Abstract
Purpose
Pulmonary nodule detection has great significance for early treating lung cancer and increasing patient survival. This work presents a novel automated computer-aided detection scheme for pulmonary nodules based on deep convolutional neural networks (DCNNs).
Methods
The proposed approach employs 3D DCNNs based on squeeze-and-excitation network and residual network (SE-ResNet) for pulmonary nodule candidate detection and false-positive reduction. Specifically, a 3D region proposal network with a U-Net-like structure is designed for detecting pulmonary nodule candidates. For the subsequent false-positive reduction, a 3D SE-ResNet-based classifier is presented to accurately discriminate the true nodules from candidates. The 3D SE-ResNet modules boost the representational power of the network by adaptively recalibrating channel-wise residual feature responses. Both models utilize 3D SE-ResNet modules to learn nodule features effectively and improve nodule detection performance.
Results
On the public available lung nodule analysis 2016 dataset with 888 scans included, the proposed method reaches high detection sensitivities of 93.6% and 95.7% at one and four false positives per scan, respectively. Meanwhile, the competition performance metric score of 0.904 is achieved. The proposed method has the capability to detect multi-size nodules, especially the extremely small nodules.
Conclusion
In this paper, a 3D DCNNs framework based on 3D SE-ResNet modules is proposed to detect pulmonary nodules in chest CT images accurately. Experimental results demonstrate superior effectiveness of the proposed approach in pulmonary nodule detection task.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Lung cancer is the leading cause of cancer death around the world with high morbidity and mortality [1]. According to relevant studies [2], the 5-year survival rate of lung cancer patients is among 10–16%. Approximately 70% of patients in these studies are diagnosed with advanced cancer without effective treatments. Early detection of pulmonary nodules is very critical for patient care, which will increase the overall 5-year survival rate to 52% [3]. As one of the most sensitive imaging modalities, chest computed tomography (CT) has been widely used for pulmonary nodule screening owing to its high image quality, resolution and rapid acquisition [4]. With the rapid increase of screening demand and CT availability, radiologists are getting much pressure to process the huge amount of image data. Hence, computer-aided detection (CADe) schemes in chest CT have been developed to help radiologists identify the pulmonary nodules and improve detection efficiency.
A number of CADe schemes have been introduced to detect pulmonary nodules in CT images. A CADe scheme typically consists of two stages: (1) pulmonary nodule candidate detection and (2) false-positive reduction. The first step aims to detect pulmonary nodule candidates at a very high sensitivity, which generates many false positives in the meantime. The second stage is to reduce the false positives by classifying whether the candidates are true nodules or not. Although many researchers have made some progresses in pulmonary nodule detection, it remains a challenging task due to the diversity and complexity of pulmonary nodules. Pulmonary nodules, as shown in Fig. 1, have large variations in size, shape and location [5], such as isolated nodules, juxta-pleural nodules, juxta-vascular nodules and ground glass opacity nodules. Additionally, some false-positive candidates are often mistaken for suspicious nodules because of the similar morphological characteristics with true nodules.
Over these years, several conventional CADe schemes have been developed based on image processing and machine learning techniques [6]. Lu et al. [7] proposed a hybrid method (dot-enhancement filters, fuzzy connectedness segmentation and regression tree classification, etc.) for pulmonary nodule detection. Saien et al. [8] used sparse field level sets to segment nodule candidates and selected 18 features based on gray-level distribution, size and shape to train RUS AdaBoost classifier for false-positive elimination. Gong et al. [9] adopted a 3D tensor filtering algorithm and local image feature analysis to detect nodule candidates. A 3D level set segmentation method was used to correct the contours of nodule candidates. Then, random forest classifier and 19 image features were used to reduce false positives. Since the design of handcrafted features of pulmonary nodules is based on prior knowledge (shape, intensity, size, etc.), there are obvious limitations in the conventional CADe schemes. Firstly, pulmonary nodules’ feature extraction relies on the accurate segmentation of the nodule candidates. However, the accurate segmentation of nodules is affected by the irregular shapes of nodules, the attachments of nodules to other anatomical objects, different image quality, etc. In addition, the wide variation in lung nodules in CT scans prevents handcrafted features from fully characterizing complicated nodules, which results in the limited discriminative capacity for pulmonary nodules.
Recently, with the development of deep convolutional neural networks (DCNNs) [10, 11] and the emergence of a large amount of labeled data [12], deep learning techniques have been rapidly developed in the field of medical image analysis [13,14,15]. DCNNs with extraordinary learning power can acquire highly discriminative features from image data, eliminating the requirement of handcrafting pulmonary nodule features. Setio et al. [16] extracted multi-planner 2D patches from each candidate and used multiple streams of 2D convolutional neural networks (CNNs) for false-positive reduction. ZNET [12] used U-Net [17] on each axial slice for nodule candidate detection. In false-positive reduction, three 2D orthogonal slices of each candidate were fed to the wide residual network [18]. Due to the 3D nature of CT images, 3D CNNs have recently been proposed, encoding 3D spatial contextual information of pulmonary nodules, which are more conducive than 2D DCNNs in terms of recognizing nodules. Huang et al. [19] generated nodule candidates using a local geometric-model-based filter and classified candidates using a 3D CNN. Dou et al. [20] trained three 3D CNNs with different input sizes to merge multi-level contextual information for false-positive reduction. Besides, Dou et al. [21] adopted 3D fully convolutional network with online sample filtering for candidate screening. They introduced two residual blocks [22] into a 3D CNN for false-positive reduction. Jin et al. [23] constructed a deep 3D residual CNN with spatial pooling and cropping (SPC) layers for false-positive reduction stage. Ding et al. [24] used 2D Faster R-CNN [25] with VGG-16 model to generate suspicious candidates and adopted a 3D DCNN to remove false-positive nodules. Zhu et al. [26] used a 3D Faster R-CNN with dual path networks (DPNs) [27] and a U-Net-like encoder–decoder structure for nodule detection, where DPN benefited from the advantage of ResNet [22] and DenseNet [28]. Khosravan et al. [29] modeled pulmonary nodule detection as a cell-wise classification problem and designed a single 3D DCNN with dense connections [28]. Besides, Zhu et al. [30] utilized weakly labeled data in electronic medical records and adopted 3D DCNNs with expectation–maximization for weakly supervised pulmonary nodule detection.
Even though CADe schemes based on DCNNs have achieved good results, most of the networks (ResNet, Wide ResNet, DenseNet, DPN, etc.) applied in CADe schemes only improve networks performance by changing spatial dimensions of networks. For example, the famous ResNet relieves the problem of gradient disappearing while deepening the network depth and brings remarkable accuracy enhancement from the network depth increase. Squeeze-and-excitation network (SENet) [31] is proven to have performed well in many object detection and image classification applications by considering channel relationships of the convolutional features. Moreover, SENet has also been developed in medical image analysis. Zhu et al. [32] incorporated squeeze-and-excitation (SE) residual blocks in the U-Net to improve segmentation accuracy of organs-at-risks from head and neck CT. However, to our best knowledge, the effectiveness of SE-ResNet on pulmonary nodule detection has not been extensively explored.
This paper proposes a novel CADe scheme based on 3D DCNNs framework for automated pulmonary nodule detection. The proposed framework mainly follows two stages: (1) nodule candidate detection that adopts 3D region proposal network (RPN) using a U-Net-like structure and (2) false-positive reduction using a 3D DCNN classifier. In addition, both models utilize SE-ResNet modules to boost the representational power of the network and improve detection performance. The proposed method has been validated on lung nodule analysis 2016 (LUNA16) dataset and achieved good performance in pulmonary nodule detection.
Methods
Nodule candidate detection
3D SE-ResNet module
The SE-ResNet module integrates the advantages of the two state-of-the-art networks: residual learning for feature reuse and squeeze-and-excitation operations for adaptive feature recalibration.
In deep networks, the phenomenon of vanishing/exploding gradients and network degradation become more obvious with the increased depth in the plain network. The identity-based shortcut connection in residual learning is an efficient way to enhance information flow over feature propagation and is able to mitigate the above problems in deeper networks. The SE blocks put the focus on channel-wise information, not spatial, to improve the representational power of the network. The SE blocks perform dynamic channel-wise feature recalibration by explicitly modeling the interdependencies between channels. Spurred by the effectiveness of SE block and residual learning on general image processing tasks, the proposed method designs 3D SE-ResNet modules in the framework for pulmonary nodule detection. The 3D SE-ResNet module introduces the SE block into residual learning to adaptively recalibrate channel-wise residual feature responses. Through the feature recalibration strategy, the network is able to acquire the importance degree of each residual feature channel, which can enhance useful channel features according to the importance degree and suppress less useful ones. The SE blocks enhance the representational capacity of the basic modules throughout the network. Considering that pulmonary nodule detection in volumetric CT scans is a 3D object detection problem, the proposed 3D SE-ResNet module makes the most of 3D spatial contextual information of pulmonary nodules by extending the 2D SE block and residual block to the 3D form. The 3D SE-ResNet module extracts more valuable 3D features from CT scans than those extracted from the 2D form. It can be formulated as
where \( {\mathbf{X}} \) is the input feature. The function \( F_{\text{res}} \left( {\mathbf{x}} \right) \) is the 3D residual mapping to be learned. \( {\mathbf{X}}^{\text{res}} \) is the residual feature.
where \( {\mathbf{z}} = \left[ {z_{1} , \, z_{2} , \ldots ,z_{\text{c}} } \right] \) and \( z_{c} \) is the \( c \)th element of \( {\mathbf{z}} \in {\mathbb{R}}^{C} \). \( F_{\text{sq}} \) represents the squeeze function that aggregates global spatial information into channel-wise statistics by global average pooling. \( C \) is the number of channels of the residual mapping, and \( L \times H \times W \) is the spatial dimensions of \( {\mathbf{X}}^{\text{res}} \). \( {\mathbf{x}}_{c}^{\text{res}} \in {\mathbb{R}}^{L \times H \times W} \) refers to the feature map of \( c \)th channel from the residual feature \( {\mathbf{X}}^{\text{res}} \).
where \( F_{\text{ex}} \) is the excitation function that generates scale values \( {\mathbf{s}} \in {\mathbb{R}}^{C} \) for residual feature channels. It is parameterized by two fully connected (FC) layers with parameters \( {\mathbf{W}}_{1} \in {\mathbb{R}}^{{\tfrac{C}{r} \times C}} \) and \( {\mathbf{W}}_{2} \in {\mathbb{R}}^{{C \times \tfrac{C}{r}}} \), the ReLU function \( \delta \) and the sigmoid function \( \sigma \). To reduce computation costs, the reduction ratio is set to \( r = 16 \).
where \( F_{\text{scale}} \left( {{\mathbf{X}}_{c}^{\text{res}} ,s_{c} } \right) \) refers to channel-wise multiplication between the feature map \( {\mathbf{X}}_{c}^{\text{res}} \in {\mathbb{R}}^{H \times W \times L} \) and the learned scale value \( s_{c} \). The scale value \( s_{c} \) represents the importance degree of \( c \)th channel. After the squeeze-and-excitation operations, the calibrated residual feature \( \widetilde{{\mathbf{X}}}^{\text{res}} \) is obtained.
where the formulation \( \widetilde{{\mathbf{X}}}^{\text{res}} {\mathbf{ + X}} \) is realized by a shortcut connection and element-wise addition. We obtained the output feature \( {\mathbf{Y}} \) after the ReLU function \( \delta \). The basic 3D ResNet module and the basic 3D SE-ResNet module are illustrated in Fig. 2a, b, respectively.
The 3D SE-ResNet module easily optimizes the deep network and helps DCNNs extract expressive features of pulmonary nodules. Additionally, it can selectively emphasize informative nodule features. By taking advantage of the 3D SE-ResNet modules, the proposed method learns the features with high discrimination capability, which is favorable to identify the pulmonary nodules from the complicated environment.
Nodule candidate detection network architecture
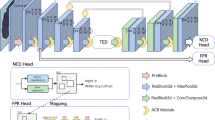
The network for nodule candidate detection adopts 3D RPN [25] based on a U-Net-like structure, as shown in Fig. 3. The 3D RPN utilizes multi-scale anchors for object detection, and the U-Net-like structure with skip connections effectively combines the features from shallow and deep layers. This integration is conducive for detection of multi-scale nodules.
3D DCNN architecture for pulmonary nodule candidate detection. Each cube in the figure represents a 4D tensor, where the number inside the cube represents the spatial size and the number outside the cube represents the number of channels. The model employs three anchors and multitask learning loss, including coordinates \( (x,y,z) \) and diameter d regression, and candidate box classification
The inputs of this network are cropped cubes of CT images. Firstly, there are two convolutional layers with \( 3 \times 3 \times 3 \) kernel size in the feedforward path. And four 3D SE-ResNet blocks interleaved with four 3D max-pooling layers. The first two 3D SE-ResNet blocks consist of two SE-ResNet modules, and the rest have three modules. The backward path is constructed by two deconvolutional layers (kernel size of 2 and stride of 2) and two 3D SE-ResNet blocks. After each deconvolutional operation, the feature maps are merged with the corresponding layers from the feedforward path. It is followed by dropout with a probability of 0.5 and two convolution layers with a kernel size of 2. Three different sizes of anchors are applied to the final feature map in order to generate a set of object proposals, each with a 5 * 1 vector that represents the location of \( (x,y,z) \), diameter and objectness score. In accordance with the size distribution of nodules, the anchors are designed with the sizes of 5, 15, and 35 mm, respectively.
Multitask learning loss function
The binary class label of each anchor box is assigned based on its intersection over union (IoU) with the target nodule. If the IoU overlap of an anchor box is higher than 0.5, it is assigned as a positive sample. Anchors that have IoU lower than 0.02 are labeled as negative samples. Other anchors that are neither positive nor negative will be neglected in the training process. The contribution of multitask learning in RPN had been validated by Ren et al. [25]. The ablation experiments in [25] indicate that multitask learning in RPN outperforms classification learning (object vs. not object) or regression learning alone and improves detection accuracy. Therefore, we minimize an objective function following the multitask loss in RPN. Our loss function is composed of classification loss for the anchor box (nodule vs. not nodule) and regression loss for nodule coordinate \( (x,y,z) \), and nodule size \( d \). For each labeled nodule, the multitask function is defined by:
where \( p \) and \( p^{ * } \) represent the predicted probability and label for an anchor box, respectively. The label for the positive sample is \( p^{ * } { = }1 \) and for the negative sample is \( p^{ * } { = }0 \). \( t_{\beta } \) and \( t_{\beta }^{ * } \) represent the four parameters of the predicted bounding box and ground-truth bounding box, respectively. \( \lambda \) is weight parameter of the two loss terms, set to 0.5. \( L_{\text{cls}} \left( {p,p^{ * } } \right) \) is the classification loss calculated by binary cross-entropy loss function, and \( L_{\text{reg}} \left( {t_{\beta } ,t_{\beta }^{ * } } \right) \) is regression loss of location information calculated by smooth \( L1 \) loss function. The equation suggests that only positive sample labeled as \( (p^{ * } = 1) \) is considered in regression loss.
The total regression loss of location information is defined by:
The four parameters of the predicted bounding box and ground-truth bounding box are given by:
where \( \left( {x,y,z,d} \right) \) are the coordinates and the side length of the predicted bounding box, \( \left( {x^{ * } ,y^{ * } ,z^{ * } ,d^{ * } } \right) \) are parameters of the ground-truth bounding box and \( \left( {x_{\text{a}} ,y_{\text{a}} ,z_{\text{a}} ,d_{\text{a}} } \right) \) are parameters of the anchor bounding box.
Online hard negative mining
There are more negative samples than positive samples in the training process of nodule candidate detection stage. Most negative samples could be easily classified by the network except a few hard ones which have similar morphological appearance with nodules. The hard negative samples including more valuable information than the simple ones help networks improve the ability to distinguish negatives. Hence, online hard negative mining (OHNM) strategy is adopted to deal with the sample imbalance problem. Firstly, the network generates a set of proposed bounding boxes with different confidences after forward propagation. Then, \( N \) negative samples are selected randomly from a candidate pool. According to the classification confidence scores before the sigmoid function, these negative samples are sorted in descending order and the top \( n \) samples are selected as the hard negatives. Other negative samples are discarded and not considered in the loss computation.
False-positive reduction
After the step of nodule candidate detection, it usually results in many false positives (pleural). In order to accurately discriminate the true nodules from a large number of candidates, a 3D DCNN is designed for nodule candidate classification, as shown in Fig. 4. By reducing false positive, more pulmonary nodules can be detected at a lower false positive rate. Candidate crops are extracted according to predicted coordinates from the detector. They are first fed into two convolutional layers with a kernel size of 3 and a max-pooling layer. Three 3D SE-ResNet blocks are employed to learn high-level features, where each block contains three SE-ResNet modules. Each block is followed by a max-pooling layer. Finally, 3D average pooling layer and an FC layer are used for nodule or nonnodule classification. Furthermore, dropout layers with a probability of 0.2 are added after max-pooling layers and the FC layer to avoid overfitting. The classification loss is measured by binary cross-entropy error.
Experiments and results
Dataset
The proposed method was evaluated on the LUNA16 dataset including 888 CT scans with 1186 nodules in total. The dataset was randomly and equally split into ten subsets for tenfold cross-validation. For each fold, one subset was selected for testing, seven subsets for training and two subsets for validation. The LUNA16 dataset was refined from the large public LIDC-IDRI dataset. In this dataset, the scans with slice thickness greater than 2.5 mm and missing slices were excluded. The pulmonary nodule annotations in the dataset were collected during a two-phase image annotation process performed by four professional radiologists. The reference standard of LUNA16 challenge consists of all nodules ≥ 3 mm accepted by at least three out of four radiologists.
Preprocessing
Lung segmentation images provided by LUNA16 were used to obtain lung region mask. Convex hull and dilation were used to make sure the segmented lung includes all nodules. The final image pixel values were clipped to \( [ - 1200, \, 600] \) and normalized to \( [0, \, 255] \). Pixels for nonlung regions were padded with 170. As the scans in the dataset had different spatial resolutions, all the slices were resampled to an isotropic resolution of \( 1 \times 1 \times 1\;{\text{mm}} \).
Implementation details
In the stage of nodule candidate detection, the size of 3D patches was \( 128 \times 128 \times 128 \) because of the GPU memory limitation. Data augmentation of positive samples was carried out to alleviate the overfitting problem. The patches were rescaled between \( [0.8, \, 1.2] \), left–right flipped and rotated randomly. The model was trained for 100 epochs and the initial learning rate was 0.01, which was decreased to 0.001 after 40 epochs and 0.0001 after 80 epochs, respectively. The batch size parameter was determined by GPU memory. A large number of nodule proposals would be produced during the detection stage, but many of them overlapped with each other. Therefore, nonmaximum suppression (NMS) with IoU threshold as 0.1 was used in the test phase. In addition, nodules with a probability of less than 0.1 were discarded. For false-positive reduction stage, the cropped inputs were \( 48 \times 48 \times 48 \) 3D patches. The positive samples were augmented in the same way as those in the detection stage and translated of \( \pm 1\;{\text{mm}} \) along each axis. The mini-batch size for the model was 128, and training was stopped when the accuracy on the validation set did not improve after 40 epochs. The learning rate initialized as 0.01 and 0.001 after ten epochs, and 0.0001 at the halfway of the training. Both models adopted stochastic gradient descent with a momentum of 0.9. Besides, batch normalization was applied to improve the regularization ability of models. The framework was implemented with PyTorch deep learning library using 6 NVIDIA TITAN Xp 12 GB GPUs.
Evaluation metrics
The performance of the proposed scheme was evaluated by measuring the detection sensitivity and the corresponding false positive rate per scan (FPs/scan). The sensitivity is the percentage of lung nodules that are correctly identified, which is defined as:
where TP is the number of true positive cases and FN is the number of false-negative cases. The analysis is performed by using free receiver operating characteristic (FROC) analysis according to LUNA16 challenge. The competition performance metric (CPM) score is used in this paper, which is defined as the average sensitivity at seven predefined false-positive rates: 0.125, 0.25, 0.5, 1, 2, 4 and 8 FPs/scan. Besides, the 95% confidence interval using bootstrapping with 1000 bootstraps is calculated. According to the LUNA16 official evaluation system, a candidate is regarded as a true nodule if the location of the candidate is within a distance R of the nodule center, where R refers to half the diameter of the nodule annotation. Although LUNA16 challenge ended on January 3, 2018, the evaluation script of LUNA16 is still available online. Therefore, we evaluated the performance by the publicly available evaluation script of LUNA16.
Nodule detection results
During nodule candidate detection stage, a total of 1157 nodules were detected on the whole dataset. The high sensitivity of 97.55% with 33.79 candidates per scan on average is achieved by the proposed approach. Meanwhile, the average diameter difference between detected nodules and ground truth is 1.21 mm. After false-positive reduction, the proposed CADe scheme achieves superior sensitivity and high CPM score of 0.904. The FROC curve of the scheme is presented in Fig. 5. It suggests that our method yields sensitivities of 93.6% at 1 FPs/scan and 95.7% at 4 FPs/scan, respectively. Figure 6 shows the comparisons of the number of detected nodules with the number of ground truths in different sizes at 8 FPs/scan, which illustrates that the proposed method has the ability to detect multi-size nodules with a high detection rate, especially the extremely small nodules less than 5 mm.
Ablation study
To further explore the performance of the proposed method, ablation experiments about false-positive reduction are performed. In the ablation experiments, we used one subset for testing, seven subsets for training and two subsets for validation. Three different models are compared with the proposed classifier to assess the effectiveness of SE-ResNet, and the results are shown in Fig. 7. The first model named as Plain-DCNN is a plain deep network with the same number of convolutional layers as the proposed one. The second one named as ResNet is identical to the proposed one except for replacing each SE-ResNet module with a residual module. The last one named as DeeperNet is implemented by using four SE-ResNet modules in each block to make the network deeper. For a fair comparison, all the models use the same preprocessing strategy. Compared with the plain deep network, the proposed method significantly improves the CPM score from 0.861 to 0.916. The residual learning used in ResNet can increase sensitivity to a certain extent. Adding more SE-ResNet modules to the proposed model does not further improve performance. The SE-ResNet combining channel-wise operations with residual learning not only makes the deep network easier to be optimized but also adaptively calibrates residual feature maps, thus further improving the performance of the scheme.
Cases analysis
Examples of detected true nodules, false positives and false negatives are shown in Fig. 8. The examples presented in the first row are true samples with complex morphology. Note that the proposed method has the ability to detect extremely small nodules less than 5 mm. As shown in the second row, the false positives (such as pulmonary vessels, bronchus, etc.) have quite similar characteristics with nodules. As for the undetected nodules, most of them are irregular nodules or subsolid nodules with extremely small sizes. Besides, the lump-like nodules attached to organs and abnormal nodules with cavities are difficult to be found. These missed nodules are underrepresented in the LUNA16 dataset, and it is potential to improve the scheme performance by special data augmentations for these complicated nodules.
Discussion
In this paper, a novel CADe scheme for pulmonary nodule detection in CT scans based on 3D DCNNs framework is presented. The proposed method achieves promising performance for the nodule detection task. The success of the proposed approach mainly lies in three aspects. Firstly, the detector and classifier in the framework are all based on 3D DCNNs, which are more suitable for volumetric medical image processing. Secondly, a 3D RPN, based on U-Net-like structure, that is trained with OHNM strategy is employed for nodule candidate detection, and a 3D DCNN classifier is used for false-positive reduction. Thirdly, both models introduce SE-ResNet modules to accelerate the training process and improve the accuracy of nodule detection. Experimental results on LUNA16 dataset demonstrate that the proposed method can detect the delitescent pulmonary nodules accurately.
To evaluate the performance of the proposed method in a broader context, we reported the state-of-the-art methods using the LIDC-IDRI dataset in Table 1. As can be seen from Table 1, the proposed CADe scheme achieves compelling performance in comparison with other CADe schemes, indicating that the method based on 3D DCNNs framework is applicable and effective for automatic pulmonary nodule detection.
Generally, there are two types of existing CADe schemes: conventional method-based and deep learning method-based schemes. Lu et al. [7] and Saien et al. [8] adopted conventional methods to detect pulmonary nodules and achieved sensitivities of 85.2% and 83.98%, respectively. These methods depend enormously on handcrafted features, which might affect the generalization ability of classifiers. Additionally, only 198 nodules used in [8] were less persuasive. Although conventional methods achieved good results, their performance might decline sharply on large datasets due to the variability of nodules and the limitations of handcrafted features, such as Gong et al. [9] (79.3% sensitivity). Besides, the rest methods are all based on deep learning techniques. The biggest difference between deep learning method and conventional method is that the former automatically learns highly discriminative features from a large amount of medical image data instead of relying on handcrafted features.
For deep learning method-based schemes, Setio et al. [8] and ZNET [12] used multi-stream 2D CNNs for pulmonary nodules detection with CPM scores of 0.827 and 0.811, respectively. Although both of them used multiple cross-sectional image patches for training, 2D CNNs could not include enough 3D spatial information of nodules, especially for nodules with irregular shapes. Compared with the methods that used 2D CNNs, the proposed 3D DCNNs can directly capture and extract 3D nodule features and obtain better results than them. Hung et al. [19] and Dou et al. [21] underperform the proposed method because their 3D CNNs structures are relatively shallow compared to our 3D DCNNs. The 3D DCNNs benefit from deep structure and boost the ability to extract high-level nodule features. Ding et al. [24] generated nodule candidates with 2D Faster R-CNN and reduced false positives with 3D DCNNs, achieving a CPM score of 0.891. Faster R-CNN is an advanced two-stage detection framework for object recognition, including two subnets: RPN and region-of-interest (ROI) classifier. However, there are only two classes (nodule vs. not nodule) in the pulmonary nodule detection task and the design of these two subnets is relatively redundant as they produce a similar output (binary classification and bounding box regression). The proposed candidate detection network uses 3D RPN that can perform end-to-end detection for nodule candidates. In addition, the application of SE-ResNet modules in our framework improves the network representative capability compared with the plain DCNNs. Jin et al. [23] proposed an effective false-positive reducer, but it is only one part of a typical CADe system. Both Khosravan et al. [29] and Zhu et al. [26] adopted a single network for detection without further post-processing achieved CPM scores of 0.897 and 0.842, respectively. Although the single-stage methods are fast and have less number of parameters, the proposed two-stage method is more accurate in the detection of pulmonary nodules.
According to the latest leaderboard of LUNA16 challenge, the proposed method outperforms most teams such as iDST-VC (CPM score: 0.897), qfpxfd [24], 3DCNN_NDET (CPM score: 0.882), MEDICAI (CPM score: 0.862), resnet [21]. Additionally, PATech (CPM score: 0.951), LUNA16FONOVACAD (CPM score: 0.947) and zhongliu_xie (CPM score: 0.922), etc. achieve better detection performance. In accordance with the limited descriptions that they provided online, most teams used 3D DCNNs and residual learning for pulmonary nodule detection. The residual learning, dense connection and dual path blocks used in [21, 23, 26, 29] and LUNA16 teams only consider the connections between convolutional layers. Different from these methods, the 3D SE-ResNet module used in our method concentrates on the channel-wise relationships of convolutional features. The 3D SE-ResNet module adaptively recalibrates residual feature maps using channel-wise operations, which enhances the representational capability of the basic modules and efficiently extracts more representative nodule features.
In this study, we proposed an effective pulmonary nodule detection method based on 3D DCNNs framework with SE-ResNet modules. The proposed method achieves a high CPM score of 0.904 and has the ability to detect multi-scale nodules. Considering the complex characteristics of pulmonary nodules, 3D networks can capture more valuable spatial information and extract more representative features than 2D networks. Although 3D networks take up large storage space and are limited by GPU memory, they are more appropriate for the pulmonary nodule detection task. As for nodule candidate detection network, this work adopts 3D RPN that can be trained end-to-end specifically for the task of generating suspicious nodule candidates. RPN utilizes the convolutional feature map to simultaneously regress region bounds and objectness scores. In addition, the multi-scale anchor-based method in RPN is vital for detecting multi-scale nodules. Compared with nature images, medical images have a wide range of grayscale and vague boundaries. The U-Net is suitable for processing highly complex medical images. The U-Net-like structure is employed in the network to combine the high-resolution features from the down-sampled path with the low-resolution features from the up-sampled path, which makes network not only have strong recognition ability but also enhance the accuracy of localization. Moreover, the 3D SE-ResNet module allows the deep network easier to be optimized and dynamically recalibrates channel-wise features of residual learning, which is beneficial to boost nodule feature discriminability. The scheme is able to estimate the diameter of nodules while detecting them, which could assist radiologists in the diagnosis of benign and malignant nodules. In future work, we will pay attention to the diagnosis of pulmonary nodules.
Conclusion
In this paper, an automated CADe scheme based on deep learning has been presented for pulmonary nodule detection. The scheme is composed of nodule candidate detection stage and false-positive reduction stage, where both of them using SE-ResNet modules achieved superior results. A 3D RPN based on U-Net-like structure is used for detecting nodule candidates. Then, a 3D DCNN classifier is developed for false-positive reduction. The proposed method exhibits promising performance by the validation experiment on LUNA16 dataset; thus, the scheme will be a powerful clinical tool for lung cancer screening in the near future.
References
Torre LA, Siegel RL, Jemal A (2016) Lung cancer statistics. Adv Exp Med Biol 893:1–19. https://doi.org/10.1007/978-3-319-24223-1_1
Henschke CI (2000) Early lung cancer action project: overall design and findings from baseline screening. Cancer 89(11):2474–2482. https://doi.org/10.1002/1097-0142(20001201)89:11+%3c2474:aid-cncr26%3e3.0.co;2-2
Siegel RL, Miller KD, Jemal A (2018) Cancer statistics, 2018. CA Cancer J Clin 68(1):7–30. https://doi.org/10.3322/caac.21442
National Lung Screening Trial Research T, Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM, Sicks JD (2011) Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 365(5):395–409. https://doi.org/10.1056/nejmoa1102873
Karki A, Shah R, Fein A (2017) Multiple pulmonary nodules in malignancy. Curr Opin Pulm Med 23(4):285–289. https://doi.org/10.1097/MCP.0000000000000393
Zhang G, Jiang S, Yang Z, Gong L, Ma X, Zhou Z, Bao C, Liu Q (2018) Automatic nodule detection for lung cancer in CT images: a review. Comput Biol Med 103:287–300. https://doi.org/10.1016/j.compbiomed.2018.10.033
Lu L, Tan Y, Schwartz LH, Zhao B (2015) Hybrid detection of lung nodules on CT scan images. Med Phys 42(9):5042–5054. https://doi.org/10.1118/1.4927573
Saien S, Moghaddam HA, Fathian M (2018) A unified methodology based on sparse field level sets and boosting algorithms for false positives reduction in lung nodules detection. Int J Comput Assist Radiol Surg 13(3):397–409. https://doi.org/10.1007/s11548-017-1656-8
Gong J, Liu JY, Wang LJ, Sun XW, Zheng B, Nie SD (2018) Automatic detection of pulmonary nodules in CT images by incorporating 3D tensor filtering with local image feature analysis. Phys Med 46:124–133. https://doi.org/10.1016/j.ejmp.2018.01.019
Schmidhuber J (2014) Deep learning in neural networks: an overview. Neural Netw 61:85–117. https://doi.org/10.1016/j.neunet.2014.09.003
Szegedy C, Liu W, Jia YQ, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. IEEE CVPR 2015:1–9. https://doi.org/10.1109/CVPR.2015.7298594
Setio AAA, Traverso A, de Bel T, Berens MSN, Bogaard CVD, Cerello P, Chen H, Dou Q, Fantacci ME, Geurts B, Gugten RV, Heng PA, Jansen B, de Kaste MMJ, Kotov V, Lin JY, Manders J, Sonora-Mengana A, Garcia-Naranjo JC, Papavasileiou E, Prokop M, Saletta M, Schaefer-Prokop CM, Scholten ET, Scholten L, Snoeren MM, Torres EL, Vandemeulebroucke J, Walasek N, Zuidhof GCA, Ginneken BV, Jacobs C (2017) Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: the LUNA16 challenge. Med Image Anal 42:1. https://doi.org/10.1016/j.media.2017.06.015
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M et al (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88. https://doi.org/10.1016/j.media.2017.07.005
Murphy A, Skalski M, Gaillard F (2018) The utilisation of convolutional neural networks in detecting pulmonary nodules: a review. Br J Radiol 91(1090):20180028. https://doi.org/10.1259/bjr.20180028
Hu Z, Tang J, Wang Z, Zhang K, Zhang L, Sun Q (2018) Deep learning for image-based cancer detection and diagnosis—a survey. Pattern Recognit 83:134–149. https://doi.org/10.1016/j.patcog.2018.05.014
Setio AA, Ciompi F, Litjens G, Gerke P, Jacobs C, van Riel SJ, Wille MM, Naqibullah M, Sanchez CI, van Ginneken B (2016) Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Trans Med Imaging 35(5):1160–1169. https://doi.org/10.1109/TMI.2016.2536809
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. MICCAI 2015:234–241. https://doi.org/10.1007/978-3-319-24574-4_28
Zagoruyko S, Komodakis N (2016) Wide residual networks. In: BMVC 2016
Huang XJ, Shan JJ, Vaidya V (2017) Lung nodule detection in CT using 3D convolutional neural networks. IEEE ISBI 2017:379–383. https://doi.org/10.1109/ISBI.2017.7950542
Dou Q, Chen H, Yu LQ, Qin J, Heng PA (2017) Multilevel contextual 3-D CNNs for false positive reduction in pulmonary nodule detection. IEEE Trans Bio Med Eng 62(7):1558–1567. https://doi.org/10.1109/TBME.2016.2613502
Dou Q, Chen H, Jin YM, Heng PA (2017) Automated pulmonary nodule detection via 3D ConvNets with online sample filtering and hybrid-loss residual learning. MICCAI 2017(10435):630–638. https://doi.org/10.1007/978-3-319-66179-7_72
He K, Zhang XY, Ren SQ, Sun J (2016) Deep residual learning for image recognition. IEEE CVPR 2016:770–778. https://doi.org/10.1109/CVPR.2016.90
Jin HS, Li ZY, Tong RF, Lin LF (2018) A deep 3D residual CNN for false-positive reduction in pulmonary nodule detection. Med Phys 45(5):2097–2107. https://doi.org/10.1002/mp.12846
Ding J, Li AX, Hu ZQ, Wang LW (2017) Accurate pulmonary nodule detection in computed tomography images using deep convolutional neural networks. MICCAI 2017(10435):559–567. https://doi.org/10.1007/978-3-319-66179-7_64
Ren SQ, He KM, Girshick R, Sun J (2016) Faster R-CNN: towards real-time object detection with region proposal networks. IEEE TPAMI 2016:1137–1149. https://doi.org/10.1109/TPAMI.2016.2577031
Zhu WT, Liu CC, Fan W, Xie XH (2018) DeepLung: 3D deep convolutional nets for automated pulmonary nodule detection and classification. IEEE WACV 2018:673–681. https://doi.org/10.1101/189928
Chen YP, Li JN, Xiao HX, Jin XJ, Yan SC, Feng JS (2017) Dual path networks. In: NIPS 2017
Huang G, Liu Z, van der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. IEEE CVPR 2017:2261–2269. https://doi.org/10.1109/CVPR.2017.243
Khosravan N, Bagci U (2018) S4ND: single-shot single-scale lung nodule detection. MICCAI 2018(11071):794–802. https://doi.org/10.1007/978-3-030-00934-2_88
Zhu W, Vang YS, Huang Y, Xie X (2018) DeepEM: deep 3D ConvNets with EM for weakly supervised pulmonary nodule detection. MICCAI 2018(11071):812–820. https://doi.org/10.1007/978-3-030-00934-2_90
Hu J, Shen L, Albanie S, Sun G, Wu EH (2018) Squeeze-and-excitation networks. In: IEEE CVPR 2018, pp 7132–7141. https://doi.org/10.1109/cvpr.2018.00745
Zhu WT, Huang YF, Zeng L, Chen XM, Liu Y, Qian Z, Du N, Fan W, Xie XH (2018) AnatomyNet: deep learning for fast and fully automated whole-volume segmentation of head and neck anatomy. Med Phys 46(2):576–589. https://doi.org/10.1002/mp.13300
Acknowledgements
This research was supported by the National Natural Science Foundation of China (Grant No. 81871457), the National Natural Science Foundation of China (Grant No. 51775368), the National Natural Science Foundation of China (Grant No. 51811530310) and the Science and Technology Project of Tianjin (Grant No. 18YFZCSY01300). We are grateful to the LUNA16 challenge organizers for their efforts in collecting and sharing chest CT scan data for comparing pulmonary nodule detection algorithms.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gong, L., Jiang, S., Yang, Z. et al. Automated pulmonary nodule detection in CT images using 3D deep squeeze-and-excitation networks. Int J CARS 14, 1969–1979 (2019). https://doi.org/10.1007/s11548-019-01979-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-019-01979-1