Abstract
A lipase-producing bacterium was isolated and identified as Pseudomonas monteilii TKU009. A lipase (F2) and lipase-like materials (F1) were purified from the culture supernatant of P. monteilii TKU009 with soybean powder as the sole carbon/nitrogen source. The molecular mass of F1 and F2 was estimated to be 44 kDa by SDS-PAGE and gel filtration. The optimum pH, optimum temperature, and pH and thermal stabilities of F2 were 7, 40°C, 8–11, and 50°C; and of F1 were 6, 40°C, 6–7, and 50°C, respectively. F2 was completely inhibited by EDTA and slightly by Mg2+, Fe2+, Mn2+, and SDS. F1 was completely inhibited by EDTA and Fe2+ and strongly by Zn2+, Mn2+, Ca2+, Mg2+, and SDS. The activities of both the enzymes were enhanced by the addition of non-ionic surfactants Triton X–100 and Tween 40, especially for F1. F2 preferably acted on substrates with a long chain (C10–C18) of fatty acids, while F1 showed a broad spectrum on those with chain length of C4–C18. The marked activity of F2 in organic solvents makes it an ideal choice for application in a water-restricted medium including organic synthesis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The activities of enzymes in mixed water-organic solvents are of particular importance for the conversion of those substrates that are moderately soluble or insoluble in water. Lipases are among the most promising and important biocatalysts for carrying out reactions in both aqueous and non-aqueous media [1]. Lipases are ubiquitously produced by animals, plants, and microorganisms. Recently, microbial lipase has been widely used as a catalyst for the biosynthesis [2, 3]. Increasing attention has been paid to microbial lipases due to their wide range of biochemical properties and relative easiness to produce and isolate in bioreactors [2, 4]. The catalytic properties of microbial lipases have rendered many biotechnological applications in the food, cosmetic, detergent, pharmaceutical, and energy industries [3, 5–8]. This is primarily due to their ability to utilize a broad spectrum of substrates in the synthesis or hydrolysis of lipid compounds, as well as their activity in organic solvents [1, 9].
Lipases have been reported to be from several Pseudomonas species, including P. cepacia [6], P. aeruginosa [9, 10], P. fluorescens [11], and P. mendocina [12]. Among them, those from P. aeruginosa, P. cepacia, and P. fluorescens have been manufactured and extensively used in organic synthesis, which includes manipulation in non-aqueous solutions [9, 13]. Soybean oil has been used as an enzymatic substrate for biodiesel fuel production by immobilized P. cepacia lipase [6]. Enzymatic transesterification of soybean oil offers an environmentally more attractive option to the conventional physiochemical process. The production scale process of biodiesel using lipase might be worked out in the future. Taking into account the feasibility of bioconversion of soybeans to biodiesel fuel by fermentation, directly with a lipase-producing strain, the screening of lipase producing strains from Taiwan soils by the use of soybeans as the sole carbon/nitrogen source has been performed. In this paper, we report the purification and characterization of a lipase and a lipase-like material from the culture supernatant of the newly isolated strain P. monteilli TKU009 using soybean as the sole carbon/nitrogen source. We report herein the isolation and characterization of a unique and useful enzyme from P. monteilli.
Materials and methods
Materials
Yeast extract and nutrient broth were obtained from Difco (France) and the substrates p-nitrophenyl (pNP) palmitate (pNPP), pNP acetate, pNP butyrate, pNP caprylate, pNP caprate, pNP myristate, pNP palmitate, and pNP stearate were from Sigma Chemical Co. (Milwaukee, USA). BSA and dye reagent concentrate for protein determination, and reagents of SDS-PAGE were purchased from Bio-Rad Co. (Richmond, CA, USA) and the gels DEAE-Sepharose CL-6B, Sephacryl S-200, and Sephacryl S-100 from Amersham Pharmacia Biotech AB (Uppsala, Sweden). All other chemicals used were of analytical grade.
Identification of strain TKU009
The bacterial strain TKU009 was identified on the basis of morphological, physiological and biochemical parameters as well as on the basis of 16S rDNA-based sequence analysis after PCR amplification with primers and cloning. The identification was carried out by Food Industry Research and Development Institute, Taiwan. Nucleotide bases of the DNA sequence obtained were compiled and compared with sequences in the GenBank databases using BLAST program. From the morphological observation and from the physiological and biochemical characteristics, the microorganisms were further identified according to the description in Bergey’s Manual of Systematic Bacteriology [14] (identified by Food Industry Research and Development Institute).
Microorganism and enzyme production
Pseudomonas monteilii TKU009 was isolated from the soil at Taipei in Taiwan and maintained on nutrient agar plates at 30°C. Growing of this bacterium was carried out in a basal medium containing 0.1% K2HPO4 and 0.05% MgSO4·7H2O (pH 7), and supplemented with 1–3% (w/v) of soybean powder as carbon source. Various volumes and pH of the resultant medium in a 250-mL Erlenmeyer flask were aerobically cultured at 25, 30 and 37°C for 1 to 5 days on a rotary shaker (150 rpm). After centrifugation (12,000g, at 4°C for 20 min), the supernatants were collected for measurement of lipase activity.
Purification of the enzyme
Production of lipase
For the production of lipase, P. monteilii TKU009 was grown in 125 mL liquid medium in an Erlenmeyer flask (250 mL) containing 2% soybean powder, 0.1% K2HPO4, 0.05% MgSO4·7H2O at pH 10. One milliliter of the seed culture was transferred into 125 mL of the same medium and grown in an orbital shaking incubator for 4 days at 25°C and pH 10. After incubation, the culture broth was centrifuged (12,000g, at 4°C for 20 min), and the supernatant collected.
DEAE-Sepharose CL-6B chromatography
To the supernatant (450 mL), ammonium sulfate was added to 30% saturation. The mixture was kept at 4°C overnight and the precipitate was collected by centrifugation at 4°C for 20 min at 12,000g. The precipitate was then dissolved in a small amount of 50 mM sodium phosphate buffer (pH 7), and dialyzed against the buffer. The dialysate (25 mL) was loaded onto a DEAE-Sepharose CL-6B column (3.8 cm × 30 cm) equilibrated with 50 mM sodium phosphate buffer (pH 7). One lipase (F1) was washed from the column with the same buffer and another lipase (F2) was eluted with a linear gradient of 0–1 M NaCl in 50 mM sodium phosphate buffer (pH 7). The lipase-containing fractions were combined and concentrated by ammonium sulfate precipitation. The precipitate was collected by centrifugation and dissolved in 5 mL of 50 mM sodium phosphate buffer (pH 7).
Sephacryl S-100 chromatography
The enzyme solution was loaded onto a Sephacryl S-100 gel filtration column (1.5 cm × 70 cm), equilibrated with 50 mM sodium phosphate buffer (pH 7) and eluted with the same buffer. The fractions of the peak that exhibit lipase activity were combined and concentrated by means of ammonium sulfate precipitation. The precipitate was collected by centrifugation and dissolved in 50 mM sodium phosphate buffer (pH 7).
Protein determination
Protein content was determined by the method of Bradford using Bio-Rad dye reagent concentrate and bovine serum albumin as the standard. After column chromatography, the protein concentration was estimated by measuring the absorbance at 280 nm [15].
Measurement of enzyme activity
Lipase activity was determined by spectrophotometer using p-nitrophenyl palmitate (p-NPP) as a substrate. One milliliter of 99% ethanol containing 50 mg of p-NPP was mixed with 9 mL of 50 mM sodium phosphate buffer, pH 7, containing gum arabic (0.11%) and triton X-100 (0.44%). Diluted enzyme solution (0.1 mL) was added to 1 mL sodium phosphate buffer (50 mM, pH 7) and the reaction mixture was pre-warmed to 30°C and then mixed with 1 mL of freshly prepared substrate solution. The reaction mixture was incubated at 30°C for 3 min, and subjected to colorimetric assay at 410 nm. One unit of enzyme activity was defined as the amount of enzyme that liberated 1 μM p-nitrophenol per minute under the assay conditions. Under the conditions described, the extinction coefficient of p-nitrophenol is 1.46 × 105 cm2 M−1.
Determination of molecular mass
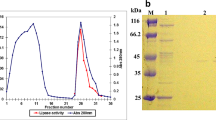
The molecular mass of the purified lipase was determined by sodium dodecyl sulfate-polyacryamide gel electrophoresis (SDS-PAGE) according to the method of Laemmli. The standard proteins (Geneaid, Taiwan) used for calibration were phosphorylase b (molecular mass, 97.4 kDa), albumin (66.2 kDa), ovalbumin (45 kDa), carbonic anhydrase (29 kDa), trypsin inhibitor (20.1 kDa), and α-lactabumin (14.4 kDa). Before electrophoresis, samples were incubated overnight in 10-mM phosphate buffer (pH 7) containing β-mercaptoethanol. The gels were stained with Coomassie Brilliant Blue R-250 in methanol–acetic acid–water (5:1:5, v/v), and decolorized in 7% acetic acid. The molecular mass of the TKU009 lipase in the native form was determined by a gel filtration method. The sample and standard proteins were applied to a Sephacryl S-100 column (1.5 cm × 70 cm, Amersham Pharmacia), equilibrated with 50 mM phosphate buffer (pH 7). Bovine serum albumin (molecular mass, 67 kDa), Bacillus sp. α-amylase (50 kDa), and hen egg white lysozyme (14 kDa) were used as molecular mass markers [15].
Effect of pH on enzyme activity and stability
To investigate the optimal pH, lipase activity was determined at 30°C at various pH values (4.0–11.0) of buffer. The stability of the enzyme in the range of pH 4.0–11.0 was examined by incubating the enzyme solution for 1 h at 25°C at different pH, and then the residual activity of lipase was performed according to the standard assay protocol described above.
Effect of temperature on enzyme activity and stability
The enzyme activity was measured in the range of 25–90°C using the standard activity assay procedure. As to the effect of temperature on enzyme stability, the lipase was incubated at different temperatures (25–90°C) for 1 h in 50 mM sodium phosphate buffer (pH 7.0) and then the residual activity measured at pH 7 and 30°C, according to the standard assay protocol described above.
Effect of metal ions, surfactants and lipase inhibitors on the enzymatic activity
The effects of various metal ions (1 mM) including Mg2+, Cu2+, Fe2+, Ca2+, Zn2+, Mn2+, and Ba2+ were investigated. The effects of lipase inhibitors were studied using phenylmethylsulfonyl fluoride (PMSF) and ethylenediaminetetraacetic acid (EDTA). The effects of the surfactants were analyzed against 0.1% Tween-40 and Triton X-100, and 1 mM SDS. In these experiments, the enzyme was pre-incubated with metal ions, surfactants, or inhibitors for 60 min at 25°C and then the residual activity was tested by the use of p-NPP as the substrate.
Substrate specificity
Substrate specificities toward different pNP esters freshly prepared as above were determined at 30°C and 50 mM sodium phosphate buffer (pH 7.0) by means of the spectrophotometric assay described above. The synthetic pNP esters with a chain length between C2 and C18 were pNP acetate (C2), pNP butyrate (C4), pNP caprylate (C8), pNP caprate (C10), pNP myristate (C14), pNP palmitate (C16), and pNP stearate (C18), respectively.
HPLC analysis
Purified lipases were used for the hydrolysis and alcoholysis of soybean oil. The reactions were carried out at a flow-rate of 1 mL/min using a solvent gradient from 0 to 50% B in 30 min, and 50–0% B in 35 min. Mobile phase A was methanol and mobile phase B consisted of hexane and isopropanol (4:5, v/v). The UV detection was performed at 295 nm. For each condition, experiments were repeated three times.
Results and discussions
Identification of strain TKU009
Strain TKU009 is a Gram-negative bacterium, showing catalase and oxidase activities, which grows in both aerobic and anaerobic environments. The identification of strain TKU009 was carried out by the Bioresource Collection and Research Center (formerly named the Culture Collection and Research Center), Taiwan. On the basis of 16S rDNA studies, it was found that strain TKU009 was closer to Pseudomonas monteilii, P. plecoglossicida, P. parafulva, P. mosselii, P. oryzihabitans, P. putida, P. cremoricolorata, P. jessenii, P. asplenii, P. stutzeri, P. fuscovaginae, P. fulva, P. flavescens, P. graminis, P. lutea, P. pseudoalcaligenes, P. umsongensis and P. koreensis. Phylogenetic relationships could be inferred through the alignment and cladistic analysis of homologous nucleotide sequences of known bacteria, and the approximate phylogenetic position of the strain is shown in Fig. 1. According to the composition of fatty acid assay, the fatty acids of strain TKU009 were mostly sum in feature 3 (16:1 ω7c and/or 15:0 ISO 2OH) (31.0%), 18:1 ω7c (25.1%) and 16:0 (24.0%), close to those of P. monteilii, P. putida, P. asplenii, P. lutea, P. umsongensis, P. koreensis, P. fulva, P. graminis, P. parafulva, and P. plecoglossicida. The API identification system assay showed that strain TKU009 was closer to P. monteilii, P. umsongensis, P. fulva, P. parafulva, and P. plecoglossicida. The Biolog identification system assay indicated that strain TKU009 was closer to P. monteilii. The DNA G + C content of strain TKU009 was 61.3 mol%, close to that of P. monteilii. The hybridization analysis indicated that the hybridization of strain TKU009 and P. monteilii was 77.7% and the similarity of DNA was more than 70%. Taken together, strain TKU009 was identified as a strain of P. monteilii.
Optimization of lipase production
To study the effect of carbon/nitrogen sources on the production of lipases by TKU009, soybean powder was used as sole carbon/nitrogen source because it contained mostly protein and oil. Besides, soybean is a very common and cheap material in Taiwan. Soybean oil has also been used as an enzymatic substrate for biodiesel fuel production by immobilized Pseudomons cepacia lipase [6]. Taking into account the feasibility of bioconversion of soybean to biodiesel fuel by fermentation with a lipase-producing strain, soybean was thus an ideal choice as the sole carbon/nitrogen source and as an oil substitute in this study. Growth was carried out in basal medium containing additional carbon/nitrogen sources of 1–3% (w/v) soybean powder. The result showed that 2% soybean powder (1.36 U/mL, the third day) was an optimal concentration for lipase production. Investigating the effect of cultivation volume on the production of lipase, we found that 125 mL of medium was suitable for lipase production. Therefore, 125 mL of basal medium containing 2% soybean powder was used for further investigation. The results showed that higher lipase activity exhibited at incubation time of 4 days (Fig. 2) decreased after 5 days. Of the three different temperatures (25, 30, and 37°C) and initial pH used for incubation, the optimal conditions were found to be 25°C and pH 9.5. During the process of incubation, lipase activity, cell growth, and pH in the broth were measured. The time courses of cell growth and lipase activity are shown in Fig. 2. The lipase activity increased along with the cell growth as judged by OD660 and reached a maximum (1.81 unit/mL) when the cell growth reached a peak on the fourth day of incubation. Bacillus coagulans BTS-3 [16] and B. thermoleovorans CCR11 [17] were reported as lipase producing strains. The former produced lipase by the use of yeast extract, peptone, NaCl, CaCl2, and olive oil as a cultivation medium at 55°C for 48 h while the latter by the use of nutrient broth, gum arabic, CaCl2, and olive oil as a cultivation medium at 55°C, pH 6.5 for 48 h. In comparison, TKU009 lipase was produced by the use of cheaper medium under milder conditions.
Time courses of growth and lipase production of P. monteilii TKU009 in a soybean-containing medium: lipase activity (U/mL) (filled circle); pH (filled square); and growth (filled triangle). For the production of lipase, P. monteilii TKU009 was grown in 125 mL liquid medium in an Erlenmeyer flask (250 mL) containing 2% soybean powder, 0.1% K2HPO4, and 0.05% MgSO4·7H2O (pH 9.5)
Isolation and purification of lipases F1 and F2
In the presence of soybean powder as a carbon/nitrogen source, P. monteilii TKU009 released enzymes into the culture broth, some displayed lipase activity as shown in Table 1. The purification of lipases from the culture supernatant (450 mL) was as described in “Materials and methods”. By the application of ion exchange chromatography with DEAE-Sepharose CL-6B, two protein peaks (F1 and F2) exhibiting lipase activities were resolved in the fractions eluted by buffer containing 0–1 M NaCl. Each protein peak was pooled separately. The purification procedures are summarized in Table 1. Since lipase F1 contained insoluble compound, it is difficult to continue recovering the enzyme in an emulsion system. According to HPLC assay (Fig. 3), the composition of F1 contained triglycerides that were supposed to be from soybeans and not from TKU009. Therefore, F1 is herein called as lipase-like material. For F2, the purification steps were very effective and combined to give overall purifications of 31-fold. The overall activity yield of the purified lipase (F2) was 1.1%, with specific lipase activity of 12.2 unit/mg and a final amount of 0.9 mg. The purified enzyme F2 was also confirmed to be homogeneous by the use of SDS-PAGE and its molecular weight was estimated to be 44 kDa by the use of a standard curve established with proteins of known molecular weight (Fig. 4). Gel filtration on a Sephacryl S-100 column gave a molecular weight of 40 kDa for lipase F2. These results indicate that the enzyme is monomeric.
The molecular mass of TKU009 lipase F2 was obviously different from most of the other microbial lipases, such as those from Pseudomonas aeruginosa (59.4 kDa) [10], Aspergillus carneus (27 kDa) [18], Penicillium camembertii Thom PG-3 (28.18 kDa) [19], Pseudomonas sp. strain S5 (60 kDa) [20], B. coagulans BTS-3 (31 kDa) [16], B. thermoleovorans CCR11 (11 kDa) [17], and Acinetobacter sp. ES-1 (32 kDa) [21]. Only B. cereus C71 (42 kDa) [22] has a molecular weight similar to lipase F2 of P. monteilii TKU009.
Effect of pH and temperature
The effect of pH on the catalytic activity was studied by the use of p-NPP as a substrate under the standard assay conditions. The pH activity profile of the lipases F1 and F2 show maximum around pH 6 and pH 7, respectively. The optimal pH range of most microbial lipases falls in the alkaline region [22]. The optimal pH of TKU009 lipases is thus quite different from those of A. carneus (pH 9) [18], P. aurantiogriseum lipase (pH 8) [23], B. coagulans BTS-3 lipase (pH 8.5) [16], B. thermoleovorans CCR11 lipase (pH 9–10) [17], B. cereus C71 lipase (pH 9) [22], Yarrowia lipolytica lipase (pH 8) [24], P. aeruginosa lipase (pH 11) [9], and Pseudomonas sp. strain S5 lipase (pH 9) [20]. The pH stability profiles of the lipases F1 and F2 were determined by the measurement of the residual activity at pH 7 after incubation at various pH values at 25°C for 1 h. The lipases F1 and F2 were stable in the range pH 6–7 and pH 7–11, respectively.
The effect of temperature on the activity of lipase F1 and F2 was studied with p-NPP as the substrate. The optimal temperature for both F1 and F2 was 40°C. To examine the heat stability of F1 and F2, the enzyme in 50 mM phosphate buffer (pH 7) was allowed to stand for 1 h at various temperatures, and then the residual activity was measured. F1 and F2 retained their activities from 25 to 50°C and showed 70% residual activities at 50°C for F1 and at 60°C for F2, but became completely inactive at 60°C for F1 and 90°C for F2. The loss of activity increased with increasing temperature, indicating that the lipases from TKU009 were not stable at high temperature when compared to other lipases from microorganisms [25, 26]. Moreover, it was found that the thermostability of TKU009 lipases was related to the purity of the enzymes, with the crude enzymes displaying higher thermal stability than the purified enzymes (data not shown).
Effects of various chemicals
To further characterize F1 and F2, the effects of some known enzyme inhibitors and various metal ions on their activities were examined by pre-incubating the enzymes with chemicals in 50 mM phosphate buffer (pH 7) for 60 min at 25°C and then residual lipase activity was determined with p-NPP as the substrate. As shown in Table 2, lipase activity of F2 was enhanced by Ca2+ and Zn2+, but strongly inhibited by EDTA; while F1 was completely inhibited by EDTA and Fe2+ and strongly influenced by Zn2+, Mn2+, Ca2+, and Mg2+. The Ca2+-binding site and the activation mechanism by Ca2+ of the bacterial lipase have been clarified [27]. The metal-chelating agent EDTA was a strong inhibitor, indicating that the TKU009 lipases F1 and F2 were metalloenzymes. The catalytic activity of lipase F2 from P. monteilii TKU009 was enhanced by Ca2+, indicating this enzyme is calcium-dependent. Similar result was reported by Lee et al. [21] for the lipase from Acinetobacter sp. ES-1.
Effect of various surfactants
Many enzymes are inactivated by the addition of surfactants to the reaction solution, such as the common surfactant SDS. The effects of different surfactants on stability of the lipases F1 and F2 were thus studied. These lipases were incubated with various surfactants at 25°C for 60 min and the remaining enzymatic activity was determined under the assaying conditions described in the “Materials and methods”. The enzyme activity without any surfactants (control) was taken as 100%. The results are summarized in Table 3. In the presence of 0.1% Tween 40 and Triton X-100 (nonionic surfactant), the activity of F1 was determined to be 132 and 354%, respectively, of its original activity; while F2 retained 107 and 113% activity, respectively. In the presence of the anionic surfactant of SDS (1 mM), the activity of F1 and F2 showed 35 and 68% activity, respectively. In accordance to our results, loss of activity in the presence of SDS but an enhanced activity in the presence of Triton X-100 and Tween 40 [9, 22, 24] was also found. Surfactants are known to increase the lipid water interfacial area, which in turn enhances the observed rate of lipase-catalyzed reactions [22]. Thus, addition of surfactants provides a simple method to improve the reaction efficiency of lipases. Since the catalytic reactions of lipases take place at the lipid-water interface, it is reasonable to assume that some surface-activating substances can facilitate access of the substrate to the enzyme and enhance the catalytic rates.
Effect of organic solvents
Enzymes are frequently found to be inactivated by organic solvents. The effects of a few organic solvents (25%, v/v) on the activities of F1 and F2 are depicted in Table 4. The enzymes were incubated with solvents (25%, v/v) at 37°C for 30 min and the remaining enzymatic activity was determined under normal assay conditions. The lipase activity of the sample without any organic solvent was taken as the control (100%). The activity of lipase F1 was activated by 25% n-hexane, while the activity of lipase F2 was activated by 25% ethanol and 25% n-hexane. Lipase F1 showed ~50–75% remaining activity in isopropanol, acetone, and ethanol and 80% in methanol, while F2 had ~45% remaining activity in isopropanol and methanol and ~60% in isopentanol and acetone. Generally, enzymes lose their activity in organic co-solvent with concentrations higher than 10–20% [9]. A thin layer of water molecules remain bound to the enzyme molecules in organic solutions, allowing retention of the native conformation of the enzymes. When water miscible organic solvents are added, they can deprive the bound water from the enzyme and inactivate it. In fact, lipases are diverse in their sensitivity to miscible organic solvents [9, 24], e.g., the lipases from P. aeruginosa B11-1 [28], Bacillus sp. [29], B. thermoleovorans CCR11 [17], and Yarrowia lipolytica [24] showed a high stability in the presence of water miscible organic solvents.
Substrate specificity
The substrate specificity of F1 and F2 was examined by the use of p-nitrophenyl esters with various chain lengths of the acid moiety. The results (Table 5) showed that F1 has a broad spectrum toward substrates with a chain length from C4 to C18, showing the highest activity toward pNP caprylate (C8). F2 also has a wide range of substrate specificity (C10–C18) with the highest activity toward pNP myristate (C14). Although the activity decreased greatly on substrates of longer carbon chains (>14 carbon atoms), it still had 26 and 17% activity on pNP stearate (C18) for F1 and F2, respectively. Lipases are defined as the enzymes hydrolyzing long-chain acyglycerols (≧10 carbon atoms); however, it is known that most of lipases are also active on short-chain fatty acid esters [22]. In this study, the specificity of the two lipolytic enzymes from P. monteilii TKU009 toward p-nitrophenyl esters of long fatty acids indicated that they are true lipases.
In this study, we successfully purified two lipases F1 and F2 from the culture supernatant of the newly isolated strain P. monteilii TKU009. To avoid potential influences in lipase purification from the addition of external oil to the growth medium, soybean powder was chosen as the sole carbon/nitrogen source in this study. The use of soybean in the medium for the production of lipases herein is also remarkably simpler and cheaper than other reported lipase producing strains. Moreover, this study suggests that P. monteilli TKU009 may potentially be used as a biocatalyst alternative to immobilized lipase for biodiesel fuel production from soybean oil.
References
D’Annibale A, Sermanni GG, Federici F, Petruccioli M (2006) Olive-mill wastewaters: a promising substrate for microbial lipase production. Bioresour Technol 97:1823–1833
Jaeger KE, Ransac S, Dijkstra BW, Colson C, Mv Heuvel, Misset O (1994) Bacterial lipases. FEMS Microbiol Rev 15:29–63. doi:10.1111/j.1574-6976.1994.tb00121.x
Jaeger KE, Reetz MT (1998) Microbial lipases form versatile tools for biotechnology. Trends Biotechnol 16:396–403. doi:10.1016/S0167-7799(98)01195-0
Hasan F, Shah AA, Hameed A (2006) Industrial applications of microbial lipases. Enzyme Microb Technol 39:235–251. doi:10.1016/j.enzmictec.2005.10.016
Aloulou A, Rodriguez JA, Puccinelli D, Mouz N, Leclaire J, Leblond Y et al (2007) Purification and biochemical characterization of the LIP2 lipase from Yarrowia lipolytica. Biochim Biophys Acta 1771:228–237
Noureddini H, Gao X, Philkana RS (2005) Immobilized Pseudomonas cepacia lipase for biodiesel fuel production from soybean oil. Bioresour Technol 96:769–777. doi:10.1016/j.biortech.2004.05.029
Burkert JFM, Maugeri F, Rodrigues MI (2004) Optimization of extracellular lipase production by Geotrichum sp. using factorial design. Bioresour Technol 91:77–84. doi:10.1016/S0960-8524(03)00152-4
Bornscheuer UT, Kazlauskas RJ (2006) Hydrolases in organic synthesis—regio- and stereoselective biotransformations, 2nd edn. Wiley-VCH, Weinheim
Karadzic I, Masui A, Zivkovic LI, Fujiwara N (2006) Purification and characterization of an alkaline lipase from Pseudomonas aeruginosa isolated from putrid mineral cutting oil as component of metalworking fluid. J Biosci Bioeng 102:82–89. doi:10.1263/jbb.102.82
Singh S, Banerjee UC (2007) Purification and characterization of trans-3-(4-methoxyphenyl) glycidic acid methyl ester hydrolyzing lipase from Pseudomonas aeruginosa. Process Biochem 42:1063–1068. doi:10.1016/j.procbio.2007.04.006
Kim KR, Kwon DY, Yoon SH, Kim WY, Kim KH (2005) Purification, refolding, and characterization of recombinant Pseudomonas fluorescens lipase. Protein Expr Purif 39:124–129. doi:10.1016/j.pep.2004.09.014
Jinwal UK, Roy U, Chowdhury A, Bhaduri A, Roy PK (2003) Purification and characterization of an alkaline lipase from a newly isolated Pseudomonas mendoncina PK-12Cs and chemoselective hydolysis of fatty acid ester. Bioorg Med Chem 11:1041–1046. doi:10.1016/S0968-0896(02)00516-3
Reetz MT, Jaeger KE (1998) Overexpression, immobilization and biotechnological application of Pseudomonas lipases. Chem Phys Lipids 93:3–14. doi:10.1016/S0009-3084(98)00033-4
Krieg NR, Holt JG (1984) Bergey’s manual of systematic bacteriology, vol 1. Williams and Wilkins, Baltimore, MD
Wang SL, Hsu WT, Liang TW, Yen YH, Wang CL (2008) Purification and characterization of three keratinolytic metalloproteases produced by Chryseobacterium indologenes TKU014 in a shrimp shell powder medium. Bioresour Technol 99(13):5679–5686
Kumar S, Kikon K, Upadhyay A, Kanwar SS, Gupta R (2005) Production, purifcation, and characterization of lipase from thermophilic and alkaliphilic Bacillus coagulans BTS-3. Protein Expr Purif 41:38–44. doi:10.1016/j.pep.2004.12.010
Castro-Ochoa LD, Rodríguez-Gómez C, Valerio-Alfaro G, Oliart Ros R (2005) Screening, purification and characterization of the thermoalkalophilic lipase produced by Bacillus thermoleovorans CCR11. Enzyme Microb Technol 37:648–654. doi:10.1016/j.enzmictec.2005.06.003
Saxena RK, Sheoran A, Giri B, Davidson WS (2003) Purification strategies for microbial lipases. J Microbiol Methods 52:1–18. doi:10.1016/S0167-7012(02)00161-6
Tan T, Zhang M, Xu J, Zhang J (2004) Optimization of culture conditions and properties of lipase from Penicillium camembertii Thom PG-3. Process Biochem 39:1495–1502. doi:10.1016/S0032-9592(03)00296-6
Rahman RNZRA, Baharum SN, Basri M, Salleh AB (2005) High-yield purification of an organic solvent-tolerant lipase from Pseudomonas sp. strain S5. Anal Biochem 341:267–274. doi:10.1016/j.ab.2005.03.006
Lee KW, Bae HA, Shin GS, Lee YH (2006) Purification and catalytic properties of novel enantioselective lipase from Acinetobacter sp. ES-1 for hydrolysis of (S)-ketoprofen ethyl ester. Enzyme Microb Technol 38:443–448. doi:10.1016/j.enzmictec.2005.06.017
Chen S, Qian L, Shi B (2007) Purification and properties of enantioselective lipase from a newly isolated Bacillus cereus C71. Process Biochem 42:988–994. doi:10.1016/j.procbio.2007.03.010
Lima VMG, Krieger N, Mitchell DA, Fontana JD (2004) Activity and stability of a crude lipase from Penicillium aurantiogriseum in aqueous media and organic solvents. Biochem Eng J 18:65–71. doi:10.1016/S1369-703X(03)00165-7
Yu M, Qin S, Tan T (2007) Purification and characterization of the extracellular lipase Lip2 from Yarrowia lipolytica. Process Biochem 42:384–391. doi:10.1016/j.procbio.2006.09.019
Sharma R, Soni SK, Vohra RM, Gupta LK, Gupta JK (2002) Purification and characterisation of a thermostable alkaline lipase from a new thermophilic Bacillus sp. RSJ-1. Process Biochem 37:1075–1084. doi:10.1016/S0032-9592(01)00316-8
Sinchaikul S, Sookkheo B, Phutrakul S, Pan FM, Chen ST (2001) Optimization of a thermostable lipase from Bacillus stearothermophilus P1: overexpression, purification, and characterization. Protein Expr Purif 22:388–398. doi:10.1006/prep.2001.1456
Amada K, Kwon HJ, Haruki M (2001) Ca2+-induced folding of a family I.3 lipase with repetitive Ca2+ binding motifs at the C-terminus. FEBS Lett 509:17–21. doi:10.1016/S0014-5793(01)03108-8
Ogino H, Ishikawa H (2001) Enzymes which are stable in the presence of organic solvents. J Biosci Bioeng 91:109–116. doi:10.1263/jbb.91.109
Torres S, Castro G (2004) Non-aqueous biocatalysis in homogenous systems. Food Technol Biotechnol 42:271–277
Acknowledgment
This work was supported in part by a grant from the National Science Council, Taiwan (NSC94-2313-B-212-003).
Author information
Authors and Affiliations
Corresponding author
Additional information
Li-June Ming is a visiting Professor at the National Cheng Kung University.
Rights and permissions
About this article
Cite this article
Wang, SL., Lin, YT., Liang, TW. et al. Purification and characterization of extracellular lipases from Pseudomonas monteilii TKU009 by the use of soybeans as the substrate. J Ind Microbiol Biotechnol 36, 65–73 (2009). https://doi.org/10.1007/s10295-008-0473-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-008-0473-z