Abstract
Glioblastoma (GBM) is the most aggressive primary brain tumor. The standard radiotherapeutic treatment for newly diagnosed GBM patients is Temozolomide (TMZ). O6-methylguanine-DNA-methyltransferase (MGMT) gene methylation status is a genetic biomarker for patient response to the treatment and is associated with a longer survival time. The standard method of assessing genetic alternation is surgical resection which is invasive and time-consuming. Recently, imaging genomics has shown the potential to associate imaging phenotype with genetic alternation. Imaging genomics provides an opportunity for noninvasive assessment of treatment response. Accordingly, we propose a convolutional neural network (CNN) framework with Bayesian optimized hyperparameters for the prediction of MGMT status from multimodal magnetic resonance imaging (mMRI). The goal of the proposed method is to predict the MGMT status noninvasively. Using the RSNA-MICCAI dataset, the proposed framework achieves an area under the curve (AUC) of 0.718 and 0.477 for validation and testing phase, respectively.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The most prevalent malignant primary brain tumor in adults is glioblastoma (GBM) which accounts for 48.3% of all malicious brain tumors [1]. The first line radiotherapeutic treatment for GBM patients is Temozolomide (TMZ). The methylation state of the O6-methylguanine-DNA-methyltransferase (MGMT) gene promoter has been a significant biomarker for tumor response to TMZ treatment [2]. MGMT methylation status is associated with prolonged survival time in GBM patients [3]. The standardized method for evaluation of MGMT status is surgical resection which is invasive and time-consuming. Recently, different studies [4, 5] have found that genetic alternations are linked to phenotypic changes and can be detected using magnetic resonance imaging (MRI) features. Predicting MGMT status using MRI features can be broadly categorized into two categories: application of machine learning (ML) or deep learning (DL) models. Several studies focused on quantitative and qualitative feature extraction from pre-operative MRI, and then predicting MGMT status utilizing the extracted features in different ML models. V. G. Kanas et al. [6] apply quantitative and qualitative features extracted from segmented tumors, and then apply several dimensionality reduction methods for multivariate analysis of MGMT status. Another study by T. Sasaki et al. [7], the authors apply supervised principal component analysis to predict MGMT status utilizing shape and texture features. On the other hand, Authors in [8] apply different architectures of residual CNN to predict MGMT status. P. Chang et al. [9] focus on 2D CNN filters to extract features, and then apply principal component analysis (PCA) for dimensionality reduction of features that is used in the classification of genetic mutations. The study by E. Calabrese et al. [10] has cascaded deep learned based tumor segmentation with MGMT classification. After 3D tumor segmentation on multiparametric MRI sequence, pyradiomics is utilized for extraction of image features followed by random forest method to analyze the likelihood of MGMT status in patients. Most of the studies require extensive image pre-processing, feature extraction, and tumor segmentation before classifying MGMT methylation status.
In this work, we propose a deep learning-based approach with CNN that does not require tumor segmentation and feature extraction. In addition, to find the optimal hyperparameters in CNN we utilize Bayesian Optimization method.
2 Method
2.1 Description of Dataset
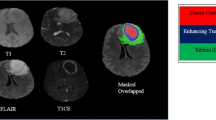
The dataset [11,12,13,14,15] is divided into three cohorts as follows: training, validation and testing. The training dataset consists of 585 patients. Each patient has four modalities of MRI scans in DICOM (Fig. 1): Fluid Attenuated Inversion Recovery (FLAIR), T1-weighted pre-contrast (T1w), T1-weighted post-contrast (T1wCE), and T2-weighted (T2). The MGMT status distribution of the training cohort is as follows: 307 patients are methylated, and 278 patients are unmethylated. The validation cohort consists of 87 patients. Note that the ground truth of validation data and testing data are kept private by the challenge organizers.
2.2 Radiogenomic Classification Model
A convolution neural network is a stacking of convolutional layers. The layers can be categorized into different stages where each stage has a same type of convolutional layer. Each convolution filter is made up of image pixel values that are modified throughout training. The model’s parameters are adjusted variables that can be approximated or learned from the data and incorporated within the learning process [16]. The model parameters are not selected manually such as weights in neural network. Hyperparameters are factors that impact the model’s training or behavior. Hyperparameters are non-model parameters that cannot be anticipated from the data set but can be customized by subject matter experts or via trial and error until an acceptable accuracy is attained [17].
To obtain high accuracy in classification tasks, users must properly handle the hyperparameter setting procedure, which varies depending on the algorithm and data collection. This procedure can be carried out by using the algorithm’s default values or manually configuring them. Another alternative is to use data-dependent hyperparameter-tuning approaches, which aim to reduce the algorithm’s estimated generalization error over a hyperparameter search space [17]. There are different methods such as random search, grid search, and Bayesian optimization to automatically configure the hyperparameters. Grid Search evaluates the learning algorithm using all possible hyper-parameter combinations [18]. Every parameter has the same chance of influencing the process. In Grid Search’s there are a lot of hyper-parameters to set, and the algorithm’s evaluation phase is highly expensive. Random searches utilize the same search space as Grid search using a random process. This method does not produce as accurate results as Grid Search, but it takes less time to compute. The first two approaches are computationally expensive because random combination of hyperparameters are considered. Bayesian Optimization use sequential method for global optimization of objective function [19].
The main idea behind Bayesian Optimization method is to limit the objective function’s evaluation phase by devoting more effort to select the next set of hyper-parameter values. This method is specifically designed to solve the problem of determining the maximum of an objective function \( f :X \rightarrow \mathbb {R} \), given as,
where, X corresponds to hyperparameter space which can be considered as three-dimensional hypermeter space [19].
The two basic component of Bayesian Optimization is statistical modeling and acquisition function to determine the next sampling of hyperparameters [20]. The statistical model generates a posterior probability distribution that specifies the objective function’s possible values at potential locations [21]. With the increment in data observations the algorithm performance improves and a potential region on hyperparameters space is determined using Gaussian Process. A Gaussian Process (GP) is a robust prior distribution on functions and a stochastic process [23]. A multi-variate gaussian process is completely defined by its mean and covariance matrix. The acquisition function determines the maximum of objective function at a particular point. The aquistion function consists of exploration and exploitation phase. The objective of exploration phase is sampling to optimize the search space and the target of exploitation phase to focus on reduced search space to select optimum samples [24].
The Bayesian optimization method employs a surrogate model that is fitted to the real model’s observations [25]. In this case, the real model is the Convolutional Neural Network (CNN) with hyperparameters selected for an iteration. In each iteration, it quantifies the uncertainty in the surrogate model using Gaussian process [26]. Then for next iteration, hyperparameter set is chosen using an acquisition function that balances the need to explore the whole search space versus focusing on high-performing sections of the search space.
The proposed pipeline in the Fig. 3 shows data augmentation steps in input data. After the input is provided in the CNN model (Fig. 2), hyperparameter optimization is performed utilizing the Bayesian Optimization method which provides the tunned model with optimized hyperparameters [27]. The tunned model is utilized for a classification model of the methylation status.
2.3 Training Stage
In the training phase, we randomly split the data into 80% training and 20% validation on the basis on methylation and unmethylation status. In training phase, we consider only the T1-weighted post-contrast (T1wCE) modality of MRI. The pre-processing steps consist of selecting fixed number of slices for each patient. At first, we sort out the middle slice number from the list of slices for each patient. Afterwards, the slices that are in the range of 25% above the middle slice and 75% below the middle slice is selected with a fixed interval. The interval is three but if a patient data contains less than 10 slices then the interval is changed to one. The selected slices are resized into 32 \(\times \) 32-pixel value and normalized between 0 and 255.
The pre-processed data is augmented prior to feeding into CNN model. The data augmentation includes random flip along the horizontal axis and random rotation with scale of 0.1. For hyperparameters optimization the number of trials is 20 and objective function is to minimize the validation loss. During parameter optimization, the bounded learning rate is between 0.0003 and 0. 003. The tunned model is trained for 200 epochs with Adam optimizer. Figure 4 shows the area under curve in the training phase with hyperparameter parameter optimized model.
3 Online Evaluation Results
We apply our proposed method to the online validation data which consists of 87 patients. The data is available in the RSNA-MICCAI Brain Tumor Radio-genomic classification competition under the Kaggle Platform [15]. We apply the pre-processing steps that are discussed in the previous section. The area under curve (AUC) value in the validation phase is 0.718. In addition, the proposed method is utilized on testing data which is kept private by the challenge organizers. The area under (AUC) value in the testing phase is 0.477. The performance of classification model is shown in the Table 1.
4 Discussion
In this study, we have applied CNN with Bayesian hyperparameters optimization for classification of MGMT status in Glioblastoma (GBM) patients from MRI. As it can be seen the model performance on testing data drops compared to validation data which might be caused by the lack of generalization of model in the testing data. Moreover, the predictive feature (MGMT) is not directly visible in imaging data which is rather a genomic biomarker identified by molecular analysis in tissue specimens. However, several studies [28,29,30] have shown that genetic alterations express themselves as phenotypic changes that can be identified by MRI features extracted from the segmented tumor. In our proposed framework, we do not consider tumor segmentation to extract features from the segmented tumor. We utilize the pre-processed DICOM directly to evaluate the methylation status in GBM patient. The goal of the proposed method is to simulate the real-world clinical scenario and asses how well the model generalizes the data obtained from different sources.
The selected slices contain the middle slice and slices selected at a regular interval for each patient. The CNN with Bayesian optimized hyperparameters assist the model to obtain better classification accuracy as depicted in Fig. 4 and in the validation phase of the challenge as shown in Table 1. The training phase contains a simple architecture of CNN and tunned with hyperparameters that is obtained by reducing an objective function in Bayesian process. Therefore, the tunning of the hyperparameters improve the performance when the data distribution is similar in training and validation phase. The dataset consists of multiple image samples from 18 institutions [31]. The source of data and distribution of test data is completely different [31] and hence the classification model is unable to generalize the new data distribution and the selected hyperparameters are not the optimized ones in testing data.
Bayesian Optimization is included in the pipeline to obtain hyperparameters that assist in better classification performance by optimizing the search space. The objective of the Bayesian approach is to reduce the validation loss in each trial and shrink the hyperparameter space after each trail. After completion of pre-defined trials on training data, best hyperparameters are included in the final training of the classification model. However, such hyperparameter optimization may not be able to generalize to data from different source.
5 Conclusion
In this paper, we propose a CNN model with Bayesian Optimized hyperparameters to classify MGMT methylation status in glioblastoma patients. Bayes Optimization is applied to obtain the hyperparameters for the CNN model. The goal of this work is to predict the MGMT status using non-invasive MRI features. The proposed approach does not require extensive tumor segmentation, image pre-processing, and feature extraction steps to predict MGMT status. The proposed method is evaluated on validation and testing dataset provided by RSNA-MICCAI. The validation and testing area under curve (AUC) were 0.718 and 0.477 respectively.
References
Ostrom, Q.T., et al.: CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012–2016. Neuro. Oncol. 21(Suppl 5), 1–100 (2019)
Liu, D., et al.: Imaging-genomics in glioblastoma: combining molecular and imaging signatures. Front. Oncol. 11, 2666 (2021)
Nam, J.Y., De Groot, J.F.: Treatment of glioblastoma. J. Oncol. Pract. 13(10), 629–638 (2017)
Korfiatis, P., et al.: MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Med. Phys. 43(6), 2835–2844 (2016)
Hajianfar, G., et al.: Noninvasive O6 Methylguanine-DNA methyltransferase status prediction in glioblastoma multiforme cancer using magnetic resonance imaging radiomics features: univariate and multivariate radiogenomics analysis. World Neurosurg. 132, 140–161 (2019)
Kanas, V.G., et al.: Learning MRI-based classification models for MGMT methylation status prediction in glioblastoma. Comput. Methods Programs Biomed. 140, 249–257 (2017)
Sasaki, T., et al.: Radiomics and MGMT promoter methylation for prognostication of newly diagnosed glioblastoma. Sci. Rep. 9(1), 1–9 (2019)
Korfiatis, P., Kline, T.L., Lachance, D.H., Parney, I.F., Buckner, J.C., Erickson, B.J.: Residual deep convolutional neural network predicts MGMT methylation status. J. Digital Imaging 30(5), 622–628 (2017). https://doi.org/10.1007/s10278-017-0009-z
Chang, P., et al.:Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. Am. J. Neuroradiol. 39(7), 1201–1207 (2018)
Calabrese, E., et al.: A fully automated artificial intelligence method for non-invasive, imaging-based identification of genetic alterations in glioblastomas. Sci. Rep. 10(1), 1–11 (2020)
Baid, U., et al.: The RSNA-ASNR-MICCAI BraTS 2021 Benchmark on Brain Tumor Segmentation and Radiogenomic Classification. https://arxiv.org/abs/2107.02314. Accessed 09 Aug 2021
Menze, B.H., et al.: The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging. 34(10), 1993–2024 (2015). https://doi.org/10.1109/TMI.2014.2377694
Bakas, S., et al.: Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Nat. Sci. Data. 4, 170–171 (2017). https://doi.org/10.1038/sdata.2017.117
Bakas, S., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-GBM collection. In: The Cancer Imaging Archive (2017). https://doi.org/10.7937/K9/TCIA.2017.KLXWJJ1Q
Bakas, S., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-LGG collection. In: The Cancer Imaging Archive (2017). https://doi.org/10.7937/K9/TCIA.2017.GJQ7R0EF
Luo, G.: A review of automatic selection methods for machine learning algorithms and hyper-parameter values. Netw. Model Anal. Health Inform. Bioinform. 5, 1–6 (2016)
Bergstra, J., Bengio, Y.: Random search for hyper-parameter optimization. J. Mach. Learn. Res. 13, 281–305 (2012)
Mersmann, O., Trautmann, H., Weihs, C.: Resampling methods for metamodel validation with recommendations for evolutionary computation. Evol. Comput. 20, 249–275 (2012)
Alibrahim, H., Ludwig, S.A.: Hyperparameter optimization: comparing genetic algorithm against grid search and Bayesian optimization. In: 2021 IEEE Congress on Evolutionary Computation (CEC), Kraków, Poland (2021). https://doi.org/10.1109/CEC45853.2021.9504761
Dewancker, I., McCourt, M.J., Clark, S.C.: Bayesian Optimization for Machine Learning : A Practical Guidebook. arXiv:abs/1612.04858 (2016)
Bergstra, J., Bardenet, R., Bengio, Y., Kégl, B.: Algorithms for hyper-parameter optimization. In: 24th International Conference on Neural Information Processing Systems (NIPS 2011), Red Hook, NY, USA (2011)
Frazier, P.: A Tutorial on Bayesian Optimization. arXiv:abs/1807.02811 (2018)
Rasmussen, C.E.: Gaussian processes in machine learning. In: Bousquet, O., von Luxburg, U., Rätsch, G. (eds.) ML -2003. LNCS (LNAI), vol. 3176, pp. 63–71. Springer, Heidelberg (2004). https://doi.org/10.1007/978-3-540-28650-9_4
Borgli, R.J., Kvale Stensland, H., Riegler, M.A., Halvorsen, P.: Automatic hyperparameter optimization for transfer learning on medical image datasets using Bayesian optimization. In: 13th International Symposium on Medical Information and Communication Technology (ISMICT), Oslo, Norway (2019)
Fraccaroli, M., Lamma, E., Riguzzi, F.: Automatic setting of DNN hyper-parameters by mixing Bayesian optimization and tuning rules. In: Nicosia, G., et al. (eds.) LOD 2020. LNCS, vol. 12565, pp. 477–488. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-64583-0_43
Guillemot, M., Heusèle, C., Korichi, R., Schnebert, S.: Maxime petit and liming Chen: tuning neural network hyperparameters through Bayesian optimization and Application to cosmetic formulation data (2019)
Snoek, J., Larochelle, H., Adams, R.P.: Practical Bayesian optimization of machine learning algorithms. Adv. Neural Inf. Process. Syst. 2012, 2951–2959 (2012)
Liu, D., et al.: Imaging-genomics in glioblastoma: combining molecular and imaging signatures. Front. Oncol. 11, 2666–2021 (2021). https://www.frontiersin.org/article/10.3389/fonc.2021.699265
Hajianfar, G., et al.: Noninvasive O6 Methylguanine-DNA methyltransferase status prediction in glioblastoma multiforme cancer using magnetic resonance imaging radiomics features: univariate and multivariate radiogenomics analysis. World Neurosurg. 132, 140–161 (2019). https://doi.org/10.1016/j.wneu.2019.08.232
Korfiatis, P., et al.: MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Med. Phys. 43(6), 2835–2844 (2016). https://doi.org/10.1118/1.4948668
RSNA-MICCAI Brain Tumor Radiogenomic Classification-Kaggle. https://www.kaggle.com/c/rsna-miccai-brain-tumor-radiogenomic-classification. Accessed 09 Aug 2021
Acknowledgements
We acknowledge partial support from National Institutes of Health grant # R01 EB020683.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Farzana, W., Temtam, A.G., Shboul, Z.A., Rahman, M.M., Sadique, M.S., Iftekharuddin, K.M. (2022). Radiogenomic Prediction of MGMT Using Deep Learning with Bayesian Optimized Hyperparameters. In: Crimi, A., Bakas, S. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2021. Lecture Notes in Computer Science, vol 12963. Springer, Cham. https://doi.org/10.1007/978-3-031-09002-8_32
Download citation
DOI: https://doi.org/10.1007/978-3-031-09002-8_32
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-09001-1
Online ISBN: 978-3-031-09002-8
eBook Packages: Computer ScienceComputer Science (R0)