genes, sdaA, iclR, arcA, and aceB, which encoded the l-serine dehydratase, isocitrate lyase regulator, aerobic respiration control protein, and malate synthase, respectively, were knocked out in DH5α sequentially to obtain strains YF-3, YF-4, YF-5, and YF-6. For genes sdaA and aceB, one-step inactivation method was carried out [15]. Primers sdaA-F/sdaA-R and aceB-F/aceB-R, and template plasmids pKD4 and pKD3, were used to obtain the linearized DNA for the deletion of sdaA and aceB, respectively. The other two genes, iclR and arcA, were knocked out by the linearized DNA fragments with extending homologous sequence [16]. Primers iclR-F/iclR-R and arcA-F/arcA-R, and chromosomal DNA of strains QZ1111 and LMJ-1 were applied to amplify the linearized DNA fragments for iclR and arcA, separately. The PCR products were then purified and electroporated into the electrocompetent strains containing the plasmid pKD46. Transformant cells were selected in solid Luria–Bertani medium (1 % tryptone, 0.5 % yeast extract, 1 % NaCl, and 1.5 % agar powder) containing chloramphenicol (17 mg/L) or kanamycin (25 mg/L). Candidate clones were screened by PCR employing primers sdaA-JF/sdaA-JR, aceB-JF/aceB-JR, iclR-F/iclR-R, and arcA-F/arcA-R, separately. The chloramphenicol or kanamycin cassette was removed with the helper plasmid pCP20. Finally, strain YF-6 was transformed with pYF-1 and pYF-2 to generate strains YF-7 and YF-8, respectively.
Growth conditions
Strains for cloning and inoculums were grown in Luria–Bertani media (1 % tryptone, 0.5 % yeast extract, and 1 % NaCl) at 37 °C for 8–12 h supplemented with the appropriate antibiotic (ampicillin (100 mg/L), chloramphenicol (17 mg/L), kanamycin (25 mg/L), spectinomycin (50 mg/L)) when necessary. For batch and fed-batch fermentation, the mineral AM1 medium [17] supplemented with 1 g/L yeast extract and 20 g/L glucose was used. A single clone was pre-cultured in 5 mL LB medium at 37 °C with 250 rpm shaking overnight. 1 mL overnight cells were inoculated into 50 mL AM1 medium for batch fermentation, and flasks were incubated at 37 °C with 250 rpm shaking. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was supplemented at the final concentration of 0.2 mM. For fed-batch fermentation, a stirred 5-L glass vessel with the BioFlo310 modular fermentor system (New Brunswick Scientific, Edison, NJ, USA) was applied, and the inoculum ratio was 10 % (v/v). When glucose concentration in the medium was below 10 g/L, feeding solution containing 500 g/L glucose was supplemented into the medium. Temperature was maintained at 37 °C, pH was controlled at 6.8 with NH3·H2O, and dissolved oxygen concentration was kept at 30 % via changing the fermentor agitation speed and aeration rate.
Analytical methods
Cell growth was monitored by OD600 with a spectrophotometer (Shimazu, Japan). Glucose, succinate, acetate, lactate, pyruvate, and 2-hydroxyglutaric acid (2-HGA) were quantitatively analyzed by high-performance liquid chromatography (HPLC; Shimazu, Japan) equipped with a column of Aminex HPX-87H ion exclusion particles (300 mm × 7.8 mm, Bio-Rad, USA). Samples were centrifuged at 12,000 rpm for 5 min and then filtrated with a 0.22-μm aqueous membrane. Except for 2-HGA, the mobile phase was 5 mM sulfuric acid (in Milli-Q water), with the flow rate of 0.6 mL/min and the column was maintained at 65 °C. Particularly, 2-HGA was determined by use of 2.75 mM sulfuric acid as the mobile phase. For measurement of l-serine, the supernatant obtained by centrifugation was pre-column derived as follows: 200 μL samples of supernatant, 100 μL phenyl isothiocyanate (PITC), and 100 μL triethylamine (TEA) from a Venusil AA analysis kit (Bonna-Agela Technologies, China) were supplemented into a 1.5-mL micro-centrifuge tube. After placing at room temperature for 1 h, 400 μL of n-hexane was supplemented into the tube to implement phase separation. Finally, the heavier phase (PTC-AA) was measured by HPLC with a UV detector (Shimadzu SPD-10A) according to the manufacturer.
Results and discussion
Deletion of sdaA for reducing the l-serine catabolism
l-Serine is a central intermediate which is involved in a number of cellular reactions that form glycine, l-tryptophan, glycerophospholipid, and other important mediates [18–20]. l-Serine dehydratase, encoded by sdaA gene, degrades l-serine to pyruvate, and reduces the accumulation of l-serine. It was reported that knocking out of sdaA gene slightly increased l-serine production in C. glutamicum [8, 21]. Therefore in this study, we firstly knocked out sdaA in E. coli DH5α, generating strain YF-3. Inactivation of sdaA increased the l-serine accumulation from 1.35 to 1.52 g/L, which is not very significant (Fig. 2a, b). Meanwhile, the pyruvate accumulation in YF-3 decreased from 2.23 to 1.95 g/L, which may lead to 20.5 % reduced acetate secretion (Fig. 2f).
Activation of glyoxylate pathway increased the l-serine accumulation
In previous study, it was confirmed that the activation of glyoxylate pathway increased the amount of l-serine that originated from the glycolysis pathway [22, 23]. In this study, we also knocked out iclR gene encoding an isocitrate lyase regulator [24], and constructed strain YF-4. As shown in Fig. 2c, YF-4 grew very poorly and consumed glucose slowly. The final OD600 was only 2.64 after 32 h cultivation, but it produced 2.03 g/L l-serine, which was 33.6 % higher than YF-3. This result indicated that glyoxylate cycle is an effective target for l-serine synthesis.
To improve the flux of glyoxylate pathway and TCA cycle during the microaerobic condition, the arcA gene, encoding an aerobic respiration control protein which negatively regulated the aceBAK operon [25, 26], was also inactivated to generate YF-5. Similar to YF-4, YF-5 also exhibited weak cell growth and slow glucose consumption, but the l-serine production in YF-5 increased to 2.29 from 2.03 g/L (Fig. 2d). This phenomenon was not consistent with the previous report of Waegeman et al. [22], in which the deletion of iclR and arcA increased the biomass accumulation in E. coli. This may be due to the difference in genetic background of the host. We suggested that in E. coli the activation of glyoxylate cycle accelerated the consumption of acetyl-CoA, which is the key intermediate involved in the central metabolic pathway (Fig. 1). The insufficient supply of acetyl-CoA may result in slow glucose consumption and poor growth of the host. This suggestion can also be partly validated by the reduced acetate and pyruvate secretion in YF-4 and YF-5 (Fig. 2f).
Deletion of aceB restored the cell growth of iclR and arcA mutant
Apart from generating malate, which is catalyzed by malate synthase, glyoxylate is also converted to glycerate-3-phosphate, the precursor of l-serine, via glyoxylate degradation pathway [27]. Therefore, the accumulation of glyoxylate may be beneficial for l-serine production. In order to reduce the consumption of acetyl-CoA and to save more glyoxylate for l-serine production, aceB, encoding the malate synthase, was knocked out in YF-5 to obtain YF-6. In batch cultivation, YF-6 exhibited restored cell growth, of which OD600 reached 10.3 after 32 h fermentation (Fig. 2e), but the accumulation of acetate, succinate, pyruvate, and lactate were also increased (Fig. 2f). This was probably because a part of carbon flux re-entered the by-products synthetic pathway. Nevertheless, YF-6 produced 2.51 g/L l-serine, highest among all the aforementioned gene-deletion strains. Interestingly, as shown in Fig. 2b, e, after glucose was consumed, the l-serine production in YF-3 and YF-6 continued for a few hours in batch fermentation. We guessed that these amounts were derived from glycine, pyruvate and other intermediates, which were the precursors of l-serine.
Overexpression of l-serine synthetic genes serA RT, serB, and serC
To direct more carbon flux into l-serine synthetic pathway in E. coli, serA FR, serB, and serC, encoding the deregulated 3-phosphoglycerate dehydrogenase, phosphoserine phosphatase, and phosphoserine aminotransferase, respectively, were overexpressed. Two different vectors, medium-copy vector pTrc99a with trc promoter, and low-copy vector pBBR1MCS2 with lac promoter, were employed to investigate the proper expression level of the three genes. The resulting plasmids pYF-1 and pYF-2 were then transformed into strain YF-6 and were cultivated to evaluate the l-serine production in batch fermentation. As shown in Fig. 3a, b, strain YF-8 containing pYF-2 exhibited faster glucose consumption rate and more biomass accumulation than YF-7. However, YF-7 accumulated 4.5 g/L l-serine, 21.6 % higher than that of YF-8 after 48 h cultivation, indicating medium expression of l-serine synthetic genes were benefit for l-serine accumulation. In addition, comparing to YF-8, YF-7 generated less by-products, only 1.73 g/L acetate, 1.09 g/L lactate, 0.08 g/L pyruvate, and 0.53 g/L succinate; while YF-8 secreted 2.02 g/L acetate, 1.38 g/L lactate, 0.38 g/L pyruvate, and 0.86 g/L succinate (Fig. 3c). Nevertheless, comparing with the host strain YF-6, the l-serine production in YF-7 and YF-8 increased by 79.8 and 47.9 %, respectively. In addition, YF-7 and YF-8 accumulated fewer by-products than that of YF-6, demonstrating overexpression of serA FR, serB, and serC was necessary for increasing l-serine production and decreasing by-products secretion.
Fed-batch fermentation of YF-7 in 5-L fermentor
To further evaluate the l-serine production capability in recombinant strain YF-7, fed-batch cultivation in 5-L fermentor was then performed (Fig. 4). In fed-batch cultivation, strain YF-7 exhibited a fast initial growth at first 18 h and reached its maximum OD600 of 39.5 at 46 h. The l-serine production in YF-7 continuous increased at an exponential phase, and the maximum titer reached was 8.34 g/L after 52 h cultivation.
Accumulation of 2-hydroxyglutarate in l-serine producing strains
During the analysis of the metabolites in the medium, we surprisingly identified a five-carbon dicarboxylic acid with a hydroxyl group at the alpha position, 2-hydroxyglutaric acid (2-HGA) (Fig. 5). It was reported 3-phosphoglycerate dehydrogenase encoded by serA gene had α-ketoglutarate reductase activity which could catalyze the reduction of α-ketoglutarate to 2-HGA [28]. In strain YF-3 with deletion of sdaA, 0.4 g/L 2-HGA was detected, similar to that of wild DH5α strain. When the glyoxylate cycle was activated, 2-HGA accumulation in strains YF-4 and YF-5 was decreased to 0.28 and 0.15 g/L, separately. In YF-4 and YF-5, a part of isocitrate was recruited to generate glyoxylate and succinate. Consequently, the generation of α-ketoglutarate decreased, leading to reduced 2-HGA. While in YF-6, glyoxylate cycle was blocked by deleting aceB gene, and the 2-HGA accumulation re-increased to 0.35 g/L. Since wild SerA had α-ketoglutarate reductase activity, overexpression of serA RT could also increase the accumulation of 2-HGA. Consistent with our conjecture, in YF-7 and YF-8, the 2-HGA accumulation was 1.92 and 1.54 g/L, approximately 5.5- and 4.4-fold to that of YF-6, respectively. In a previous study, Zhao et al. [28] confirmed 2-HGA is a competing substrate of glycerate-3-phosphate, the direct precursor of l-serine. Accordingly, reduction of 2-HGA by engineering the SerA may be beneficial for l-serine production in E. coli.
In summary, by overexpressing the three l-serine synthetic genes with suitable expression level, blocking the degradation of l-serine to pyruvate, and regulating the glyoxylate pathway, an l-serine producing strain was constructed. This strain produced 8.34 g/L l-serine in 5-L fed-batch fermentation, which suggested a potential application. To further improve the yield and productivity, other target genes need to be regulated.
References
Jiang W, Xia B, Liu Z (2013) A serine hydroxymethyltransferase from marine bacterium Shewanella algae: Isolation, purification, characterization and l-serine production. Microbiol Res 168(8):477–484
Lai S, Zhang Y, Liu S, Liang Y, Shang X, Chai X, Wen T (2012) Metabolic engineering and flux analysis of Corynebacterium glutamicum for l-serine production. Sci China Life Sci 55(4):283–290
Remesy C, Fafournoux P, Demigne C (1983) Control of hepatic utilization of serine, glycine and threonine in fed and starved rats. J Nutr 113(1):28–39
Pizer LI, Potochny ML (1964) Nutritional and regulatory aspects of serine metabolism in Escherichia coli. J Bacteriol 88:611–619
Stolz M, Peters-Wendisch P, Etterich H, Gerharz T, Faurie R, Sahm H, Fersterra H, Eggeling L (2007) Reduced folate supply as a key to enhanced l-serine production by Corynebacterium glutamicum. Appl Environ Microbiol 73(3):750–755
Hsiao HY, Wei T (1986) Enzymatic production of l-serine with a feedback control system for formaldehyde addition. Biotechnol Bioeng 28(10):1510–1518
Izumi Y, Yoshida T, Miyazaki SS, Mitsunaga T, Ohshiro T, Shimao M, Miyata A, Tanabe T (1993) l-serine production by a methylotroph and its related enzymes. Appl Microbiol Biotechnol 39(4–5):427–432
Peters-Wendisch P, Stolz M, Etterich H, Kennerknecht N, Sahm H, Eggeling L (2005) Metabolic engineering of Corynebacterium glutamicum for l-serine production. Appl Environ Microbiol 71(11):7139–7144
Gu P, Yang F, Kang J, Wang Q, Qi Q (2012) One-step of tryptophan attenuator inactivation and promoter swapping to improve the production of l-tryptophan in Escherichia coli. Microb Cell Fact 11(1):30
Lee SY, Park JH (2010) Integration of systems biology with bioprocess engineering: l-threonine production by systems metabolic engineering of Escherichia coli. Adv Biochem Eng Biotechnol 120:1–19
Park JH, Kim TY, Lee KH, Lee SY (2011) Fed-batch culture of Escherichia coli for l-valine production based on in silico flux response analysis. Biotechnol Bioeng 108(4):934–946
Park JH, Lee KH, Kim TY, Lee SY (2007) Metabolic engineering of Escherichia coli for the production of l-valine based on transcriptome analysis and in silico gene knockout simulation. Proc Natl Acad Sci USA 104(19):7797–7802
Li Y, Chen GK, Tong XW, Zhang HT, Liu XG, Liu YH, Lu FP (2012) Construction of Escherichia coli strains producing l-serine from glucose. Biotechnol Lett 34(8):1525–1530
Peters-Wendisch P, Netzer R, Eggeling L, Sahm H (2002) 3-Phosphoglycerate dehydrogenase from Corynebacterium glutamicum: the C-terminal domain is not essential for activity but is required for inhibition by l-serine. Appl Microbiol Biotechnol 60(4):437–441
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97(12):6640–6645
Li M, Gu P, Kang J, Wang Y, Wang Q, Qi Q (2012) Extending homologous sequence based on the single gene mutants by one-step PCR for efficient multiple gene knockouts. Folia Microbiol (Praha) 57(3):209–214
Martinez A, Grabar TB, Shanmugam KT, Yomano LP, York SW, Ingram LO (2007) Low salt medium for lactate and ethanol production by recombinant Escherichia coli B. Biotechnol Lett 29(3):397–404
Aboulwafa M, Hvorup R, Saier MH Jr (2004) Dependency of sugar transport and phosphorylation by the phosphoenolpyruvate-dependent phosphotransferase system on membranous phosphatidylethanolamine in Escherichia coli: studies with a pssA mutant lacking phosphatidylserine synthase. Arch Microbiol 181(1):26–34
Ikeda M (2006) Towards bacterial strains overproducing l-tryptophan and other aromatics by metabolic engineering. Appl Microbiol Biotechnol 69(6):615–626
Lorenz E, Stauffer GV (1996) MetR-mediated repression of the glyA gene in Escherichia coli. FEMS Microbiol Lett 144(2–3):229–233
Netzer R, Peters-Wendisch P, Eggeling L, Sahm H (2004) Cometabolism of a nongrowth substrate: l-serine utilization by Corynebacterium glutamicum. Appl Environ Microbiol 70(12):7148–7155
Waegeman H, Beauprez J, Moens H, Maertens J, De Mey M, Foulquié-Moreno M, Heijnen J, Charlier D, Soetaert W (2011) Effect of iclR and arcA knockouts on biomass formation and metabolic fluxes in Escherichia coli K12 and its implications on understanding the metabolism of Escherichia coli BL21 (DE3). BMC Microbiol 11(1):70
Wendisch VF, Bott M, Eikmanns BJ (2006) Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for biotechnological production of organic acids and amino acids. Curr Opin Microbiol 9(3):268–274
Yamamoto K, Ishihama A (2003) Two different modes of transcription repression of the Escherichia coli acetate operon by IclR. Mol Microbiol 47(1):183–194
Alexeeva S, Hellingwerf KJ, Teixeira de Mattos MJ (2003) Requirement of ArcA for redox regulation in Escherichia coli under microaerobic but not anaerobic or aerobic conditions. J Bacteriol 185(1):204–209
Malpica R, Franco B, Rodriguez C, Kwon O, Georgellis D (2004) Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc Natl Acad Sci USA 101(36):13318–13323
Ornston LN, Ornston MK (1969) Regulation of glyoxylate metabolism in Escherichia coli K-12. J Bacteriol 98(3):1098–1108
Zhao G, Winkler ME (1996) A novel alpha-ketoglutarate reductase activity of the serA-encoded 3-phosphoglycerate dehydrogenase of Escherichia coli K-12 and its possible implications for human 2-hydroxyglutaric aciduria. J Bacteriol 178(1):232–239
Kang Z, Gao C, Wang Q, Liu H, Qi Q (2010) A novel strategy for succinate and polyhydroxybutyrate co-production in Escherichia coli. Bioresour Technol 101(19):7675–7678
Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM 2nd, Peterson KM (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166(1):175–176
Cherepanov PP, Wackernagel W (1995) Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158(1):9–14
Acknowledgments
This work was financially supported by a Grant from the National Natural Science Foundation of China (31370085), a Grant from National High-Tech Research and Development Plan of China (2012AA022104).
Author information
Authors and Affiliations
Corresponding author
Additional information
P. Gu and F. Yang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Gu, P., Yang, F., Su, T. et al. Construction of an l-serine producing Escherichia coli via metabolic engineering. J Ind Microbiol Biotechnol 41, 1443–1450 (2014). https://doi.org/10.1007/s10295-014-1476-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-014-1476-6