Abstract
To effectively manage rare populations, accurate monitoring data are critical. Yet many monitoring programs are initiated without careful consideration of whether chosen sampling designs will provide accurate estimates of population parameters. Obtaining accurate estimates is especially difficult when natural variability is high, or limited budgets determine that only a small fraction of the population can be sampled. The Missouri bladderpod, Lesquerella filiformis Rollins, is a federally threatened winter annual that has an aggregated distribution pattern and exhibits dramatic interannual population fluctuations. Using the simulation program SAMPLE, we evaluated five candidate sampling designs appropriate for rare populations, based on 4 years of field data: (1) simple random sampling, (2) adaptive simple random sampling, (3) grid-based systematic sampling, (4) adaptive grid-based systematic sampling, and (5) GIS-based adaptive sampling. We compared the designs based on the precision of density estimates for fixed sample size, cost, and distance traveled. Sampling fraction and cost were the most important factors determining precision of density estimates, and relative design performance changed across the range of sampling fractions. Adaptive designs did not provide uniformly more precise estimates than conventional designs, in part because the spatial distribution of L. filiformis was relatively widespread within the study site. Adaptive designs tended to perform better as sampling fraction increased and when sampling costs, particularly distance traveled, were taken into account. The rate that units occupied by L. filiformis were encountered was higher for adaptive than for conventional designs. Overall, grid-based systematic designs were more efficient and practically implemented than the others.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rare species are routinely monitored in conjunction with conservation efforts to ensure the long-term viability of populations. Rare species often exhibit high temporal or spatial variability in population size or distribution (McDonald 2004). Obtaining accurate estimates of population parameters when faced with such high variability can be difficult, especially when the reality of limited budgets determines that only a small fraction of the population can be sampled. Thus, careful consideration of candidate sampling designs is advisable before the initiation of any monitoring program.
Conventional sampling methods may be inefficient when a population is patchily distributed in space, and the relative efficiencies of sampling methods could change if the overall abundance of the population varies over time. In either case, many sampling units may not contain individuals of the species of interest, resulting in a relatively high cost for the information content yielded by the sampling effort. In recent years, modified sampling designs involving adaptive selection of sample units (i.e., adaptive cluster sampling) have been proposed as a way to increase the efficiency of sampling such populations (e.g., Thompson 1990, 1991a, 1991b, 2002, 2004; Thompson and Seber 1996; Salehi and Seber 1997; Smith et al. 2004). Because the lack of precision in conventional methods is greatest when sampling rare populations with aggregated distribution patterns (Christman 2000), theory suggests adaptive designs should be superior to conventional methods in such cases. Yet there are many factors that may play a role in determining the best overall sampling strategy.
Evaluating conventional and adaptive sampling designs may involve pilot surveys or field trials. The variety of candidate designs that can be tested, however, is limited in such field efforts. Another approach is to use computer simulations to evaluate candidate designs. In this approach, the number and variety of designs is limited only by what can be readily coded into software. Whether field trials or computer simulations are involved, the effort is improved when reliable preliminary information on the population distribution is available. Yet evaluations preceding sampling are not frequently done, even though such evaluations could result in a more efficient and effective monitoring program (e.g., Khaemba et al. 2001; Pooler and Smith 2005). Here we evaluate simulations for five different sampling designs based on 4 years of field data. The purpose of this study was to identify efficient sampling designs (i.e., designs that provided precise density estimates, as measured by variance, for low cost, as measured by sample size).
Methods
Population and study area
The population of interest was the Missouri bladderpod, Lesquerella filiformis Rollins, a small (10–20 cm tall) winter annual in the mustard family (Brassicaceae) (Rollins 1956; Rollins and Shaw 1973). Lesquerella filiformis is listed as threatened and known only from a few counties in Missouri and Arkansas. Individual plants usually have multiple stems arising from a basal rosette, 1–4 cm in diameter. Lesquerella filiformis grows in open cedar glades, barrens, limestone outcrops, and rock pastures (Rollins and Shaw 1973). Yellow flowers and globose siliques are produced in mid-April to May; the plants die by late June and seeds lie dormant through the summer. Those seeds that break dormancy germinate in the late summer or fall. Rosettes remain green throughout the winter and bolt in the spring.
Lesquerella filiformis populations may fluctuate dramatically from year to year (Thomas 1996, Fig. 1). Factors that affect population size appear to operate independently of density, suggesting a combination of edaphic factors, climate, and disturbance control population size (Thomas 1996). In other winter annuals similar to L. filiformis, a persistent seed bank allows the population to survive unfavorable environmental conditions and reestablish very rapidly under more favorable conditions (Baskin and Baskin 1985).
Lesquerella filiformis populations may also exhibit relatively large amounts of spatial heterogeneity (Thomas 1996). This aggregated spatial distribution likely results from an interaction of microhabitat factors including substrate, soil depth, soil moisture, canopy shading, competition from other plants, and disturbance (Thomas and Willson 1992; Ware 2002).
This study was conducted on a population of L. filiformis inhabiting a limestone glade, Bloody Hill Glade (BHG), at Wilson’s Creek National Battlefield near Republic, Missouri.
Field surveys
BHG was divided into 963 5 × 5 m cells; the entire array covered 2.4075 ha. Exhaustive sampling of L. filiformis at BHG was conducted in each of 4 years (1997, 1998, 2003, and 2005). Surveys were conducted in mid- to late April, when most plants were in flower. In each 5 × 5 m cell, the density of L. filiformis plants was estimated using the following abundance class scale (0 = none, 1 = 1–9 plants, 2 = 10–49 plants, 3 = 50–99 plants, 4 = 100–499 plants, 5 = 500–999 plants, 6 = ≥ 1,000 plants) (Kelrick 2001).
Computer simulations
We used the software program SAMPLE (http://www.lsc.usgs.gov/AEB/davids/acs/), which was developed as a tool to simulate conventional and adaptive designs. SAMPLE accepts sample or population data, depending on whether a simulation or an analysis is desired. Auxiliary data can be input for designs, such as GIS-based adaptive sampling, that incorporate such information in the probability of selection. SAMPLE can also be used to generate spatially clustered populations for simulation, graph scatterplots, and analyze existing data from adaptive and conventional surveys.
Five sampling designs were evaluated by simulation, based on the L. filiformis population densities and distributions documented in the field surveys. For each design, one or more design factors were varied. Three of the designs evaluated involved adaptive cluster sampling (Thompson and Seber 1996). Adaptive cluster sampling, which may prove useful for sparse but aggregated populations, requires that when the variable of interest satisfies a given condition, additional units in the neighborhood of that unit are added to the sample (Thompson 1990; Seber and Thompson 1994). For example, a condition to trigger adaptive cluster sampling may be that 10 or more plants are present in a cell. When this condition is met, all adjacent cells sharing a contiguous border with the primary sampling cell (the neighborhood) are sampled. While the definition of a “neighborhood” among adaptive cluster sampling applications may vary, in this study neighborhoods were cross-shaped. Sampling continued for all cells that met the condition (e.g., ≥ 10 plants) until either: (1) no neighboring cells met the condition or (2) the boundary of the sampling region was reached.
The five designs simulated were:
-
(1)
Simple random sampling (SRS). This was the ‘null’ sampling model used for comparison with other approaches. Sample size was the only design factor.
-
(2)
Adaptive simple random sampling (ASRS). This was an adaptive cluster sampling approach with the initial sample plot selected by simple random sampling (Thompson 1990; Seber and Thompson 1994). The design factors were initial sample size, condition to adapt, and neighborhood. The condition to adapt was the count within a sampling unit; this triggered adaptive sampling in the neighborhood of the sampling unit. The neighborhood was defined as all units that shared a side with the unit that met the condition to adapt; this neighborhood definition was used in all adaptive designs. The modified Horvitz-Thompson estimator was used to estimate density (Thompson 1990).
-
(3)
Grid-based systematic sampling (GSS). A random plot was initially selected, and then additional plots were selected at equal intervals in the x and y directions. The design allowed for multiple random starts (1, 2, or 3) within the study site. The design factors were total sample size and number of random starts.
-
(4)
Adaptive grid-based systematic sampling (AGSS). The initial sample plot was selected as in GSS, and then sampling proceeded adaptively. The design factors were total initial sample size, number of random starts, condition to adapt, and neighborhood.
-
(5)
GIS-based adaptive sampling (GISA). In this adaptive design, the condition to adapt is based on the value of auxiliary information available in a geodatabase. The design factors were the auxiliary information used to trigger adaptive sampling, initial sample size, and condition to adapt. Thus, a unit’s neighborhood is adaptively sampled if the unit’s auxiliary variable meets the condition. The condition can take a number of forms depending on the expected relationship between the variable of interest (e.g., plant density) and the auxiliary variable in the ith unit (x i ). For example, condition (C) could be any of these three forms: x i > C, x i < C, or CL < x i < CU. In this study, plant density from previous years provided the auxiliary information. (In the results section, the previous year’s information is appended to the design’s name [e.g., GISA98] to indicate that counts from 1998 were incorporated into a future year’s sample selection.) GIS-based adaptive sampling was not simulated for the 1997 data, as auxiliary information was not available.
For all designs, the primary variable of interest was the density of plants (number/area) across the glade. A secondary variable of interest was the proportion of area occupied. We compare performance of the designs based on estimator precision for fixed sample size and the rate that plants were encountered in the sample.
The field surveys conducted within the comprehensive grid produced data in abundance categories. We converted categories of abundance into counts by generating a Poisson random deviate with expected value equal to the category midpoint. Thus, we randomly generated a single population for each year based on the observed abundance categories. Each population was then resampled using the candidate designs in each year. Number of times that each population was resampled (i.e., replications) was 1,000.
To account for sampling costs, we used field experience to determine typical times required to setup the site, travel within the site, and search a plot, and we incorporated these times into the simulations. Setup time required 48 h and involved locating and marking reference points throughout the site. Rate of travel (distance per time) was 10 m per min and included navigating among plots. Search time (time per plot) was 15 min per 25 m2 plot. Total cost was calculated by
where c is total cost (time), c 0 = setup time, c 1 = search time, ν = final sample size, d = distance traveled, and r = rate of travel (i.e., distance per time). Distance traveled (d) was calculated by starting at the southwest corner of the site and computing the distance (d 1) to the nearest sample plot (plot 1). Then from plot 1, the distance (d 2) to the nearest sample plot (plot 2) was calculated. This was continued for the remainder of the sample, and distance traveled was the sum of the plot-to-plot distances, i.e., \( d = \sum\nolimits_{{i = 1}}^{\nu } {d_{i} } \). Distance and cost were calculated for each replication and then averaged to generate an expected cost.
In this paper we present results for 2003, because our main purpose is to demonstrate the value of simulating sampling as a guide for sampling design, and because patterns observed in 2003 were qualitatively similar to results from other years. The full results of all simulations for all 4 years are available as Electronic supplementary material (ESM) (S1–S4).
Results
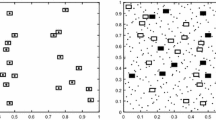
The density of the L. filiformis population, as determined by exhaustive grid-based sampling, varied over an order of magnitude over the 4 years, while the proportion of area occupied almost doubled from 1998 to 2005 (Table 1). The 2005 population was the third largest recorded in 18 years (Fig. 1). A moderate amount of spatial heterogeneity was observed across the glade (Fig. 2).
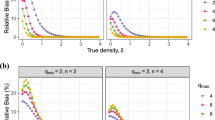
The simulation results revealed that sampling fraction (i.e., the final sample size/the number of units in the population) was the most important factor determining precision of density estimates (Table 2). The coefficient of variation (CV) decreased as sampling fraction increased for all designs (Fig. 3). A linear model showed that ln(sampling fraction), design, and their interaction were strong predictors of the coefficient of variation (F 11,46 = 138.96, P < 0.0001, R 2 = 0.98). Least square means from the linear model summarize how relative performance of designs changed across sampling fraction (Table 3). At a sampling fraction of 0.1, GSS had a lower CV than SRS, ASRS, AGSS, or GISA. At a sampling fraction of 0.2, CVs for GSS and AGSS were low, CVs for SRS and GISA were high, and the CV for ASRS was intermediate. At a sampling fraction of 0.3, CVs for ASRS and AGSS were the lowest, and CVs for SRS, GSS, and GISA were similar.
When total sampling costs (time in hours) were taken into account, relative precision changed modestly (Fig. 4a, b). However, when costs were restricted to distance traveled, adaptive cluster sampling designs (AGSS and ASRS) had much lower CVs than other designs across a range of distances (Fig. 4c). The reason that differences were modest for total sampling costs, but large for distance traveled, was because of the high setup cost, which was equal among all designs. The condition to adaptively sample neighboring units also affected sampling fraction. Sampling fraction declined when the condition was increased (e.g., from 10 to 100), because fewer cells existed that supported the higher plant densities.
Adaptive designs sampled a greater proportion of cells containing plants compared to conventional designs (Fig. 5). For example, the rate that plants were encountered during sampling was as much as 90% greater for adaptive than for conventional designs (Table 2). The encounter rate was higher for GIS98 compared to GIS97, suggesting that the plant distribution for the more recent year was a better predictor of plant distribution in 2003 (Fig. 5).
Box plots of the proportion of sampling units occupied by at least one Lesquerella filiformis plant in 2003 when population occupancy was 0.38. For conventional designs (GSS, SRS), the sample proportion is an estimate of the population occupancy. For adaptive designs (GISA97, GISA98, AGSS, ASRS), the sample proportion is higher than the population occupancy because adaptive sampling increases the probability of selecting occupied units, but adaptive sampling estimators result in unbiased estimation of population occupancy. Each box encloses 50% of the data with the median value of the variable displayed as a line. The lines extending from the top and bottom of each box mark the minimum and maximum values of the data set
There was some deviation between population parameters (e.g., density and proportion of area occupied) and the estimates averaged across the 1,000 replications (Table 2). Percent relative bias averaged −0.06% and ranged from −3.2 to 2.6% for density estimates and averaged −0.12% and ranged from −1.6 to 1.1% for occupancy estimates. These deviations were attributed to the random nature of the simulations. All the candidate designs and estimators are theoretically unbiased for density and proportion of area occupied, assuming perfect detection within the sampled units. Thus, as the number of replications increase the average estimate will approach the population parameter.
Overall, grid-based systematic designs would probably be more practically implemented than the others for this system. For example, to achieve a coefficient of variation of 24% required a sampling fraction of 11% when using a GSS design (Table 2).
Discussion
The problem of sampling rare, spatially clustered populations is widely recognized, and a number of sampling designs have been evaluated for this purpose (see review by Christman 2000). The performance of sampling designs is difficult to predict, however, based on generic studies, because the performance of sampling designs (as measured by precision, for example) depends on the spatial distribution and density of the population of interest. Ultimately, there is high practical value in being able to simulate sampling on an actual or approximate population; doing so can save time and money.
Although the theory of adaptive cluster sampling was introduced in 1990 (Thompson 1990), application has lagged behind, and this approach has only recently seen wide use. A number of recent studies have revealed adaptive cluster sampling to be an efficient method (e.g., Acharya et al. 2000; Vasudevan et al. 2001; Conners and Schwager 2002; Philippi 2005; Talvitie et al. 2006), although others have expressed some reservations in the use of adaptive methods over more conventional designs (e.g., Hanselman et al. 2003; Smith et al. 2003; Noon et al. 2006; Goldberg et al. 2007). The relative efficiency of adaptive sampling will ultimately depend upon a number of factors, and vary among different populations (Smith et al. 2004).
Adaptive sampling can be efficient when target organisms occur in clusters, and the clusters are relatively rare across the landscape (Thompson and Seber 1996; Smith et al. 2004). Although adaptive designs increased the likelihood of sampling where plants were, rather than where they were not, these adaptive designs did not uniformly result in more precise density estimates compared to conventional designs. This is possibly because L. filiformis was widespread throughout the site and clustered only to a moderate degree (Fig. 2).
We compared the designs based on the precision of density estimates for fixed sample size, cost, and distance traveled. Relative performance changed little by fixing on total costs compared to fixing sample size because, on average, setup costs comprised over 50% of total costs, and setup costs were shared by all designs. When only travel related costs were considered, adaptive sampling designs (particularly AGSS and ASRS) had much higher precision for fixed distance traveled compared to the other designs. In this situation, travel costs were low compared to the time required to setup and search a unit. However, when sampling a large site or one that is difficult to negotiate, travel costs can become high and minimizing travel can become relatively important.
When candidate designs perform similarly in terms of precision for fixed cost, the choice of which design to apply will depend on factors such as practicality and ease of use. Conventional designs allow sampling fraction to be determined prior to sampling, unlike adaptive designs. In contrast to simple random sampling, systematic sampling is often easier to apply in field surveys. In the adaptive versions of both designs, relatively more cells containing the target organisms will be sampled, but the final sample fraction will be unknown until sampling is complete. A high fraction could lead to the expenditure of more time and resources than planned to complete the survey.
There are several practical benefits of the GIS-based adaptive sampling design. First, units and networks to be sampled can be selected within a GIS before starting the survey. Thus, sample size will be known prior to fieldwork (although the resulting sample size could be quite large in relation to available resources). Second, because the networks or patch of sampled units are defined with existing data, ‘edge’ units (i.e., neighboring cells that do not meet the criteria for adaptive sampling) need not be sampled during fieldwork unless they are part of the initial sample. Third, the effect of the condition to adapt on final sample size can be investigated prior to fieldwork using a GIS-based analysis. The success of the GIS-based adaptive design is, however, contingent on the availability of predictive information prior to sampling. We used previous counts from the grid survey as a surrogate, which is artificial in this case because such information was generated for this simulation study. Alternatively, habitat and landscape variables that are predictive of organism distribution and available in GIS can be used in a GIS-based adaptive design (van Manen et al. 2005).
Contrasting our results to similar studies underlines the importance of evaluating conventional and adaptive sampling designs on a case-by-case basis. For example, Khaemba et al. (2001) simulated sampling for aerial surveys of African wildlife and found no significant differences between estimates obtained under random and systematic designs. An increase in precision was observed, however, for an adaptive design (Khaemba et al. 2001). In contrast, Smith et al. (2003) evaluated adaptive sampling for rare freshwater mussel populations and found that whereas encounter rates of rare individuals and species were higher for adaptive than for conventional designs, adaptive sampling did not increase precision of density estimates (Smith et al. 2003). Lesquerella filiformis was neither as clustered as the African wildlife populations nor as rare as the freshwater mussels.
Implications
In this case study, no single design was uniformly superior in terms of precision of density estimates across all sample sizes, over the years and population densities evaluated. Consideration of the simulation results and logistics of sampling, however, indicated that grid-based systematic designs would be more efficient logistically and more practically implemented than the others. Thus, simulations proved valuable as they indicated an appropriate sample fraction and allowed an evaluation of the practical advantages and disadvantages of the various sampling approaches.
Ultimately, many potential sampling designs exist, and design choice may depend upon conservation objectives, population characteristics, and available resources. The performance of various sampling designs may be difficult to predict, however, because such performance depends on the spatial distribution and density of the population of interest. Evaluation of candidate designs by computer simulation programs, such as SAMPLE, prior to sampling is likely to provide information critical to choosing an efficient overall design. Simulations can be an inexpensive decision tool for tailoring the evaluations to the particular populations under study.
References
Acharya B, Bhattarai G, de Gier A, Stein A (2000) Systematic adaptive cluster sampling for the assessment of rare tree species in Nepal. For Ecol Manage 137:65–73
Baskin JM, Baskin CC (1985) Life cycle ecology of annual plant species of cedar glades of southeastern United States. In: White J (ed) The population structure of vegetation. Dr. W. Junk Publishers, Dordrecht, pp 371–398
Christman MC (2000) A review of quadrat-based sampling of rare, geographically clustered populations. J Agric Biol Environ Stat 5:168–201. doi:10.2307/1400530
Conners ME, Schwager SJ (2002) The use of adaptive cluster sampling for hydroacoustic surveys. ICES J Mar Sci 59:1314–1325. doi:10.1006/jmsc.2002.1306
Goldberg NA, Heine JN, Brown JA (2007) The application of adaptive cluster sampling for rare subtidal macroalgae. Mar Biol (Berl) 151:1343–1348. doi:10.1007/s00227-006-0571-2
Hanselman DH, Quinn TJII, Lunsford C, Heifetz J, Clausen D (2003) Applications in adaptive cluster sampling of Gulf of Alaska rockfish. Fish Bull (Wash DC) 101:501–513
Kelrick MI (2001) Missouri bladderpod monitoring protocol for Wilson’s Creek National Battlefield. In: U.S. Geological Survey, Northern Prairie Wildlife Research Center. Jamestown
Khaemba WM, Stein A, Rasch D, De Leeuw J, Georgiadis N (2001) Empirically simulated study to compare and validate sampling methods used in aerial surveys of wildlife populations. Afr J Ecol 39:374–382. doi:10.1046/j.0141-6707.2001.00329.x
McDonald LL (2004) Sampling rare populations. In: Thompson WL (ed) Sampling rare or elusive species: concepts, designs, and techniques for estimating population parameters. Island Press, Covelo, pp 11–42
Noon BR, Ishwar NM, Vasudevan K (2006) Efficiency of adaptive cluster and random sampling in detecting terrestrial herpetofauna in a tropical rainforest. Wildl Soc Bull 34:59–68. doi:10.2193/0091-7648(2006)34[59:EOACAR]2.0.CO;2
Philippi T (2005) Adaptive cluster sampling for estimation of abundances within local populations of low-abundance plants. Ecology 86:1091–1100. doi:10.1890/04-0621
Pooler PS, Smith DR (2005) Optimal sampling design for estimating spatial distribution and abundance of a freshwater mussel population. J North Am Benthol Soc 24:525–537
Rollins RC (1956) On the identity of Lesquerella angustifolia. Rhodora 58:199–202
Rollins RC, Shaw EA (1973) The genus Lesquerella (Cruciferae) in North America. Harvard University Press, Cambridge
Salehi MM, Seber GAF (1997) Two stage adaptive cluster sampling. Biometrics 53:959–970. doi:10.2307/2533556
Seber GAF, Thompson SK (1994) Environmental adaptive sampling. In: Patil GP, Rao CR (eds) Handbook of statistics, vol 12. Environmental sampling. Elsevier, New York, pp 201–220
Smith DR, Villella RF, Lemarie DP (2003) Application of adaptive cluster sampling to low-density populations of freshwater mussels. Environ Ecol Stat 10:7–15. doi:10.1023/A:1021956617984
Smith DR, Brown JA, Lo NCH (2004) Application of adaptive cluster sampling to biological populations. In: Thompson WL (ed) Sampling rare or elusive species. Island Press, Covelo, pp 93–152
Talvitie M, Leino O, Holopainen M (2006) Inventory of sparse forest populations using adaptive cluster sampling. Silva Fenn 40:101–108
Thomas LP (1996) Population ecology of a winter annual (Lesquerella filiformis Rollins) in a patchy environment. Nat Areas J 16:216–226
Thomas LP, Willson GD (1992) Effects of experimental trampling on the federally endangered species, Lesquerella filiformis Rollins, at Wilson’s Creek National Battlefield, Missouri. Nat Areas J 12:101–105
Thompson SK (1990) Adaptive cluster sampling. J Am Stat Assoc 85:1050–1059. doi:10.2307/2289601
Thompson SK (1991a) Adaptive cluster sampling: designs with primary and secondary units. Biometrics 47:1103–1115. doi:10.2307/2532662
Thompson SK (1991b) Stratified adaptive cluster sampling. Biometrika 78:389–397. doi:10.1093/biomet/78.2.389
Thompson SK (2002) Sampling. John Wiley & Sons, New York
Thompson SK, Seber GAF (1996) Adaptive sampling. John Wiley, New York
Thompson WL (ed) (2004) Sampling rare or elusive species. Island Press, Covelo
van Manen FT, Young JA, Thatcher CA, Cass WB, Ulrey C (2005) Habitat models to assist plant protection efforts in Shenandoah National Park, Virginia, USA. Nat Areas J 25:339–350
Vasudevan K, Kumar A, Chellam R (2001) Structure and composition of rainforest floor amphibian communities in Kalakad-Mundanthurai Tiger Reserve. Curr Sci 80:406–412
Ware S (2002) Rock outcrop plant communities (glades) in the Ozarks: a synthesis. Southwest Nat 47:585–597. doi:10.2307/3672662
Acknowledgments
M. I. Kelrick developed field methods and performed data collection in 1997 and 1998. D. Beaulac collected data in 2003. Except for data collected in 2003, the NPS Inventory and Monitoring Program funded this project. Software development was partially supported through the USGS Status and Trends of Biological Resources Program, and we acknowledge P. Geissler for his guidance and support. J. Young and W. Cass provided helpful comments on an earlier draft.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Morrison, L.W., Smith, D.R., Young, C.C. et al. Evaluating sampling designs by computer simulation: a case study with the Missouri bladderpod. Popul Ecol 50, 417–425 (2008). https://doi.org/10.1007/s10144-008-0100-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10144-008-0100-x