Abstract
Reservoir souring in offshore oil fields is caused by hydrogen sulphide (H2S) produced by sulphate-reducing bacteria (SRB), most often as a consequence of sea water injection. Biocide treatment is commonly used to inhibit SRB, but has now been replaced by nitrate treatment on several North Sea oil fields. At the Statfjord field, injection wells from one nitrate-treated reservoir and one biocide-treated reservoir were reversed (backflowed) and sampled for microbial analysis. The two reservoirs have similar properties and share the same pre-nitrate treatment history. A 16S rRNA gene-based community analysis (PCR-DGGE) combined with enrichment culture studies showed that, after 6 months of nitrate injection (0.25 mM NO3 −), heterotrophic and chemolithotrophic nitrate-reducing bacteria (NRB) formed major populations in the nitrate-treated reservoir. The NRB community was able to utilize the same substrates as the SRB community. Compared to the biocide-treated reservoir, the microbial community in the nitrate-treated reservoir was more phylogenetically diverse and able to grow on a wider range of substrates. Enrichment culture studies showed that SRB were present in both reservoirs, but the nitrate-treated reservoir had the least diverse SRB community. Isolation and characterisation of one of the dominant populations observed during nitrate treatment (strain STF-07) showed that heterotrophic denitrifying bacteria affiliated to Terasakiella probably contributed significantly to the inhibition of SRB.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
During secondary oil production in North Sea oil fields, sea water is often injected in order to maintain reservoir pressure and enhance oil recovery. The introduction of sea water alters the physical and chemical conditions in the reservoir as sulphate-rich sea water gradually cools the reservoir formation [1, 2] and blends with the warm (60–200°C) indigenous reservoir water (formation water) which is low in sulphate and rich in organic acids [3–5]. The cooling of the reservoir formation creates a new biotope for mesophilic sea water bacteria in the near injector area. Traditionally, oxygen is removed from the sea water before injection, so aerobic microbial activity in the reservoir is limited. The anoxic conditions combined with the high sulphate content of sea water give favourable conditions for sulphate-reducing bacteria (SRB). SRB produce the highly toxic gas hydrogen sulphide (H2S) during anaerobic respiration with sulphate, an activity that leads to reservoir souring [2, 6]. Biogenic souring of oil reservoirs is of great concern to the oil industry as it leads to corrosion, plugging and deterioration of oil and gas quality [2, 6, 7]. Water treatment with biocides is commonly used as a souring inhibitor but the effect varies. This may be due to the selection towards biocide-resistant populations [8] and biocide resistance of biofilm associated SRB [9].
Nitrate treatment has been shown to inhibit souring in reservoir model systems [10–12] and in the field [13–18]. The method is based on shifting the microbial activity from sulphate reduction to nitrate reduction by injection of nitrate. According to the biofilm model, H2S production is generated in the near injector area of the reservoir and is dependent on carbon, sulphate and bacterial biomass [19]. The latter is again regulated by the availability of mineral nutrients. In North Sea oil reservoirs, hydrocarbons from residual oil are accessible in surplus and may serve as carbon and energy sources for both SRB and nitrate-reducing bacteria (NRB) [20]. The organic acids from formation water also serve as potential substrates as they are readily degradable by bacteria and water injection additives containing organic components may also support microbial growth [21]. Growth on inorganic components is also possible; H2 and CO2 generated in the Earth’s crust drive a deep biosphere primary production involving SRB, iron-reducing bacteria and methanogens [22]. Sulphide-oxidizing bacteria (SOB) are also part of the microbial community in oil reservoirs [23, 24] and some isolates use nitrate as electron acceptor [25].
Due to the more favourable energy potential of nitrate reduction compared to sulphate reduction, NRB will out compete SRB for common substrates, but this mechanism is believed to be valid only in carbon limited systems [18]. It has been suggested that the inhibitory effect of nitrite is a key mechanism in nitrate treatment [11, 14] in addition to the increase in redox potential due to NRB activity [26, 27]. Nitric oxide and nitrous oxide, intermediates of denitrification, have also been shown to inhibit bacterial growth in general [28, 29]. The activity of nitrate-reducing sulphide-oxidizing bacteria (NR-SOB) will contribute to increased redox potential by biological oxidation of sulphide.
For offshore application, it is necessary to limit the quantity of nitrate injected due to logistic concerns. Laboratory and field experiments have shown that continuous injection of a low concentration of nitrate (0.25–0.71 mM) is sufficient to reduce reservoir souring [11, 12, 17]. Concomitant with enrichment of NRB, a reduction in numbers and activity of SRB was observed in the water injection systems at Veslefrikk and Gullfaks oil fields during nitrate treatment [13, 17, 18]. A reduction in H2S produced from the Gullfaks field also showed effect at reservoir level [17].
The aim of the present study was to characterise the microbial community near injector in a nitrate-treated North Sea oil reservoir and to identify NRB contributing to the inhibition of SRB. The microbial community of a biocide-treated reservoir was analysed for comparison. Sampling was done by reversing the water flow at the injector wells (backflowing) and collecting samples at different time intervals. The microbial communities of the two reservoirs were studied by 16S rRNA gene-based PCR-DGGE analysis followed by DNA sequencing. An extensive enrichment culture study was performed in order to survey NRB and SRB activities in the two reservoirs. PCR-DGGE analysis of enrichment cultures was performed in an attempt to connect dominating environmental populations to specific activity.
Materials and methods
Sampling
The Statfjord field is located in the Tampen Spur area in the Norwegian sector of the North Sea. The reservoirs consist of sandstone from the Lower Brent formation. Commercial oil production started in 1979 and deaerated sea water (gas stripped by methane) treated with biocides was injected for pressure support from the start. The wells included in this study, A-42 and B-26, are comparable regarding both formation properties and treatment history. At well B-26, biocide treatment was replaced by nitrate injection and had been treated with nitrate for the duration of 6 months at the time of sampling. Nitrate was added continuously in the form of [Ca(NO3)2] to a final concentration of 0.25 mM NO3 −. The present biocide regime at well A-42 is tetrakis hydroxymethyl phosphonium sulphate (THPS) dosed three times a week (250 ppm) upstream deaerator. In addition, oxygen scavenger is continuously added into the deaerator and antifoam is added upstream deaerator if needed.
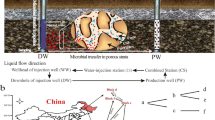
In the Statfjord reservoirs, the distance between injectors and producers is usually large and it takes between 2 and 7 years to flood one pore volume. In order to assess the effect of nitrate injection on the microbial community in the reservoir, water from the near injector area was sampled and analysed. The two injection wells, A-42 and B-26, were shut down for 4 days and the water flow was reversed (backflowed) and sampled at different time intervals (Fig. 1). Samples for microbial analysis were collected in 500 ml sterilised glass Duran bottles after 0, 2, 6, 12, 30, 48, 60, 78 and 96 h backflowing from well A-42 and after −0.5, 1, 4, 8, 24, 36, 54, 72 and 95 h backflowing from well B-26. The sample collected at the time −0.5 h contained water that had reached well-bottom but not the reservoir. The bottles were completely filled before capping to minimise exposure to air.
Illustration of the sandstone reservoir at Statfjord with injector and producer wells. The stippled arrows show the water direction during water injection. The solid arrows show the water flow during reverse flooding (backflowing) and the potential for collection of water from non-flooded areas of the reservoir
The sulphide content of the backflowed water showed that SRB activity was lower in the nitrate-treated reservoir (<3 mg H2S/l) compared to the biocide-treated reservoir (25–30 mg H2S/l) (Table 1). The biocide-treated well, A-42, is henceforth referred to as STA and the nitrate-treated well, B-26, is referred to as STB.
Enrichment culture study
NRB were enriched in a marine mineral medium with resazurin as redox indicator [30]. The medium was prepared in two different redox states: one was made anoxic by flushing with N2:CO2 (90:10) targeting facultative anaerobic NRB (F-NRB), the other was in addition reduced with 2 mM Na2S (final concentration) targeting obligate anaerobic NRB (O-NRB). SRB were enriched in a marine mineral medium [31] modified and designated W20 as previously described [11]. The following mixed vitamin solution (5 ml) was added per litre media (modified after [31, 32]): d(+)biotin (2 mg/l); 4-aminobenzoic acid (8 mg/l); Ca-pantothenate, cyanocobalamin (10 mg/l); nicotinic acid, thiamine dichloride (20 mg/l); pyridoxamine·2HCl (30 mg/l). The media were dispensed in aliquots of 15 ml into nitrogen-flushed 20 ml tubes and sealed with butyl rubber stoppers and aluminium crimp seals. Growth was assessed on substrates that reflect the range of carbon and energy sources accessible under in situ conditions in the reservoir. The substrate amount and concentrations used were as follows: H2:CO2 (80:20% headspace, added after inoculation); formate, propionate, butyrate, valerate and caproate (10 mM); acetate and lactate (20 mM); palmitate (15 mM); benzoate (0.1 mM); toluene, ethanol, methanol (0.1%, v/v); mixed acids C3–C6 (2.5 mM of each) and crude oil (0.15 ml). Lactate was included for the purpose of targeting Desulfovibrio spp.
A medium for enrichment of aerobic sulphide-oxidizing bacteria (A-SOB) was prepared by placing 5 ml sulphide-rich 3% agar in the bottom of a 20 ml N2-flushed tube. The agar was topped with 2 ml W20 and sealed as described above. The agar was left to polymerize overnight before the tubes were topped with 10 ml W20 medium. After inoculation, a syringe needle connected to a 0.2 μm filter was pierced through the rubber stopper for air supply. The medium for NR-SOB was prepared in the same way but with reduced NRB medium instead of W20 and without air supply.
In order to ensure anoxic conditions during inoculation, subsamples of the backflowed water were prepared in N2-flushed 100 ml serum bottles sealed with butyl rubber stoppers and aluminium crimp seals. The enrichment cultures were inoculated by transferring 1 ml of the subsamples using a N2-flushed syringe. When needed, the subsamples were injected with N2 to compensate for the reduction in water volume. The cultures were incubated at 30°C for 3 months.
Growth was monitored by visually observing the cultures for turbidity and was confirmed by phase microscopy. Activity of NRB was determined by measuring the reduction in nitrate concentration by ion exchange chromatography (IC25 Chromatograph/AS11-HC4x250 Column, Dionex, CA, USA) using UV detection (Spectra-physics UV150, Thermo Separation Products, CA, USA). Activity of SRB was determined by accumulation of H2S in the cultures according to Cord-Ruwisch [33]. Activity of NR-SOB was determined by decrease in nitrate concentration and by visual assessment of sulphur production (formation of white precipitate). Activity of A-SOB was assessed by production of sulphur.
Bacterial PCR-DGGE analysis and the subsequent sequence analysis of dominant bands were performed on enrichment cultures from sample 6 h from STA and sample 4 h from STB.
Characterisation of isolate
Salt and temperature requirements were determined during aerobic growth on pyruvate (10 mM) in a medium (D20) containing: NaCl, 20.0 g; Na2SO4, 4.0 g; MgCl2·6H2O, 3.0 g; NH4Cl, 1.4 g; K2HPO4·3H2O, 0.84 g; KH2PO4, 0.5 g; KCl, 0.5 g; CaCl2·2H2O, 0.15 g; 1 ml trace element solution SL 10 [34] per litre distilled water. The medium was autoclaved before adding 5 ml mixed vitamin solution and 30 ml 1 M NaHCO3. The medium was dispensed in aliquots of 15 ml into 20 ml tubes and sealed with rubber stoppers (Apotekproduksjon AS, Oslo, Norway) and aluminium crimp seals. Growth was determined by the increase in OD600.
The ability to utilize the following electron donors under nitrate-reducing conditions was determined by culturing in F-NRB medium: H2 (80:20% H2:CO2 headspace); H2S (1–20 μM); formate, propionate, butyrate, caproate (10 mM); acetate (20 mM); toluene (0.1%, v/v p.a. quality), crude oil (0.2 ml); n-dodecane (0.15 ml). Growth was determined by increase in OD600 and by phase microscopy (crude oil and n-dodecane), in three successive transfers. The mode of nitrate reduction was determined by measuring NO3 − and NO2 − by ion exchange chromatography and ammonium fluorometrically [35] during anaerobic growth with nitrate as electron acceptor. Aerobic growth on hydrocarbons was assessed in D20 medium amended with toluene, crude oil and n-dodecane. All incubations were performed at 30°C, except for the temperature experiment.
Molecular analysis
From both environmental samples and enrichment cultures, a volume of 1.5 ml was centrifuged at 12,000×g for 20 min. The supernatant was discharged and the pellets were frozen at −20°C until further analysis. The pellets were suspended in 50 μl molecular biology grade water before analysis (Eppendorf, Hamburg, Germany). Amplification of the V3 region of bacterial 16S rRNA genes was performed by whole-cell PCR using the forward Bacteria primer pA8f with GC clamp [36] and reverse universal primer PRUN518r [37]. Amplification of archaeal V3 region 16S rRNA genes was performed by whole-cell PCR using forward Archaea primer Arc931f [38] with GC clamp [37] and reverse primer UA1406r [39]. Per reaction, the PCR mixture contained: 3 μl cell suspension, 0.5 μM of each primer, 12.5 μl HotStarTaq Master Mix (Qiagen, Hilden, Germany), 2.5 μg BSA, and molecular biology grade water to a final volume of 25 μl. Bacterial PCR was performed in a GeneAmp2400 thermal cycler (Applied Biosystems, CA, USA) as follows: 95°C for 15 min; 35 cycles at 94°C for 30 s, 55°C for 30 s, 72°C for 1 min; 72°C for 10 min. The same program was used for archaeal PCR, except the annealing temperature was set at 64°C and amplification was run for 40 cycles. Positive amplification was determined by electrophoresis of 5 μl PCR sample in 1.5% agarose gel stained with ethidium bromide.
DGGE analysis of PCR products was performed as described earlier [40] but modified using a denaturing gradient ranging from 20 to 60% and by running the gel for 18 h at 70 V. The gels were photographed after staining with SYBR Gold (Invitrogen, Carlsbad, CA, USA) in 1× TAE for 45 min. Bands were excised from the DGGE gel and DNA eluated with molecular biology grade water and reamplified. PCR was performed as described above with the following modifications: 1 μl of DNA eluate served as template, no BSA was added and the reaction was run for 30 cycles. In some cases, reamplified DNA was run on a second DGGE for better separation before further analysis. Reamplified DNA was purified using QIAquick PCR purification kit (Qiagen, Hilden, Germany). Sequence PCR was performed using the primer PRUN518r and BigDye V.3.1 sequencing kit in accordance with the manufacturer (Applied Biosystems, CA, USA). Sequence analysis was performed on ABI PRISM 3700 and ABI 3730xl DNA analyzer (Applied Biosystems, CA, USA) by the Sars Centre DNA sequence facility at University Research of Bergen (Unifob AS, Bergen, Norway). The DNA sequences were analysed using BLAST tool [41] for identification of the closest relative registered in the GenBank (NCBI) [42] and by the Ribosomal Database Project (RDP) classifier [43] for taxonomic classification. Sequences with low similarity to GenBank sequences were checked for chimera [44]. Similar sequence types were compared pair-wise using BLAST 2 sequences tool (NCBI) [45]. The term “sequence type” refers to a set of sequences from the same environment that share 100% sequence identity. The phylogenetic distance between sequence types was calculated using ClustalX [46]. Phylogenetic trees were constructed using the bootstrap neighbour-joining algorithm (1,000 trials) and Tree View version 1.6.6 [47].
Near complete 16S rRNA gene analysis of the bacterium isolate was performed by whole-cell PCR using the forward Bacteria primer pA8f [48, 49] and reverse Bacteria primer Hr [49]. The PCR reaction was performed as described above with 5 μl culture (~108 cells/ml) as template. The PCR product was purified, then sequenced using forward primers: pA8f, PRBA338f [50] and PRE927f [51], and reverse primers: PRUN518r, PRE944r [51] and Hr. The sequence analysis was performed as described above; the DNA analyser used was ABI 3730xl. The partial sequences obtained were assembled using the CAP3 DNA Assembly Program [52] and manually checked for gaps and undesignated bases (N). The DNA sequence was analysed using BLAST tool [41] for identification of the closest validated relatives registered in the GenBank (NCBI) [42].
Nucleotide sequence accession numbers
16S rRNA gene sequences have been deposited in the GenBank (NCBI) under accession numbers EU109512–EU109531, EU312035–EU312052, EU330894–EU330920 and EU594271.
Results
Major environmental populations
Bacterial 16S rRNA gene sequences were successfully amplified from all environmental samples, whilst no archaeal 16S rRNA genes were amplified from any of the samples.
DGGE analysis of amplified bacterial DNA from the biocide-treated reservoir (STA) showed few bands and a similar band pattern throughout the sample series (Fig. 2). The major populations were affiliated to mesophilic, heterotrophic and chemolithotrophic bacteria (Table 2). The dominant sequence type observed in all samples from 6 hours’ backflowing and onwards showed 100% sequence identity to Phaeobacter arcticus [53]. None of the sequences were affiliated to SRB.
DGGE analysis of bacterial 16S rRNA gene PCR products of environmental samples from the biocide-treated reservoir (STA) and the nitrate-treated reservoir (STB). Lanes marked “m” refer to DNA marker with different GC content. Numbers on top of lanes refer to hours of backflowing. The bands aligned with numbered arrows are referred to in Table 2
DGGE analysis of amplified bacterial DNA from the nitrate-treated reservoir (STB) revealed a diverse band pattern that varied with sampling time (Fig. 2). The first sample of the series (−0.5 h) deviated from the other samples, as it consisted of water that had reached the bottom of the well but not penetrated the reservoir. The bottom well sample shared no common populations with the reservoir samples. Amongst the major environmental populations observed in the reservoir samples was a bacterium affiliated to the denitrifier Terasakiella pusilla (sequence type STB-g190). This bacterium was observed in all samples collected between 36 and 95 hours’ backflowing. Several of the other major populations at STB also belonged to the α-Proteobacteria, and based on the information obtained from enrichment culture studies and sequence analysis, some are connected to nitrate-reducing activity (Table 2). ε-Proteobacteria constituted another major group in the nitrate-treated reservoir. Bacteria affiliated to sulphide-oxidizing Arcobacter were observed throughout the sample series, but primarily after 54 hours’ backflowing. None of the major populations from the nitrate-treated reservoir were affiliated to SRB.
Microbial processes
The enrichment culture study showed that both NRB and SRB were present in the communities of both the reservoirs. One exception was growth of NR-SOB, which was observed only in samples from STB (data not included). Activity of A-SOB was observed in samples from both the reservoirs (data not included). The diversity of substrates that sustained growth was greater in the enrichment series from STB than in the series from STA. SRB from STB grew on all substrates, but only poorly on benzoate and methanol. Enrichment cultures targeting NRB from STB yielded growth accompanied by nitrate reduction on all substrates, but cultures targeting F-NRB grew poorly on toluene and crude oil.
SRB from STA did not grow on aromatic components and methanol, and grew poorly on crude oil, formate and propionate. Cultures targeting NRB from STA grew well on fatty acids, but poorly on aromatic components, methanol and crude oil. For a detailed overview of results from the enrichment culture study, see Table A in the supplementary material.
Phylogenetic diversity
Figures 3 and 4 show the phylogenetic distance between major environmental populations and enrichment culture phylotypes observed at STA and STB, respectively, based on partial 16S rRNA gene sequences. Of the four major environmental populations at STA, two belonged to the α-Proteobacteria, one to the ε-Proteobacteria and one to the Firmicutes (Fig. 3). Overall, the enrichment culture phylotypes belonged to bacterial groups not represented amongst the major environmental populations: δ-Proteobacteria, γ-Proteobacteria and Spirochaetes. The dominant environmental population at STA affiliated to Phaeobacter (STA-k215) was retrieved in both SRB enrichment culture added benzoate and NRB enrichment cultures added H2:CO2, formate and methanol (STA-231E). The SRB enrichment culture in question did not produce H2S during incubation. The presence of NRB at STA was evident by growth of bacteria affiliated to the genera Sedimenticola and Terasakiella, where the latter had identical sequence to the major environmental population STB-g190 at STB. Also observed in NRB enrichment cultures from STA was a bacterium with identical sequence to the environmental population STB-k526 at STB (STA-236E).
Phylogenetic tree of partial 16S rRNA sequences retrieved from the biocide-treated reservoir (STA). Sequence types given in bold refer to major environmental populations. Sequence types ending with “E” refer to bacteria observed in enrichment cultures. Sequences marked with asterisk were identical to major environmental populations during nitrate treatment (STB). Reference sequences from GenBank (NCBI) were cut to the same length as the sample sequences. The scale refers to 0.1 changes per nucleotide. Bootstrap values were obtained from 1,000 bootstrap trials and are given in percentage
Phylogenetic tree of partial 16S rRNA sequences retrieved from the nitrate-treated reservoir (STB). Sequence types given in bold refer to major environmental populations. Sequence types ending with “E” refer to bacteria observed in enrichment cultures. Reference sequences from GenBank (NCBI) were cut to the same length as the sample sequences, except in cases were only partial sequence was available (dagger). The scale refers to 0.1 changes per nucleotide. Bootstrap values were obtained from 1,000 bootstrap trials and are given in percentage
Four of the major environmental populations at STB belonged to the α-Proteobacteria, including the two dominant populations STB-k190 and STB-k161 (Fig. 4). Five of the populations belonged to the γ-Proteobacteria and four belonged to the ε-Proteobacteria. In the latter group, all were affiliated to the genus Arcobacter. Only one major population at STB belonged to the Firmicutes, and it had identical sequence to the Firmicute observed at STA. Enrichment culture phylotypes belonging to the δ-Proteobacteria and Spirochaetes were also observed at STB, in addition to members of the Thermotogae and Bacteroidetes. The study revealed the presence of bacteria affiliated to both heterotrophic and chemolithotrophic NRB. Bacteria affiliated to Sulfurimonas denitrificans (94%) and Sedimenticola selenatireducens (98%) was observed growing on H2:CO2 in enrichment cultures targeting F-NRB (STB-827E and STB-828E). An unknown γ-Proteobacterium (STB-668E) with identical sequence to a bacterium observed previously in an offshore water injection system during nitrate treatment [13] was observed in enrichment cultures targeting A-SOB and NR-SOB and in an O-NRB enrichment culture with added propionate. Bacteria observed growing on fatty acids and alcohols under nitrate-reducing conditions were affiliated to the genera Terasakiella, Marinobacter, Nesiotobacter, Phaeobacter and Geotoga in addition to unknown bacteria belonging to Firmicutes (STB-832E) and Bacteroidetes (STB-657E). Some of the major environmental populations (sequence types STB-g190, STB-k526 and STB-g537) shared 100% sequence identity with NRB enrichment culture phylotypes growing on a range of substrates including fatty acids, alcohols and aromatic components. Amongst the phylotypes observed in SRB enrichments from STB were bacteria affiliated to Desulfotignum (STB-644E) and Desulfovibrio (STB-821E).
The results from the enrichment study are not conclusive and should not be construed as such. However, an overview of the metabolic and phylogenetic diversity in the two reservoir communities was obtained in addition to information about the activity of major populations. All phylotypes observed in enrichment cultures from STA and STB are listed in the supplementary material in Table B along with the media and substrates that sustained their growth.
Isolate strain STF-07
The dominant bacterium at STB (sequence type STB-g190) shared 100% sequence identity with phylotypes observed in NRB enrichment cultures from both STA and STB. A strain of this bacterium was isolated after a successive dilution series in F-NRB medium; first with formate, thereafter with pyruvate as electron donor, until a pure culture was obtained. The isolate was designated strain STF-07. Near complete 16S rRNA gene analysis of strain STF-07 revealed a 96.4% sequence similarity to its closest validated relative T. pusilla IFO 13613 (GenBank Accession No. AB006768).
The cells of STF-07 are spiral shaped, motile by polar flagellum, possibly one at each pole. The bacterium is mesophilic; growth was observed at temperatures between 15 and 40°C, with optimum growth between 30 and 33°C. No growth was observed at 9 and 45°C. Good growth was observed on acetate and fatty acids C4 to C6 under nitrate-reducing conditions, but only poor growth was observed on formate. No growth was observed on inorganic energy sources or hydrocarbons. Nitrate was reduced by mode of denitrification (nitrate and nitrite was reduced, gas was formed and no NH4 + accumulated).
Discussion
A field-related mathematical model developed by Sunde et al. [19] infers that reservoir souring is caused by biofilm-associated SRB located close to the injector wells. An effective nitrate treatment should therefore be reflected in the microbial community near the injector in the form of a shift from SRB dominance to NRB dominance. The backflowed injection water was assumed to mirror the microbial community in the reservoir by containing detached biofilm cells. Tracer studies verified not only the recollection of injected water from both STA and STB but also showed that the sampled water contained water from non-flooded areas of the two reservoirs [54]. However, by assuming that the microbial activity primarily takes place in the flooded areas, the samples should reflect the active microbial community near the injector in terms of both composition and metabolic activity.
The microbial community from the biocide-treated reservoir (STA) consisted of only a few major populations of which none were SRB. The dominant sequence type was affiliated to a Phaeobacter-like bacterium that was not associated with H2S production. The observed in situ sulphide level at STA (25–30 mg/l), however, suggested a significant SRB activity in the reservoir. Failure to detect in situ SRB may have been due to biases in the PCR amplification process. On the other hand, the PCR-DGGE analysis may well have reflected the actual in situ community, where SRB was inhibited by biocide to a level below the detection limit of the PCR-DGGE method. A recent study on the biofilm community in the water injection system at Veslefrikk (North Sea) showed that Desulfovibrio spp. associated with H2S generation and corrosion in the system during biocide treatment were low in numbers [13]. Viable counts suggested that they constituted less than 1% of the total microbial community, at which point they were not detected by the PCR-DGGE method. Thus, experience from other systems shows that H2S generation may well originate from a small non-dominant SRB community.
The combined results of culture-independent community structure analysis and enrichment culture studies showed that major NRB populations were established in the nitrate-treated reservoir (STB) after 6 months’ continuous nitrate injection. NRB strain STF-07 shares 100% sequence identity with the dominant population observed in all samples collected after 36 h. Based on the partial characterisation of strain STF-07, the dominant in situ NRB is heterotrophic, able to utilize a range of fatty acids as the sole carbon and energy source during anaerobic growth by denitrification. Other major environmental populations of α-Proteobacteria were also observed growing under nitrate-reducing conditions and based on their substrate range they were also heterotrophic, able to utilize fatty acids and aromatic components.
The finding of NRB dominance at STB suggests effective nitrate treatment in the sampled part of the reservoir. Furthermore, the fact that the dominant in situ NRB reduce nitrate by denitrification gives input to the fate of nitrate in the reservoir. Denitrification is energetically more favourable than nitrate reduction to nitrite or ammonium, and is generally considered to result in a better growth yield for denitrifiers. When considering the inhibitory mechanism of SRB in a system dominated by denitrifiers, nitrite accumulation will probably be transient due to further reduction to nitric oxide and nitrous oxide and finally nitrogen gas. This suggests that nitrite, nitric oxide and nitrous oxide should all be considered potential inhibitory agents of SRB at STB, in addition to an overall increase in redox potential.
Bacteria affiliated to Arcobacter were part of the community both near the injection point and further into the reservoir at STB. Members of the genus Arcobacter are described as microaerophilic, NR-SOB that utilize organic acids as carbon source [55]. The marine isolate Candidatus Arcobacter sulfidicus, however, differs from this description by growing autotrophically [56]. Arcobacter have frequently been reported as members of oil reservoir communities [23, 24, 57] and have also been observed in biofilm from an offshore water injection system during nitrate treatment [13]. Enrichment of NR-SOB has also been observed in other oil fields during nitrate treatment [14, 58] and their role in sulphide remediation as well as SRB inhibition have been addressed [14]. The importance of active NR-SOB for successful reduction in sulphide levels in oil fields is emphasized by some authors [59], but in the present study, NR-SOB were not the dominant NRB after 6 months of nitrate treatment. The presence of Arcobacter at STB was most predominant furthest from the injector. Their location far from the injector point suggests that these Arcobacter respired with nitrate in situ. Furthermore, their presence indicates a transition zone between nitrate reduction and sulphate reduction in the reservoir. Due to their specific metabolism, the degree of NR-SOB presence may be used as indicator for treatment efficiency. The Arcobacter that were observed in samples collected nearest to the injectors at both STA and STB may have been involved in microaerophilic oxidation of H2S in the reservoir. Although the injection water is deaerated, trace amounts of oxygen may reach the reservoir [2] and sustain microaerophilic activity nearest to injectors.
Besides the Terasakiella-like denitrifier STF-07, the enrichment study revealed an NRB community constituting apparently heterotrophic and chemolithotrophic NRB affiliated to the genera Marinobacter, Sedimenticola, Nesiotobacter and Sulfurimonas. The microbial community at STB exhibited a larger phylogenetic diversity than the community at STA and was also able to grow on a wider range of substrates. This was probably due to the absence of selection pressure caused by biocide and to the introduction of nitrate as an additional electron acceptor. The absence of major SRB populations and the low H2S generation at STB (Table 1) suggest inhibition of SRB by nitrate treatment. This conclusion was further strengthened by the enrichment culture study that revealed a phylogenetically less diverse SRB community at STB compared to STA, which suggest inhibition and maybe also elimination of SRB (Figs. 3, 4). A collapse in the SRB community during nitrate treatment has been observed in other studies, but SRB were reported to be present in low numbers [11, 17, 18] leading to regeneration of sulphide production after the termination of nitrate injection [11]. Some SRB are able to use nitrate as alternative electron acceptor [60–62], an ability that has given rise to speculation of SRB being active as nitrate reducers during nitrate treatment. Bacteria closely affiliated to Desulfovibrio and Desulfotignum were observed in enrichment cultures from both STA and STB targeting O-NRB, but there was no evidence that these bacteria formed major environmental populations during nitrate treatment.
None of the major environmental populations at STA or STB were affiliated to known hydrocarbon degraders. This was surprising as crude oil is the dominant source of carbon and energy in the water washed areas near injectors. Fatty acid containing formation water is dislodged from the area, and carbon that is available from sea water/water additives is probably consumed in the water injection system before it reaches the reservoir. Hence, it is reasonable to assume that major oil degrading populations are present in both reservoirs. They may have remained undetected due to a close association to the oil phase. Non-hydrocarbon degraders such as Terasakiella-like strain STF-07 were probably more loosely attached to the biofilm and thus more dominant in the sampled water. The Terasakiella-like bacterium probably grew on fatty acids produced by the oil degrading community. The theory of an oil fixed community also offers a third possible explanation for the absence of SRB in the samples from STA; the H2S generating SRB community may have been firmly attached to the oil phase and thus not sampled. Although this theory implies that only parts of the communities at STA and STB were sampled, it does not interfere with the fact that major NRB populations were established at STB as a result of nitrate treatment.
In conclusion, the current study shows that nitrate injection inhibited SRB activity and reduced the diversity of the SRB community in a North Sea oil reservoir. The NRB community enriched in the reservoir was able to utilize the same range of substrates as the SRB community, which shows potential for long-term inhibition of SRB in the system. As dominant population, mesophilic, heterotrophic denitrifying bacteria affiliated to Terasakiella probably contributed significantly to the inhibition of SRB. The ecological approach of nitrate treatment, which is stimulating in situ inhibiting agents, is clearly the advantage of the method compared to biocide treatment. Where batch biocide treatment may have a temporary effect at best, NRB may provide continuous inhibition of SRB in the microenvironment where they both inhabit.
References
Cochrane WJ, Jones PS, Sanders PF, Holt DM, Mosley MJ (1988) Studies on the thermophilic sulphate reducing bacteria from a souring North Sea oil field. Soc Petrol Eng 18368:301–316
Vance I, Thrasher DR (2005) Reservoir souring: mechanisms and prevention. In: Ollivier B, Magot M (eds) Petroleum microbiology. American Society for Microbiology, Washington, pp 123–142
Barth T (1991) Organic acids and inorganic ions in waters from petroleum reservoirs, Norwegian continental shelf—a multivariate statistical analysis and comparison with American reservoir formation waters. Appl Geochem 6(1):1–15. doi:10.1016/0883-2927(91)90059-X
Barth T, Riis M (1992) Interactions between organic acid anions in formation waters and reservoir mineral phases. Org Geochem 19(4–6):455–482. doi:10.1016/0146-6380(92)90012-M
Nilsen RK, Beeder J, Thorstenson T, Torsvik T (1996) Distribution of thermophilic marine sulfate reducers in North Sea oil field waters and oil reservoirs. Appl Environ Microbiol 62(5):1793–1798
Cord-Ruwisch R, Kleinitz W, Widdel F (1987) Sulfate-reducing bacteria and their activities in oil production. J Petrol Technol 39(1):97–106. doi:10.2118/13554-PA
Herbert BN (1987) Reservoir souring. In: Hill EC, Shennan JL, Watkinson RJ (eds) Microbial problems in the offshore industry. Wiley, London, pp 63–71
Telang AJ, Ebert S, Foght JM, Westlake DWS, Voordouw G (1998) Effects of two diamine biocides on the microbial community from an oil field. Can J Microbiol 44(11):1060–1065. doi:10.1139/cjm-44-11-1060
Whitham TS, Gilbert PD (1993) Evaluation of a model biofilm for the ranking of biocide performance against sulfate-reducing bacteria. J Appl Bacteriol 75(6):529–535
McInerney MJ, Bhupathiraju VK, Sublette KL (1992) Evaluation of a microbial method to reduce hydrogen-sulfide levels in a porous rock biofilm. J Ind Microbiol 11(1):53–58. doi:10.1007/BF01583732
Myhr S, Lillebø BLP, Sunde E, Beeder J, Torsvik T (2002) Inhibition of microbial H2S production in an oil reservoir model column by nitrate injection. Appl Microbiol Biotechnol 58(3):400–408. doi:10.1007/s00253-001-0881-8
Reinsel MA, Sears JT, Stewart PS, McInerney MJ (1996) Control of microbial souring by nitrate, nitrite or glutaraldehyde injection in a sandstone column. J Ind Microbiol 17(2):128–136. doi:10.1007/BF01570056
Bødtker G, Thorstenson T, Lillebø B-L, Thorbjørnsen B, Ulvøen R, Sunde E, Torsvik T (2008) The effect of long-term nitrate treatment on SRB activity, corrosion rate and bacterial community composition in offshore water injection systems. J Ind Microbiol Biotechnol 35(12):1625–1636. doi:10.1007/s10295-008-0406-x
Jenneman GE, Moffitt PD, Bala GA, Webb RH (1999) Sulfide removal in reservoir brine by indigenous bacteria. Soc Petrol Eng Prod Facil 14(3):219–225
Larsen J (2002) Downhole nitrate applications to control sulphate reducing bacteria activity and reservoir souring. Corrosion 2004 paper 02025. NACE International, Houston
Larsen J, Rod HM, Zwolle S (2004) Prevention of reservoir souring in the Halfdan field by nitrate injection. Corrosion 2004 paper 04761. NACE International, Houston
Sunde E, Lillebø BLP, Bødtker G, Torsvik T, Thorstenson T (2004) H2S inhibition by nitrate injection on the Gullfaks field. Corrosion 2004 paper 04760. NACE International, Houston
Thorstenson T, Bødtker G, Lillebø BLP, Torsvik T, Sunde E, Beeder J (2002) Biocide replacement by nitrate in sea water injection systems. Corrosion 2002 paper 02033
Sunde E, Thorstenson T, Torsvik T, Vaag JE, Espedal MS (1993) Field-related mathematical model to predict and reduce reservoir souring. Soc Petrol Eng 25197:449–456
Widdel F, Rabus R (2001) Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr Opin Biotechnol 12(3):259–276. doi:10.1016/S0958-1669(00)00209-3
Sunde E, Thorstenson T, Torsvik T (1990) Growth of bacteria on water injection additives. Soc Petrol Eng 20690:301–316
Pedersen K (2000) Exploration of deep intraterrestrial microbial life: current perspectives. FEMS Microbiol Lett 185(1):9–16. doi:10.1111/j.1574-6968.2000.tb09033.x
Sette LD, Simioni KCM, Vasconcellos SP, Dussan LJ, Neto EVS, Oliveira VM (2007) Analysis of the composition of bacterial communities in oil reservoirs from a southern offshore Brazilian basin. Anton Leeuw Int J G 91(3):253–266. doi:10.1007/s10482-006-9115-5
Voordouw G, Armstrong SM, Reimer MF, Fouts B, Telang AJ, Shen Y, Gevertz D (1996) Characterization of 16S rRNA genes from oil field microbial communities indicates the presence of a variety of sulfate-reducing, fermentative, and sulfide-oxidizing bacteria. Appl Environ Microbiol 62(5):1623–1629
Gevertz D, Telang AJ, Voordouw G, Jenneman GE (2000) Isolation and characterization of strains CVO and FWKOB, two novel nitrate-reducing, sulfide-oxidizing bacteria isolated from oil field brine. Appl Environ Microbiol 66(6):2491–2501. doi:10.1128/AEM.66.6.2491-2501.2000
Jenneman GE, McInerney MJ, Knapp RM (1986) Effect of nitrate on biogenic sulfide production. Appl Environ Microbiol 51(6):1205–1211
Poduska RA, Anderson BD (1981) Successful storage lagoon odor control. J Water Pollut Control Fed 53(3):299–310
Mancinelli RL, Mckay CP (1983) Effects of nitric oxide and nitrogen dioxide on bacterial growth. Appl Environ Microbiol 46(1):198–202
Thom SR, Marquis RE (1984) Microbial growth modification by compressed gases and hydrostatic pressure. Appl Environ Microbiol 47(4):780–787
Myhr S, Torsvik T (2000) Denitrovibrio acetiphilus, a novel genus and species of dissimilatory nitrate-reducing bacterium isolated from an oil reservoir model column. Int J Syst Evol Microbiol 50:1611–1619
Widdel F, Pfennig N (1981) Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. 1. Isolation of new sulfate-reducing bacteria enriched with acetate from saline environments—description of Desulfobacter postgatei gen. nov., sp. nov. Arch Microbiol 129(5):395–400. doi:10.1007/BF00406470
Pfennig N (1978) Rhodocyclus purpureus gen. nov. and sp. nov. a ring shaped, vitamin-B12 requiring member of family Rhodospirillaceae. Int J Syst Bacteriol 28(2):283–288
Cord-Ruwisch R (1985) A quick method for the determination of dissolved and precipitated sulfides in cultures of sulfate-reducing bacteria. J Microbiol Methods 4(1):33–36. doi:10.1016/0167-7012(85)90005-3
Widdel F, Kohring GW, Mayer F (1983) Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. 3. Characterization of the filamentous gliding Desulfonema limicola gen. nov., sp. nov., and Desulfonema magnum sp. nov. Arch Microbiol 134(4):286–294. doi:10.1007/BF00407804
Holms R, Aminot A, Kèrouel R, Hooker B, Peterson B (1999) A simple and precise method for measuring ammonium in marine and freshwater ecosystems. Can J Fish Aquat Sci 56:1801–1808. doi:10.1139/cjfas-56-10-1801
Fjellbirkeland A, Torsvik V, Øvreås L (2001) Methanotrophic diversity in an agricultural soil as evaluated by denaturing gradient gel electrophoresis profiles of pmoA, mxaF and 16S rDNA sequences. Anton Leeuw Int J G 79(2):209–217. doi:10.1023/A:1010221409815
Muyzer G, Dewaal EC, Uitterlinden AG (1993) Profiling of complex microbial-populations by denaturing gradient gel-electrophoresis analysis of polymerase chain reaction-amplified genes-coding for 16S ribosomal-RNA. Appl Environ Microbiol 59(3):695–700
Jackson CR, Langner HW, Donahoe-Christiansen J, Inskeep WP, McDermott TR (2001) Molecular analysis of microbial community structure in an arsenite-oxidizing acidic thermal spring. Environ Microbiol 3(8):532–542. doi:10.1046/j.1462-2920.2001.00221.x
Baker GC, Smith JJ, Cowan DA (2003) Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 55(3):541–555. doi:10.1016/j.mimet.2003.08.009
Øvreås L, Forney L, Daae FL, Torsvik V (1997) Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol 63(9):3367–3373
Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402. doi:10.1093/nar/25.17.3389
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL (2005) GenBank. Nucleic Acids Res 33:D34–D38. doi:10.1093/nar/gki063
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73(16):5261–5267. doi:10.1128/AEM.00062-07
Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, Chandra S, McGarrell DM, Schmidt TM, Garrity GM, Tiedje JM (2003) The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res 31(1):442–443. doi:10.1093/nar/gkg039
Tatusova TA, Madden TL (1999) BLAST 2 SEQUENCES, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett 174(2):247–250. doi:10.1111/j.1574-6968.1999.tb13575.x
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25(24):4876–4882. doi:10.1093/nar/25.24.4876
Page RDM (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12(4):357–358
Edwards U, Rogall T, Blocker H, Emde M, Bottger EC (1989) Isolation and direct complete nucleotide determination of entire genes—characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res 17(19):7843–7853. doi:10.1093/nar/17.19.7843
Giovannoni SJ (1991) The polymerase chain reaction. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, London, pp 177–203
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, London, pp 115–175
Giovannoni SJ, Delong EF, Olsen GJ, Pace NR (1988) Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol 170(2):720–726
Huang XQ, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9(9):868–877. doi:10.1101/gr.9.9.868
Zhang DC, Li HR, Xin YH, Liu HC, Chi ZM, Zhou PJ, Yu Y (2008) Phaeobacter arcticus sp. nov., a psychrophilic bacterium isolated from the Arctic. Int J Syst Evol Microbiol 58:1384–1387. doi:10.1099/ijs.0.65708-0
Bjørnestad EØ, Sunde E, Dinning A (2005) The effect of produced water reinjection on reservoir souring in the Statfjord field. Petroleum abstracts no. 884455
Vandamme P, Falsen E, Rossau R, Hoste B, Segers P, Tytgat R, Deley J (1991) Revision of Campylobacter, Helicobacter, and Wolinella taxonomy - emendation of generic descriptions and proposal of Arcobacter gen. nov. Int J Syst Bacteriol 41(1):88–103
Wirsen CO, Sievert SM, Cavanaugh CM, Molyneaux SJ, Ahmad A, Taylor LT, DeLong EF, Taylor CD (2002) Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl Environ Microbiol 68(1):316–325. doi:10.1128/AEM.68.1.316-325.2002
Grabowski A, Nercessian O, Fayolle F, Blanchet D, Jeanthon C (2005) Microbial diversity in production waters of a low-temperature biodegraded oil reservoir. FEMS Microbiol Ecol 54(3):427–443. doi:10.1016/j.femsec.2005.05.007
Telang AJ, Ebert S, Foght JM, Westlake DWS, Jenneman GE, Gevertz D, Voordouw G (1997) Effect of nitrate injection on the microbial community in an oil field as monitored by reverse sample genome probing. Appl Environ Microbiol 63(5):1785–1793
Telang AJ, Jenneman GE, Voordouw G (1999) Sulfur cycling in mixed cultures of sulfide-oxidizing and sulfate- or sulfur-reducing oil field bacteria. Can J Microbiol 45(11):905–913. doi:10.1139/cjm-45-11-905
Dalsgaard T, Bak F (1994) Nitrate reduction in a sulfate-reducing bacterium, Desulfovibrio desulfuricans, isolated from rice paddy soil-sulfide inhibition, kinetics, and regulation. Appl Environ Microbiol 60(1):291–297
Dunsmore B, Whitfield TB, Lawson PA, Collins MD (2004) Corrosion by sulfate-reducing bacteria that utilize nitrate. Corrosion 2004 paper 04763. NACE International, Houston
Widdel F (1988) Microbiology and ecology of sulfate- and sulfur-reducing bacteria. In: Zehnder AJB (ed) Biology of anaerobic microorganisms. Wiley, New York, pp 469–585
Acknowledgments
This work was supported by StatoilHydro and the Norwegian Research Council through the Petromaks program. Sampling for microbial analysis was performed by Aquateam AS (Oslo, Norway) for StatoilHydro. Bente E. Thorbjørnsen, Rikke Helen Ulvøen and Tove R. Leiknes are especially thanked for their work with the enrichment cultures and other technical assistance. Nirmaladevi Sivasambu is thanked for assistance with molecular analysis and Bente-Lise P. Lillebø is thanked for the nitrate measurements. Vivienne Knowles is thanked for improving the manuscript linguistically.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bødtker, G., Lysnes, K., Torsvik, T. et al. Microbial analysis of backflowed injection water from a nitrate-treated North Sea oil reservoir. J Ind Microbiol Biotechnol 36, 439–450 (2009). https://doi.org/10.1007/s10295-008-0515-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-008-0515-6