Abstract
Predicting methylation of the O6-methylguanine methyltransferase (MGMT) gene status utilizing MRI imaging is of high importance since it is a predictor of response and prognosis in brain tumors. In this study, we compare three different residual deep neural network (ResNet) architectures to evaluate their ability in predicting MGMT methylation status without the need for a distinct tumor segmentation step. We found that the ResNet50 (50 layers) architecture was the best performing model, achieving an accuracy of 94.90% (+/− 3.92%) for the test set (classification of a slice as no tumor, methylated MGMT, or non-methylated). ResNet34 (34 layers) achieved 80.72% (+/− 13.61%) while ResNet18 (18 layers) accuracy was 76.75% (+/− 20.67%). ResNet50 performance was statistically significantly better than both ResNet18 and ResNet34 architectures (p < 0.001). We report a method that alleviates the need of extensive preprocessing and acts as a proof of concept that deep neural architectures can be used to predict molecular biomarkers from routine medical images.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Background
Glioblastoma multiforme (GBM) is the most common primary brain tumor accounting for 45% of all malignant primary central nervous system tumors with a median survival of around 14.6 months [1]. GBMs are usually treated with surgical resection followed by radiation therapy (60 Gy in 30 fractions of 2 Gy) and temozolomide chemotherapy, improving median survival by 3 months versus radiotherapy alone. MRI is most commonly used to assess response due to its superior contrast compared with other imaging modalities [2, 3]. More specifically, imaging biomarkers extracted from functional MRI have been found to correlate with survival [4, 5].
Personalized medicine is an important new trend that attempts to identify genetic or other properties of tumors that allow more targeted therapy. Identification of these properties may enable more patient-specific treatment, and thus results in improved outcomes. Methylguanine methyltransferase (MGMT) is a key gene that encodes for a protein that repairs DNA. Several reports in the literature indicate that MGMT promoter methylation is associated with longer survival [6] as well as response to temozolomide [7, 8]. However, while determination of MGMT methylation status has been standard of care for some time, an accurate result is not always obtained due to the requirement of large tissue specimens. Furthermore, there are a limited number of laboratories that are able to perform these tests.
An emerging hypothesis is that genetic and/or molecular alteration within GBM manifests as specific, macroscopic, observable changes in MRI anatomical imaging [9]. Visual findings as well as texture features, originating from functional or anatomical MR imaging, have been investigated as imaging biomarkers to predict MGMT status [10,11,12,13,14,15]. However, the results from these studies are contradictory. Moon et al. [13] found that ill-defined tumor borders are associated with MGMT promoter methylation in a mixed group of WHO grade III and IV patients. However, in a similar study by Gupta et al. [15], no correlation between MGMT and either ill-defined borders or perfusion imaging-based biomarkers was found.
Most recently, Kanas et al. [16] highlighted the association of quantitative and qualitative morphologic imaging biomarkers extracted from preoperative imaging and MGMT methylation status in GBM. However, qualitative imaging feature estimation is a challenging task that depends on the individual experience. In a study of 43 patients by Ahn et al. [14], biomarkers based on ADC and fractional anisotropy (FA) parametric maps were found to be poor predictors of MGMT methylation, while capillary permeability (i.e., Ktrans) achieved an area under the receiver operating characteristic (ROC) curve of 0.75 contradicting the results reported by Moon et al. [13].
Texture features combined with classical machine learning have been proven to predict MGMT status utilizing MR imaging [11, 17]. Korfiatis et al. [17] utilizing an SVM-based classier achieved an area under the ROC curve of 0.85 (95% CI: 0.78–0.91) using four texture features (correlation, energy, entropy, and local intensity) originating from the T2 weighted images.
Recent studies [18] also highlighted the importance of using molecular biomarkers to group gliomas that have similar clinical behavior, response to therapy, and outcome. These findings highlight the significance of predicting molecular biomarkers and the potential impact in clinical practice if this could be done from MRI. One of the challenges in the texture-based approaches or morphologic-based approaches is the requirement for several preprocessing steps such as intensity standardization, skull stripping, and tumor segmentation. Segmentation of tumors is a challenging step often requiring manual correction of tumor segmentations produced by automated algorithms. Manual corrections increase the cost and time and can lead to inter-observer variation challenging algorithm which requires a segmentation mask as an input.
Deep learning has been emerging as an important technology in many different fields [19]. Convolutional neural networks (CNNs) are one form of deep learning that has been proven to be useful for both diagnosis and image analysis tasks [20,21,22,23,24,25]. In their simplest form, a CNN consists of a series of layers of convolution filters followed by data reduction layers at the end of the network.
Objective
In this study, we show that (1) CNNs provide a means to predict MGMT status and that (2) this can be done without the need for a separate tumor localization step, which is a common requirement in classical machine-learning approaches. Furthermore, there is not much theoretical basis for determining the optimal network architecture for a given task. In this work, we compare three different residual deep neural network (ResNet) architectures to evaluate their ability in predicting methylation status without the need for a distinct tumor segmentation step.
Dataset and Training, Validation, and Test-set Creation
This study was reviewed and approved as minimal risk by our institution’s Internal Review Board. Patients with newly diagnosed GBM (astrocytoma grade IV, WHO classification) treated at Mayo Clinic between January 1, 2007, and December 31, 2015, were identified. The inclusion criteria were age ≥ 18 years and preoperative MR scans that included T2 and T1 weighted post-contrast images performed at Mayo Clinic with known MGMT methylation status. All images were anonymized utilizing CTP (http://mircwiki.rsna.org/index.php?title=CTP-The_RSNA_Clinical_Trial_Processor) and the image processing pipelines were managed with MIRMAID [26].
One hundred fifty-five (155) presurgery MRI examinations were utilized in this study (66 methylated and 89 unmethylated tumors). MGMT methylation status was based on a review of histopathological analysis of tumor tissue. MRI imaging was performed on 1.5T or 3T scanners and included T2 weighted fast spin echo (TR, 4000–4800 ms; TE, 96–107 ms; slice thickness, 3 mm); axial T1 weighted images (TR, 20 ms; TE, 6 ms; slice thickness, 3 mm), with a FOV of 24 cm and a matrix size of 256 × 256; and matching T1-weighted post-contrast images. In all the exams, the contrast agent was power injected at 5 ml per second followed by a 20 cm3 saline chaser at the same flow rate. The contrast agent was gadolinium at 0.1 mmol/kg.
From the 155 presurgery MRI examinations utilized in this study, 66 patients had methylated and 89 patients had unmethylated tumors. For the methylated group, 53 scans were performed on a 1.5T scanner (40 GE and 13 Siemens), while 13 were performed on a 3T scanner (5 GE and 8 Siemens). For the unmethylated group, 76 scans were performed on a 1.5T scanner (54 GE and 21 Siemens), while 13 were performed on a 3T scanner (9 GE and 4 Siemens).
For the purpose of this study, only T2 images were used. N4 was used for the bias field correction [27]. This step is necessary since bias field signal is a low-frequency and very smooth signal that corrupts MRI images and can potentially affect image analysis steps following [28]. The slices from all examinations were divided into three categories: MGMT methylated tumors, MGMT unmethylated tumors, and the third category containing non-tumor slices.
Forty-five (45) examinations were randomly selected for testing, while the remaining 110 examinations were used to create the training and validation dataset. This resulted in 7856 images (1621 unmethylated, 934 methylated, 5301 no tumor) used for training and 2612 (335 unmethylated, 250 methylated, 2027 no tumor) used for testing. All analysis was performed on a slice-by-slice basis assigning each slice the same label for the same patient (i.e., methylated or unmethylated) with subsequent “voting” applied to also evaluate at the patient level.
Methods
Classification Scheme
Models were developed and trained using the NVidia (NVidia Corporation, Santa Clara, CA) graphical processing unit (GPU) utilizing the Keras (https://keras.io/) Python package. The model used was based on the deep residual learning network (ResNet) model [29]. ResNet enables training of deeper architectures, because the layers learn residual functions with reference to the layer inputs, instead of learning unreferenced functions. This enables the networks to be robust to the vanishing gradient problem and deals with the degradation of accuracy appearing in conventional deep networks. A shortcut connection allows for identity mapping ensuring that each subsequent layer will have the necessary information to learn additional features. ResNets consist of modularized architectures that stack building blocks of the same connecting shape. In this paper, we call these blocks “Residual Units” [30].
For the purpose of this study, three ResNet architectures were considered. The first consisted of 18 layers (referred to as ResNet18), the next consisted of 34 layers (referred to as ResNet34), and the last consisted of 50 layers (referred to as ResNet50). The He [31] weight initialization was used for all the ResNets [32]. Batch normalization (BN) was used right after each convolution and before activation [33]. As an extension to the original model, dropout was added at the fully connected layer [29]. ReLU was used as an activation function. The optimizer used was also that described by He [31].
Model Training and Assessment
The networks during the training validation phase were trained for 50 epochs utilizing stochastic gradient as optimizer with a learning rate of 0.01, minibatch of 32, momentum 0.5, and weight decay of 0.1. The learning rate was decreased by a factor of 10 when the learning rate was stable for more than 10 epochs. The training was stopped when the validation accuracy during the training phase was stable for 10 epochs.
The overall accuracy, the precision, recall, and F1 scores were used to evaluate the proposed model. A stratified cross-validation (a variation of k-fold) that returns stratified folds was utilized. The folds preserve the percentage of samples for each class. A k-fold cross-validated paired t test was performed to assess the difference in the models considered [34]. Each fold generated five different models for each one of the three ResNets considered in this study. To obtain the final result on the testing cases, the best model from the five folds was retrained utilizing all the data. Bonferroni correction was applied to account for multiple comparisons [35]. Selection of the best architecture was made from the F1 score achieved on validation set.
Results
The ResNet50 architecture was able to achieve accuracy of 94.90% (+/− 3.92%) for the test set (classification of a slice as no tumor, methylated MGMT, or non-methylated). ResNet34 achieved 80.72% (+/− 13.61%) while ResNet18 accuracy was 76.75% (+/− 20.67%) when the algorithms applied to the test set. ResNet50 performance was statistically significantly better than both ResNet18 and ResNet34 architectures (p < 0.001). Table 1 captures the results in terms of precision, recall, and F1 scores for the validation stage while Table 2 captures the results for each network trained during the cross-validation stage when applied to the test set.
Figure 1 shows the ResNet18 layer model.
Visual depiction of the ResNet50 model. Conv, pool, and fc stand for convolutional, pooling, and fully connected layer, respectively. The pooling size used was 2 (denoted by “/2.” For instance, “256/2” means that 256 filters were used and the size of pooling layer was 2). So the first box (“7 × 7 conv, 64”) means that the convolutional kernel size was 7 × 7 and 64 filters. We then explicitly describe the following layer as a 2 × 2 pooling layer, but elsewhere in this figure we use the shorthand of “/2” in the box showing the layer. Solid lines (—) indicate identity and dashed lines (- - -) indicate cross-residual weighted connections
Table 1 summarizes the results for all three ResNet models used in this study during validation, and Table 2 summarizes the results for all three ResNet models used in this study during testing. As observed from Table 2, comparing the trained models during the cross-validation stage, ResNet18 and ResNet34 had the largest variation when applied on the test sets. Table 3 depicts the confusion matrix for the final model when evaluated on the slice basis in the testing dataset. The classifier was able to correctly identify 98% of the slices with no pathology.
Table 4 shows results when a majority vote was used for all abnormal slices of a given subject, to decide whether that subject was methylated or not, using the ResNet50 algorithm. This also shows good performance, with the average values of 90% or better.
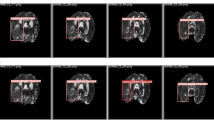
Figure 2 depicts the activation corresponding to the three first layers of the ResNet50 for two different slices.
The computationally expensive part of the proposed algorithm is the training phase, with total training time ranging from 3 to 10 h. For all networks, the inference for a patient scan takes less than 30 s for a scan including the preprocessing steps utilized while no specialized hardware is needed.
Discussion
Deep neural network architectures are able to predict MGMT status with high accuracy. Furthermore, it can also differentiate between slices containing tumor versus those that do not, minimizing the need for human intervention. By combining these into one network, we demonstrate that CNNs can learn complex tasks beyond simple yes/no classification.
Several papers have reported encouraging results regarding the importance of imaging-based features for predicting MGMT status [11, 13, 14, 16, 17], but they were limited by the significant preprocessing required to extract the imaging biomarkers, which also degraded reproducibility.
Reducing manual steps required by computer-aided diagnosis systems is a key element to enable translation of such systems into the clinical practice. The only preprocessing steps required by our system are image normalization and bias corrections, and both those steps are fully automated and the calculations require less than 2 min for a typical desktop computer. Our efforts were focused on a three-class problem rather than a binary approach (methylated versus unmethylated) enabling our algorithm to operate without the need for a tumor segmentation step.
In general, convolutional-based deep neural networks have been the main tool used for winning many competitions on publically available datasets [19]. ResNets enable training networks with hundreds of layers, while classic CNN architectures have poorer performance with addition of extra layers. ResNets consist of residual blocks that are at the early stages of the same dimensions as the data while in later stages, the data down-sampled and the number of layers are increased. This is generally attributed to the fact that early stages learn features that are relatively simple (e.g., edges). Such architectures improved the performance in many publicly available benchmarks [29]. He et al. developed a 152-layer network that outperformed the VGG net on the imagenet dataset [29]. Residual networks behave like ensembles of relatively shallow networks [36]. From these networks, the ones with shorter paths are more effective. Deeper architectures based on ResNet have been reported to yield better results [29].
Our results demonstrate that the ResNet50 (the deepest model considered in this study) outperformed all the shallower architectures (ResNet18, ResNet34) for all the metrics considered in this study with statistically significant difference. Table 3 shows performance on a patient basis; the performance on a patient basis is very high, though we expected the performance should be higher using patient-based votes (Table 4), since there are likely a few slices at the edges of tumors that may not have enough signal to produce an accurate decision. By voting all slices in a patient, these “noise” slices would be out-voted. This requires further investigation.
ResNet architectures and CNNs in general do not require calculation of texture features. Avoiding the feature selection step, which increases in complexity with the number of features available, is challenging and can lead to poor performance if the right features are not selected. With the proposed system, each layer of the network learns unique features; thus, there is no need for a feature reduction step. The proposed system’s performance, in terms of sensitivity and specificity, is better than the one reported by Korfiatis et al. [17] where conventional machine learning and second-order statistics were used.
There is widespread belief that large datasets are required to use deep-learning methods, particularly if the number of network parameters is high, and no transfer learning is used. Here we demonstrate that it is possible to achieve good results with a relatively small number of examinations (N = 155). This is particularly interesting because the “signal” is not so obvious between methylated and unmethylated tumors. While most humans can distinguish cats from dogs in a photograph, it is often viewed that thousands or more example photos are required for a deep neural algorithm to distinguish cats and dogs. Our results show the power of ResNets to identify visually subtle features for the task of predicting MGMT methylation. We also note that this was achieved without any form of data augmentation.
We applied no advanced intensity standardization technique, but a rather simple technique based on the mean value of the image, since our goal was to avoid intensity as one of the main features learned by the network [37]. This should make the network a more generalizable solution.
Future efforts will focus on utilization of larger networks hoping to achieve more accurate results. Deeper networks could potentially improve the results as well as the generalizability of the model; however, more data will be needed to minimize the effect of overfitting. Additional optimization algorithms should be investigated since the optimizer utilized here was the one proposed in the original paper by He et al.
One weakness of this study is the utilization of a dataset originating from a single site. Future effort should include dataset originating from multiple sites so the robustness of the model can be fully evaluated. The moderate variation in performance of folds in Tables 3 and 4 suggest some overfitting, and more examples should improve performance. Additionally, we considered only T2 weighted images, as prior work with texture suggested these were the most useful. The potential value of T1 post-contrast images or other image types should be investigated.
Conclusion
We report a method that alleviates the need of extensive preprocessing and acts as a proof of concept that deep neural architectures can be used to predict molecular biomarkers from routine medical images without labor-intensive user-dependent analysis.
References
Johnson DR, O’Neill BP: Glioblastoma survival in the United States before and during the temozolomide era. J Neurooncol 107:359–364, 2011
Ellingson BM, Wen PY, van den Bent MJ, Cloughesy TF: Pros and cons of current brain tumor imaging. Neuro Oncol 16(Suppl 7):vii2–vi11, 2014
Weizman L, Ben-Sira L, Joskowicz L, Aizenstein O, Shofty B, Constantini S, Ben-Bashat D: Prediction of brain MR scans in longitudinal tumor follow-up studies. Med Image Comput Comput Assist Interv 15:179–187, 2012
Law M, Young RJ, Babb JS, Peccerelli N, Chheang S, Gruber ML, Miller DC, Golfinos JG, Zagzag D, Johnson G: Gliomas: predicting time to progression or survival with cerebral blood volume measurements at dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Radiology 247:490–498, 2008
Jain R, Poisson LM, Gutman D et al.: Outcome prediction in patients with glioblastoma by using imaging, clinical, and genomic biomarkers: focus on the nonenhancing component of the tumor. Radiology 272:484–493, 2014
Zhang K, Wang X-Q, Zhou B, Zhang L: The prognostic value of MGMT promoter methylation in glioblastoma multiforme: a meta-analysis. Fam Cancer 12:449–458, 2013
Li H, Li J, Cheng G, Zhang J, Li X: IDH mutation and MGMT promoter methylation are associated with the pseudoprogression and improved prognosis of glioblastoma multiforme patients who have undergone concurrent and adjuvant temozolomide-based chemoradiotherapy. Clin Neurol Neurosurg 151:31–36, 2016
Rivera AL, Pelloski CE, Gilbert MR, Colman H, De La Cruz C, Sulman EP, Bekele BN, Aldape KD: MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro Oncol 12:116–121, 2010
Ellingson BM: Radiogenomics and imaging phenotypes in glioblastoma: novel observations and correlation with molecular characteristics. Curr Neurol Neurosci Rep 15:506, 2015
Rundle-Thiele D, Day B, Stringer B et al.: Using the apparent diffusion coefficient to identifying MGMT promoter methylation status early in glioblastoma: importance of analytical method. J Med Radiat Sci 62:92–98, 2015
Drabycz S, Roldán G, de Robles P, Adler D, McIntyre JB, Magliocco AM, Cairncross JG, Mitchell JR: An analysis of image texture, tumor location, and MGMT promoter methylation in glioblastoma using magnetic resonance imaging. Neuroimage 49:1398–1405, 2010
Levner I, Drabycz S, Roldan G, De Robles P, Gregory Cairncross J, Mitchell R: Predicting MGMT Methylation Status of Glioblastomas from MRI Texture. Med Image Comput Comput Assist Interv. 2009;12(Pt 2):522–530
Moon W-J, Choi JW, Roh HG, Lim SD, Koh Y-C: Imaging parameters of high grade gliomas in relation to the MGMT promoter methylation status: the CT, diffusion tensor imaging, and perfusion MR imaging. Neuroradiology 54:555–563, 2012
Ahn SS, Shin N-Y, Chang JH, Kim SH, Kim EH, Kim DW, Lee S-K: Prediction of methylguanine methyltransferase promoter methylation in glioblastoma using dynamic contrast-enhanced magnetic resonance and diffusion tensor imaging. J Neurosurg 121:367–373, 2014
Gupta A, Omuro AMP, Shah AD, Graber JJ, Shi W, Zhang Z, Young RJ: Continuing the search for MR imaging biomarkers for MGMT promoter methylation status: conventional and perfusion MRI revisited. Neuroradiology 54:641–643, 2012
Kanas VG, Zacharaki EI, Thomas GA, Zinn PO, Megalooikonomou V, Colen RR: Learning MRI-based classification models for MGMT methylation status prediction in glioblastoma. Comput Methods Programs Biomed 140:249–257, 2017
Korfiatis P, Kline TL, Coufalova L, Lachance DH, Parney IF, Carter RE, Buckner JC, Erickson BJ: MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Med Phys 43:2835, 2016
Eckel-Passow JE, Lachance DH, Molinaro AM et al.: Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med 372:2499–2508, 2015
Greenspan H, van Ginneken B, Summers RM: Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans Med Imaging 35:1153–1159, 2016
Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S: Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging 35:1207–1216, 2016
Dalmış MU, Litjens G, Holland K, Setio A, Mann R, Karssemeijer N, Gubern-Mérida A: Using deep learning to segment breast and fibroglandular tissue in MRI volumes. Med Phys 44:533–546, 2017
Dhungel N, Carneiro G, Bradley AP: A deep learning approach for the analysis of masses in mammograms with minimal user intervention. Med Image Anal 37:114–128, 2017
Spampinato C, Palazzo S, Giordano D, Aldinucci M, Leonardi R: Deep learning for automated skeletal bone age assessment in X-ray images. Med Image Anal 36:41–51, 2017
Yan Z, Zhan Y, Zhang S, Metaxas D, Zhou XS: Multi-Instance Multi-Stage Deep Learning for Medical Image Recognition. IEEE Transactions On Medical Imaging. doi:10.1109/TMI.2016.2524985
Rajkomar A, Lingam S, Taylor AG, Blum M, Mongan J: High-throughput classification of radiographs using deep convolutional neural networks. J Digit Imaging 30:95–101, 2017
Korfiatis PD, Kline TL, Blezek DJ, Langer SG, Ryan WJ, Erickson BJ: MIRMAID: a content management system for medical image analysis research. Radiographics 35:1461–1468, 2015
Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC: N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 29:1310–1320, 2010
Juntu J, Sijbers J, Dyck D, Gielen J: Bias Field Correction for MRI Images. In: Advances in Soft Computing. Springer. pp 543–551
He K, Zhang X, Ren S, Sun J: Deep Residual Learning for Image Recognition. arXiv [cs.CV]. 2015. https://arxiv.org/abs/1512.03385
He K, Zhang X, Ren S, Sun J: Identity Mappings in Deep Residual Networks. In: Lecture Notes in Computer Science. 2016, pp 630–645. https://springerlink.bibliotecabuap.elogim.com/chapter/10.1007/978-3-319-46493-0_38
He K, Zhang X, Ren S, Sun J: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. Las Vegas. 2016, pp 770–778
He K, Zhang X, Ren S, Sun J: Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification. 2015 I.E. International Conference on Computer Vision (ICCV), 2015. doi: 10.1109/iccv.2015.123
loffe S, Szegedy C: Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. arXiv [cs.LG]. 2015. https://arxiv.org/abs/1502.03167
Dietterich TG: Approximate statistical tests for comparing supervised classification learning algorithms. Neural Comput 10:1895–1923, 1998 1998
Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze, Vol. 8 (1936), pp. 3–62 Key: citeulike:1778138
Veit A, Wilber M, Belongie S: Residual Networks Behave Like Ensembles of Relatively Shallow Networks. arXiv [cs.CV]. 2016. https://arxiv.org/abs/1605.06431
Nyúl LG, Udupa JK: On standardizing the MR image intensity scale. Magn Reson Med 42:1072–1081, 1999
Acknowledgements
This study was supported by the NCI Grant CA160045, NVidia: Research Support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Korfiatis, P., Kline, T.L., Lachance, D.H. et al. Residual Deep Convolutional Neural Network Predicts MGMT Methylation Status. J Digit Imaging 30, 622–628 (2017). https://doi.org/10.1007/s10278-017-0009-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-017-0009-z