Abstract
The spontaneous alcoholic fermentation of grape must is a complex microbiological process involving a large number of various yeast species, to which the flavour of every traditional wine is largely attributed. Whilst Saccharomyces cerevisiae is primarily responsible for the conversion of sugar to alcohol, the activities of various non-Saccharomyces species enhance wine flavour. In this study, indigenous yeast strains belonging to Metschnikowia pulcherrima var. zitsae as well as Saccharomyces cerevisiae were isolated and characterized from Debina must (Zitsa, Epirus, Greece). In addition, these strains were examined for their effect on the outcome of the wine fermentation process when used sequentially as starter cultures. The resulting wine, as analyzed over three consecutive years, was observed to possess a richer, more aromatic bouquet than wine from a commercial starter culture. These results emphasize the potential of employing indigenous yeast strains for the production of traditional wines with improved flavour.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The dynamics of fermentation, the composition and the flavour of wine are greatly influenced by non-Saccharomyces yeasts, an ecologically and taxonomically diverse group [1]. Non-Saccharomyces yeasts usually dominate in mature grape berries, where they account for 50–75% of the total yeast population, and consequently they persist during the early stages of the fermentation process [2]. Although non-Saccharomyces are generally not very ethanol tolerant and are unable to completely ferment the sugars present in must, they markedly affect wine flavour and quality through the production of various metabolites that contribute significantly to the quality of the final product [3–6]. It is widely reported that yeasts produce over 400 compounds during fermentation, including alcohols, esters, organic acids, aldehydes, ketones and terpenes [7], which contribute to the individual character of the wine [8]. Local geography plays a role too, as vineyards in different areas are characterized by specific Saccharomyces and non-Saccharomyces florae [9] that are adapted to the local conditions. This results in the production of unique wines with characteristic flavours, aromas and alcoholic strengths [10–14]. Furthermore, the contribution of the yeast depends on several additional parameters, such as the fermentation temperature, the quality of the grape juice, and the presence and concentration of additives, including sulfur dioxide [15, 16].
In the present work, the sequential use of indigenous wild yeast strains of Metschnikowia pulcherrima and Saccharomyces cerevisiae as starters is investigated, as well as their impact on the resulting wine product. These strains were isolated and characterized from naturally fermenting Debina must originating from the area of Zitsa (Epirus, Greece). This partnership has not been investigated before at the laboratory or industrial level.
Materials and methods
Isolation of indigenous yeast strains
Indigenous yeast strains were isolated from spontaneously fermenting Debina grape must originating from the area of Zitsa (Ioannina, Epirus, Greece), one of the most prominent wine-producing areas in Epirus. Debina is a white-wine-producing variety, and all samples were collected in late September when the grapes were ready for harvest. Five grape clusters of approximately 100 g each were collected randomly in the vineyard using aseptic sampling conditions. These clusters were crushed aseptically in the laboratory and maintained (together with their must, skins and seeds) for 20 days at 18°C to allow the spontaneous fermentation of the must and the proliferation of the indigenous yeast. Samples were taken at the beginning of fermentation (first day, Sample I, expected to contain culturable yeast representatives of the initial population, mainly non-Saccharomyces species) and at the end of fermentation, when 90% of the sugar content had been consumed (18th day, Sample II). This second sample was expected to contain mainly culturable Saccharomyces strains that can tolerate elevated ethanol concentrations. Serial decimal dilutions were prepared from these samples in Ringer’s solution (Merck) and plated on yeast extract–malt extract agar (YM agar) suitable for selecting yeasts. Chloramphenicol was added to the selection medium (100 μg/ml) to minimise bacterial growth [17]. Yeast strains from both samples were isolated on the basis of colony morphology and microscopic observations and used for taxonomic identification. In addition to the indigenous isolates, two certified yeast species, Saccharomyces cerevisiae strain 188/H51960/6 (Phillip Harris, England, UK) and Metschnikowia pulcherrima/3047 (National Collection of Yeast Cultures—NCYC, UK) were purchased and used as standard prototypes.
Biochemical characterisation of the indigenous yeast strains ZY6 and Z622
The biochemical characterisation of the isolated non-Saccharomyces strain ZY6 was achieved by performing several biochemical assays according to Yarrow [17] such as fermentation of carbohydrates, assimilation of nitrogen compounds, splitting of arbutine (β-glucosidase activity), growth at 30 and 37°C, starch formation, cycloheximide resistance, tolerance of 1% acetic acid, gelatin liquefaction, and hydrolysis of urea. For the Saccharomyces strain, Z622 API tests were used (API 32C, Biomerieux, Marcy l’Etoile, France) according to the manufacturer’s instructions.
Molecular characterisation of the indigenous yeast species
This was based on cloning and sequencing the internal transcribed spacer regions (ITS) as well as the polymorphic D1/D2 domains of 26S ribosomal RNA genes (rDNA). Prototype strains were also processed in parallel with controls.
The ITS1-5.8S rDNA-ITS2 unit was amplified using the primers ITS4 [18] and ITS5.1. Primer ITS5.1 is a modification of ITS5 [18] derived by deleting the first nucleotide (a G residue) at its 5′ end. The removal of this G results in a reduction in the difference between the T M values of the original primer pair ITS4–ITS5. The PCR cycles applied here were: an initial denaturation step at 94°C for 5 min, followed by 30 cycles consisting of three steps each (denaturation at 94°C for 1 min, annealing at 51°C for 1 min and extension at 72°C for 1 min), and a final extension step at 72°C for 10 min. The polymorphic D1/D2 domain located at the 5′ end of the 26S rDNA gene was amplified using the primer pair NL1 and NL4 [19] and the amplification conditions reported by Kurtzman and Robnett [20]. In addition, specific amplification of the ITS region of S. cerevisiae spp. was achieved using the primer pair SC1/SC2 and the reaction conditions reported by Josepa et al. [21]. All of the primers used in this study are listed in Table 1.
Amplification reactions were carried out in a PTC-100 version 7.0 thermocycler (MJ Research Inc., USA) using the Expand High Fidelity PCR system (Roche, Switzerland) and 1.5 mM MgCl2 (final concentration). The amplicons produced were purified using NucleoSpin Extract 2 in 1 (Macherey-Nagel, Germany). Molecular cloning was performed using Zero Blunt Kit (Invitrogen, USA) according to the manufacturer’s recommendations. Escherichia coli strain DH5α served as the recombinant plasmid host [22]. Cloned ITS or D1/D2 fragments were sequenced by Macrogen (Korea). Sequence alignments were performed using the program BLAST2 [23] at the National Cancer Bioinformatics Institute (NCBI) website.
Cells used for DNA extraction were grown for approximately 24 h at 30°C on YM agar, and DNA was isolated according to Zambardi et al. [24]. Amplified or extracted DNAs were separated by electrophoresis in 1% agarose gels, stained with ethidium bromide, and visualised using standard procedures [25].
Alcohol tolerance tests
A cell population of 107 cells/ml was inoculated in yeast extract–malt extract medium (YM) [17] with an initial concentration of reducing sugars of 200 g/l and containing 3, 6, or 12% v/v ethanol. A similar culture that contained no ethanol served as the control. All cultures were incubated at 30°C. Cell growth was monitored by measuring the optical density of the culture at 600 nm for 30 h.
Growth kinetics of the strains ZY6 (M. pulcherrima var. zitsae) and Z622 (S. cerevisiae) in Debina grape must
Debina grape must was fermented by the strains ZY6 and Z622, each of which were added at an initial population of 107 cells/ml. Cells of the commercial strain VL1 were also used as a control. The initial concentration of reducing sugars was 200 g/l (Baume value of 11.5). Fermentations were carried out in a 10 l Bioflo 110 bioreactor equipped with a cooling system (New Brunswick Scientific, USA) without aeration and with the temperature adjusted to 18°C. Growth was followed by a count of yeast colony forming units (cfu) on YM agar plates after inoculating serial decimal dilutions of 0.1 ml samples taken every 48 h for 30 consecutive days. Sugar consumption was followed by measuring the level of reducing sugars using the Nelson method [26].
Must fermentations
The must used in fermentation experiments for wine production was derived from grapevines of the variety Debina. The sugar content of the must ranged between 11.2 and 11.8 Baumes, while pH values ranged from 3.05 to 3.45 (Table 2). 107 cells/ml were used for all inoculations, based on the routine practices of the Chateau Glinavos winery (Zitsa, Epirus, Greece).
Laboratory-scale fermentations were performed in 50 l cultures at 18°C. For fermentations on an industrial scale, stainless steel vessels with a capacity of 500 or 50,000 l and equipped with cooling jackets at the Chateau Glinavos winery (Zitsa, Epirus, Greece) were used as bioreactors. Industrial wine fermentations were carried out according to the following scheme: M. pulcherrima var. zitsae strain ZY6 isolated from Sample I was added as the first starter, and when the ethanol concentration reached 6% v/v, S. cerevisiae strain Z622 was added to continue and complete the process (yielding “Wine 1”). In parallel, a second wine fermentation of the same volume using must from the same lot and a commercially available S. cerevisiae strain (VL1, routinely used in Chateau Glinavos as starter culture) was always set up as a control (referred to as “Wine 2”). Wine fermentations at the industrial pilot scale were performed three times in three consecutive years (2006, 2007 and 2008). In the years 2006 and 2007, the volume that underwent pilot fermentation was 500 l. In year 2008 this volume was scaled up to 50,000 l. The temperature of the fermenting must was always maintained at 18°C.
Physicochemical analysis of the fermented wines
The official analytical methods of the European Community were employed to determine the pH, the total acidity and the concentrations of ethanol, reducing sugars and sulfur dioxide in a conventional manner [27].
Gas chromatography and mass spectrometry
Sample preparation for analysis was performed by diethyl ether extraction according to Lilly et al. [28]. Analyses were done on a Hewlett Packard (USA) model 6890 series gas chromatograph coupled to a MSD 5973 quadrupole mass spectrometer. The gas chromatograph was equipped with a J&W Scientific (USA) capillary column (model HP-5MS: length, 30 m; inside diameter, 0.3 mm; film width, 0.25 μm) with helium used as the carrier gas. The injection block temperature was kept constant at 200°C. The oven temperature was programmed as follows: 35°C (held for 5 min), then increased at a rate of 10°C/min to 230°C, where it was held for 5 min. The mass spectrometer was operated in the electron impact mode, and the masses were scanned over an m/z range of 29–350 amu. The MS source temperature was 230°C and the quadrupole temperature was 150°C. Each sample was injected in duplicate. The aromatic compounds, which served as standards for peak identification, were purchased from Roth (Germany). The internal standard method was employed for quantitative analysis.
Results
Isolation and identification of yeast strains originating from spontaneously fermenting must: selection of the appropriate strains as starters for industrial vinification
In order to isolate indigenous yeast strains of Metschnikowia and Saccharomyces species that are characteristic of the natural yeast flora present in the area studied, samples I and II were obtained from spontaneously fermenting crushed Debina grapes, as described in “Materials and Methods”.
-
(a)
Sample I: Several yeast strains belonging to different non-Saccharomyces genera; i.e. Hanseniaspora, Candida, Rhodotorula, Pichia and Metschnikowia were isolated and identified through intensive molecular analysis (unpublished results). According to the biochemical data, strain ZY6 was the only one that exhibited a similar phenotype to the prototype strain NCYC-3047 of M. pulcherrima [17] (Table 3). Taxonomic characterization based on ITS polymorphism confirmed the preliminary biochemical characterization. The sequence identities of the ZY6 and NCYC 3047 regions (359 and 357 bp, respectively) with the M. pulcherrima prototype sequence (NCBI GenBank accession number EU137672) were 95.2 and 96.1%, respectively (Table 4). The above results were further tested using the polymorphism of D1/D2 regions within the 26S rRNA gene. Sequence alignment of amplicons of the D1/D2 regions of strains ZY6 and NCYC 3047 with the corresponding region of M. pulcherrima (NCBI GenBank accession number U45736) revealed identities of 98.3 and 99.4%, respectively (Table 5). These results demonstrate that strain ZY6 is a representative of Metschnikowia pulcherrima, and justify its selection for further use in this work.
Table 3 Biochemical characteristics of strain ZY6 (M. pulcherrima var. zitsae) Table 4 Size, accession numbers and alignment data for the ITS regions Table 5 Size, accession numbers and alignment data for the D1/D2 domains -
(b)
Sample II: Hundreds of colonies originating from the late fermentation sample II were tested by API, which revealed that all belonged to the species S. cerevisiae with 99.9% identity. Among these, 20 were randomly selected and used as culture starters in laboratory-scale vinifications (50 l). Preliminary organoleptic evaluation (tasting by a professional wine evaluator) indicated that the resulting wines using the strains Z607, Z608, Z614, Z617 and Z622 had the best aromatic bouquet and were selected for molecular characterization. The sequence of amplified ITS fragments (1,136 or 1,137 bp) (Table 4) and D1/D2 domains (572 bp) (Table 5) exhibited almost 100% identity with the corresponding sequences of the species S. cerevisiae, with virtually no nucleotide differences within these regions (Table 5). Among these, as evaluated by GC-MS analysis (unpublished results), strain Z622 presented the highest total concentration of desired esters, such as isoamyl acetate, ethyl caproate, hexyl acetate, ethyl caprylate, phenyl ethyl acetate and ethyl caprate, in laboratory-scale vinifications (50 l). Furthermore, the concentration of higher alcohols (2-methyl-1-propanol, 3-methyl-1-butanol, 2-methyl-1-butanol, hexanol-1,2-phenyl ethanol) did not exceed the limit proposed in the literature of 300 mg/l [29]. Similar results were obtained when strain Z622 was compared with the commercial S. cerevisiae strain VL1 used routinely by the Chateau Glinavos winery (data not shown). Therefore, strain Z622 was selected as the second starter and was used for further testing.
Alcohol tolerance tests
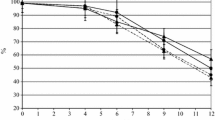
As expected, strain Z622 was capable of growing in a rich medium containing 3 and 6% v/v ethanol at the same rate as in the control medium (no ethanol) (Fig. 1A). No growth was observed in 12% v/v ethanol, but in this case the cells stayed viable for at least five days post-inoculation (data not shown). In contrast, strain ZY6 was only able to grow in 3% v/v ethanol. It retained viability in 6% v/v ethanol for 30 h post-inoculation (Fig. 1B). No viable cells were detected in 12% v/v ethanol 10 h post-inoculation.
Growth kinetics of the strains ZY6 and Z622 under fermentation conditions
In the present study, cells of the strains Z622 and ZY6 were cultivated in sterile-filtered Debina grape must at an initial cell population of 107 cells/ml. Cells of the commercial strain VL1 were also used as a control. Under these conditions, the Z622 and ZY6 cells exhibited different growth profiles and sugar consumptions in the medium used. Z622 cells consumed all of the sugar present in must within 20 days of fermentation, they doubled their population, and they maintained 30% viability 30 days post-inoculation. In contrast, ZY6 cells were unable to complete the process, consuming only up to 50% of the initial sugar content in 12 days (Fig. 2) and completely losing their viability 12 days later (24th day of fermentation). The growth kinetics of strain VL1 were comparable to those of strain Z622.
Industrial pilot scale vinifications using strain Z622 alone as the starter culture
Strain Z622 was used as a single starter culture in a industrial pilot scale vinification (500 l) (year 2005, Wine 0) and compared with the commercially available S. cerevisiae sp. strain VL1 used routinely by the Chateau Glinavos winery (year 2005, Wine 2). The physicochemical values of the grape must used are shown in Table 2. The fermentation kinetics of Z622 were similar to those of the commercial strain, as both strains completed the process within 20 days and produced wines with similar physicochemical parameters.
Quantitative analysis by GC-MS of several major volatile compounds present in the two wines revealed that, although they both presented the same concentrations of esters with pleasant fruity aromas [30], the total concentration of higher alcohols in the commercial wine (year 2005, Wine 2) exceeded the limit of 300 mg/l that is known to confer a “hard character” to wine [29] (Table 6).
Industrial pilot scale vinifications using the strains ZY6 and Z622 sequentially as starter cultures
Industrial-scale vinifications were performed in 500 l tanks containing Debina must at the Chateau Glinavos winery in Zitsa during the years 2006 and 2007. A 50,000 l tank was used during 2008. The physicochemical parameters of the grape musts used are presented in Table 2. Each year, one tank of Debina must was inoculated with strain ZY6 as the first starter, while strain Z622 (second starter) was added when the ethanol concentration reached approximately 6% (v/v) (Wine 1). This approach was used due to the results of the alcohol tolerance tests. In our hands, strain ZY6 proved to be a semi-alcohol-tolerant strain; it retained viability in 3% (v/v) ethanol for approximately 20 days and in 6% (v/v) ethanol for 12 days. A second tank containing the same must was inoculated with the commercial S. cerevisiae starter culture VL1, which is routinely used by the winery and served as the control vinification (Wine 2).
Quantitative analysis by mass spectrometry of several major volatile compounds present in the two wines revealed that, in all cases, Wine 1 presented significantly higher total concentrations of esters (known to confer fruity aromas to wine) than Wine 2 (Table 7). In addition, Wine 1 contained lower levels of higher alcohols than Wine 2 (Table 7). To statistically enhance the above results, Debina must from year 2008 was used in laboratory-scale vinifications that followed the same scheme of starter inoculations as used for the industrial pilot scale vinifications. For this purpose, 3 × 3 l laboratory vinifications were set up and inoculated separately as described above. The estimated values of the volatile compounds were similar to those obtained in the industrial-scale vinifications (Table 8). Here again, the total concentration of esters with pleasant aromas was significantly higher in Wine 1, whereas the total concentration of higher alcohols was lower in Wine 1 than in Wine 2, and was, in any case, within the acceptable limits (see the last two rows of Table 8).
Physicochemical analysis of the fermented wines
The values of the physicochemical parameters of the wines produced such as the ethanol and reducing sugar concentrations, the density, the SO2 concentration, and the total acidity were estimated. No differences were observed between any of the vinifications. All experimental and industrial vinifications gave the following results: ethanol 11% v/v, reducing sugars 0.1–0.2 g/l, density 0.990–0.991 g/l, SO2 concentration 90–100 (mg/l), total acidity 5.6–6.4 g/l.
Discussion
It is well documented in the scientific literature that, aside from the grape variety, the production of a quality wine with a local character and a traditional name of origin also requires the exclusive use of indigenous yeasts [14]. In this work, the previously uncharacterized indigenous strains ZY6 and Z622 were used as starters to produce the traditional wine of Zitsa at Chateau Glinavos (Epirus, Greece).
Initially, strain ZY6 was identified as M. pulcherrima, among several other non-Saccharomyces species isolated from the initial fermentation stage of the must used. The domination of non-Saccharomyces strains at early stages of spontaneous must fermentation has reported previously by several other investigators [2, 14]. In particular, strain ZY6 exhibited a 98.3% DNA sequence identity with the D1/D2 region, corresponding to eight nucleotide differences with other M. pulcherrima strains deposited in the NCBI database (Table 5), as well as a 95.2% identity with the ITS cluster (Table 4). On the other hand, the prototype strain NCYC 3047 exhibited a 99.4% identity with the D1/D2 region and a difference of only three nucleotides (Table 5). In general, as proposed by Kurtzman and Robnett [20], strains within a species should possess no more than three nucleotide differences in the D1/D2 domain. When differences in this domain exceed three nucleotides, the isolates should be considered different varieties, subspecies or even species. In this regard, and based on the fact that strain ZY6 differs by eight nucleotides from the D1/D2 region of the prototype M. pulcherrima species, we propose to classify strain ZY6 as a different variety named M. pulcherrima var. zitsae. According to the same molecular analysis, strain Z622 was unambiguously classified as Saccharomyces cerevisiae.
In agreement with previous reports [1, 2, 14, 16], our results demonstrated that S. cerevisiae dominates in the later stages of the spontaneous fermentation of Debina grape must. Among the S. cerevisiae strains tested, Z622 was selected as a starter because of its superior effect on wine aromatic bouquet. Furthermore, Z622 proved comparable with the commercial S. cerevisiae strain used routinely by the Chateau Glinavos winery (Zitsa, Epirus, Greece), but it has the following extra advantages: (a) a higher concentration of 2,3-butanediol, a metabolite that has been used to differentiate wine character on the basis of its possible effect on the wine bouquet [31]; and (b) a lower total higher alcohol, the concentration of which should not exceed 300 mg/l [22]. These results mirror our findings based on laboratory-scale vinifications, and verified that strain Z622 is the more suitable second starter.
The effect of the sequential use of a non-Saccharomyces (ZY6) and a Saccharomyces (Z622) strain on wine quality was investigated by comparing Wine 1 with Wine 2 during the years 2006, 2007 and 2008 in industrial- or laboratory-scale vinifications. Although the physicochemical parameters of the wines did not differ significantly, the total concentration of esters associated with pleasant fruity aromas was estimated to be 1.4- to 3.2-fold higher in Wine 1 from all years as compared to Wine 2. Additionally, in all of the batch fermentations analyzed, the concentration of 2,3-butanediol was higher in Wine 1. On the other hand, Wine 2 contained a higher total concentration of the determined higher alcohols compared to Wine 1, which in most cases exceeded the acceptable limits [22]. Surprisingly, ethyl acetate was absent in Wine 1 from all years, as well as in the test Wine 0 produced using strain Z622 alone. Perhaps Z622 possesses an esterase that hydrolyzes ethyl acetate that is absent in VL1; the existence of unknown esterases has been reported before for other S. cerevisae strains [32, 33].
Non-Saccharomyces species such as Hanseniaspora, Candida and Pichia have been used in the past as first starters, followed by S. cerevisiae, at least in laboratory-scale vinifications [3–6]. The use of M. pulcherrima as a first starter at either laboratory or industrial scale has not been reported so far. Our results support these previous findings implying that non-Saccharomyces species are primarily responsible for the production of wines with enhanced pleasant fruity aromas [3–6].
All of the metabolites reported here were detected at concentrations above their perception threshold (10−4–10−12 g/l) [29]. These results were a perfect match with the organoleptic evaluation that took place at Chateau Glinavos in Zitsa on August 2008 for the vinification batches of 2006 and 2007 (results not shown). The professional evaluators tasted the unique aromatic bouquet that characterized Wine 1. Such results strongly support the notion that the sequential use of indigenous yeast strains starting with a non-Saccharomyces strain leads to the production of wines with fine organoleptic characteristics and a local traditional character.
References
Jolly NP, Augustyn OPH, Pretorius IS (2006) The role and use of non-Saccharomyces yeasts in wine production. S Afr J Enol Vitic 27:15–39
Fleet GH, Heard GM (1993) Yeast-growth during fermentation. In: Fleet GH (ed) Wine microbiology and biotechnology. Harwood, Chur, pp 27–54
Zironi R, Romano P, Suzzi G, Battistuta F, Corni G (1993) Volatile metabolites produced in wine by mixed and sequential cultures of Hanseniaspora guilliermonddii or Kloeckera apiculata and Saccharomyces cerevisiae. Biotechnol Lett 15:235–238
Toro ME, Vazquez F (2002) Fermentation behaviour of controlled mixed and sequential cultures of Candida cantarellii and Saccharomyces cerevisiae wine yeasts. World J Microbiol Biotechnol 18:347–354
Rojas V, Gil JV, Pinaga F, Manzanares P (2003) Acetate ester formation in wine by mixed cultures in laboratory fermentations. Int J Food Microbiol 86:181–188
Clementez-Jimenez JM, Mingorance-Cazorla L, Martinez-Rodriguez S, Las Heras-Vazquez FJ, Rodriguez-Vico F (2005) Influence of sequential yeast mixtures on wine fermentation. Int J Food Microbiol 98:301–308
Abbas CA (2006) Production of antioxidants, aromas, colours, flavours, and vitamins by yeasts. In: Querol A, Fleet G (eds) Yeasts in food and beverages. Springer, Berlin, pp 285–334
Demyttenaere JCR, Dagher C, Sandra P, Kallithraka S, Verhe R, De Kimpe N (2003) Flavour analysis of Greek white wine by solid-phase microextraction-capillary gas chromatography-mass spectrometry. J Chromatogr 985:233–246
Querol A, Barrio E, Ramon D (1994) Population dynamics of natural Saccharomyces strains during wine fermentation. Int J Food Microbiol 21:315–323
Moreno JJ, Millfin C, Ortega JM, Medina M (1991) Analytical differentiation of wine fermentations using pure and mixed yeast cultures. J Ind Microbiol 7:181–190
Regodon JA, Perez F, Valdes ME, De Miguel C, Ramirez M (1997) A simple and effective procedure for selection of wine yeast strains. Food Microbiol 1997(14):247–254
Clementez-Jimenez JM, Mingorance-Cazorla L, Martinez-Rodriguez S, Las Heras-Vazquez FJ, Rodriguez-Vico F (2004) Molecular characterization and oenological properties of wine yeasts isolated during spontaneous fermentation of six varieties of grape must. Food Microbiol 21:149–155
Nikolaou E, Soufleros EH, Bouloumpasi E, Tzanetakis N (2006) Selection of indigenous Saccharomyces cerevisiae strains according to their oenological characteristics and vinification results. Food Microbiol 23:205–211
Fleet GH (2008) Wine yeasts for the future. FEMS Yeast Res 8:979–995
Longo E, Cansado J, Agrelo D, Villa GT (1991) Effect of climatic conditions on yeast diversity in grape must from Northwest Spain. Am J Enol Vitic 42:141–144
Pretorious IS, Van der Westhuizen TJ, Augustyn OPH (1999) Yeast biodiversity in vineyards and wineries and its importance to the South African wine industry. S Afr J Enol Vitic 20:61–74
Yarrow D (1998) Methods for isolation, maintenance and identification of yeast. In: Kurtzman CP, Fell JW (eds) The yeast: a taxonomic study. Elsevier, Amsterdam, pp 77–100
White TJ, Bruns T, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, New York, pp 315–322
O’ Donnell K (1993) Fusarium and its near relatives. In: Reynolds DR, Taylor JW (eds) The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. CAB International, Wallingford, pp 225–233
Kurtzman CP, Robnett CJ (1998) Identification and phylogeny of ascomycetous yeast from analysis of nuclear large subunit (26S) ribosomal DNA partial sequence. Antonie van Leeuwenhoek 73:331–371
Josepa S, Guillamon J, Cano J (2000) PCR differentiation of Saccharomyces cerevisiae from Saccharomyces bayanys/Saccharomyces pastorianus using specific primers. FEMS Microbiol Lett 193:255–259
Hanahan D (1983) Studies of transformation of Escherichia coli with plasmids. J Mol Biol 166:4895–4901
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman D (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Zambardi G, Dtruetta A, Roure C, Fougue B, Girardo P, Chyrpe C, Marchand J, Fleurette J (1995) Rapid diagnosis of Mycobacterium tuberculosis infections by an ELISA-like detection of polymerase chain reaction products. Mol Cell Probes 9:91–99
Sambrook J, Russell D (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Nelson N (1944) A photometric adaptation of the Somogyi method for the determination of glucose. J Biol Chem 153:375–380
EC (1990) Council Regulation No. 2676/90 determining Community analysis methods applicable in the wine sector. Off J Eur Commun L272:1–192
Lilly M, Lambrechs MG, Pretorious IS (2000) Effect of increased yeast alcohol acetyltransferase activity on flavor profiles of wine and distillates. Appl Environ Microbiol 66:744–753
Rapp A, Mandery H (1986) Wine aroma. Experientia 42:873–884
Lambrechts MG, Pretorious IS (2000) Yeast and its importance to wine aroma—a review. S Afr J Enol Vitic 21:97–129
Romano P, Brandolini V, Ansaloni C, Menziani E (1998) The production of 2,3-Butanediol as a differentiating character in wine yeast. World J Microbiol Biotechnol 14:649–653
Lilly M, Bauer FF, Lambrechts MG, Swiegers JH, Cozzolino D, Pretorius IS (2006) The effect of increased yeast alcohol acetyltransferase and esterase activity on the flavour profiles of wine and distillates. Yeast 23:641–659
Verstrepen KJ, Van Laere SDM, Vanderhaeghen BMP, Derdelinckx G, Dufour JP, Pretorious IS, Winderickx J, Thevelein JM, Delvaux FR (2003) Expression levels of the yeast alcohol acetyltransferase genes ATF1, Lg-ATF1 and ATF2 control the formation of a broad range of volatile esters. Appl Environ Microbiol 69:5228–5237
Acknowledgments
This work was part of the 03ED375 research project, implemented within the framework of the “Reinforcement Programme of Human Research Manpower” (PENED), and co-financed by National and Community Funds (25% from the Greek Ministry of Development—General Secretariat of Research and Technology, and 75% from the EU—European Social Fund). The authors wish to thank the Unit of Food Quality Standardization and especially Dr. A. Badeka for the mass spectra obtained using an instrument from the Horizontal Laboratory Network of the University of Ioannina, as well as E. Glinavos, owner and manager of Chateau Glinavos, for his cooperation in offering grape must and facilities for industrial-scale vinifications.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
A patent was granted for the sequential use of M. pulcherrima var. zitsae strain ZY6 and S. cerevisiae strain Z622 for wine production (Hellenic Industrial Property Organisation, License No.1005847). Organoleptic evaluation was performed by the experts Anestis Haitidis and Dimitris Katsouras.
Rights and permissions
About this article
Cite this article
Parapouli, M., Hatziloukas, E., Drainas, C. et al. The effect of Debina grapevine indigenous yeast strains of Metschnikowia and Saccharomyces on wine flavour. J Ind Microbiol Biotechnol 37, 85–93 (2010). https://doi.org/10.1007/s10295-009-0651-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-009-0651-7