Abstract
Microbial colonisation in the forestomach of a ruminant is one of the most crucial factors in determining many of its physiological developments and digestive capabilities. The present study attempts to identify establishment pattern of microbes in relation to food, age and rumen development in the buffalo calves at every fortnight interval from birth to 6 months of age, followed by every month till animals became 1 year of age. Diversity study based on 16S rRNA gene sequencing identified rapidly changing bacterial population during initial 60 days of life, which got assemblage as rumen became physiologically mature with increasing age of animals. A lactate fermenting aerobic to facultative anaerobic genera found during initial 30 days of life were expeditiously replaced by strict anaerobic cellulolytic bacterial population with increasing age. The study confirms that initial colonisation mainly depends on the oral cavity and skin of the mother, followed by the surrounding environment and feed offered, which is reversed in order once animal gets older. Some of the well-described genera based on culture-dependent studies like Ruminococcus spp. were found to be in lesser proportion suggesting an additional role of other microbes or niche in cellulose degradation. We report the presence of Porphyromonas spp. and Mannheimia glucosidal for the first time in bovine infants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The mammalian body is accommodating more than 1000 billion microorganisms, with digestive tract solely nurturing more than 70% of these commensal microbes responsible for health and productive performance. In an adult bovine, the rumen is an important site of anaerobic fermentation executed by the partially characterised complex group of anaerobic microbiota mainly consisting of bacteria followed by protozoa, archaea and fungi (Jami et al. 2014). Physiological development of functional rumen, capable of digesting plant material is a unique process which runs parallel with microbial colonisation process, especially during early phase of life. During the initial 3–4 weeks of life, when the calf is mainly ingesting milk through sucking, the rumen is rudimentary and does not participate in digestion as a result of closure of the esophageal groove by reflex action (Van Soest 1994). As per most accepted scientific research, foetuses are sterile in utero in most of the mammals (Tissier 1900), rumen bacterial colonisation also begins at birth and is extremely important for the physiological development of fully functional rumen capable of anaerobic fermentation (Connor et al. 2013; McCann et al. 2014). In tropical subcontinents like India, water buffalo (Bubalus bubalis) contributes 55% of total milk production of the country and is one of the most important livestock animals. The classical culture-dependent research concluded that, a group of bacteria having cellulolytic capability shows their presence in the first week of age itself and gradually occupy a major portion of total community by the end of the first month (Dehority 1991; Fonty et al. 1983; Hungate et al. 1964). Very limited study to identify rumen microbial diversity has been done using culture-independent methods. A very limited number of studies have been done to understand and identify establishment pattern of ruminal microbes in bovines, that too with different animals of different age groups raising a question of genotype dependent bias in the establishment of rumen microbiome (Jami et al. 2013; Li et al. 2012; Rey et al. 2014). For many years, the 16S rRNA genes were used as the primary tool for identifying bacterial diversity in many niches (Ghattargi et al. 2018; Land et al. 2015). It would be fascinating to understand the initial competitive inhibition between aerobic, anaerobic and facultative anaerobic bacteria to establish themselves in rumen. On that account, we aimed to study establishment pattern of rumen bacterial community using genotype as well as culture-independent next generation sequencing (NGS) based amplicon sequencing of hypervariable regions of 16S rRNA gene in buffalo calves from birth to 1 year of age.
Materials and methods
Animal, location and diet
Ten male calves of Surti breed of buffalo (Bubalus bubalis) with birth weight of 34 kg were maintained from birth to 1 year of age at Animal Nutrition Research Centre, College of Veterinary Science and Animal Husbandry, Anand Agricultural University, Anand, Gujarat, India (22.535634 N, 72.972544 E) from March, 2015 to February, 2016. All animal ethics guidelines were followed and complied as per permission (IAEC 525-2015) from Ethical Committee norms. Whole experimental protocols were approved by same Institutional Animal Ethics Committee, Anand Agricultural University, Anand, Gujarat, India. Calves were maintained separately avoiding contact from other adult animals except his mother. For the first week of life, calves were allowed to suck colostrum from mother without any additional feeding. From second week, pelleted concentrate (CP 18%; EE 3%; fibre 22%; starch 38% on a dry matter basis) and hay (CP 8%; EE 1.0%; fibre 60% on a dry matter basis) in addition to colostrum were given as feed. Each calf had free access to feed as well as deep well water of drinking quality. No diseases were noted during the experiment except few sporadic incidence of loose faeces which got cured without any treatment.
Sample collection from rumen
Rumen liquor samples from each calf were collected 2 h post feeding at 15 days interval from 1 day to 6 months and thereafter, 30 days interval till 1 year of age. The initial five samplings were done using two-way Foley’s catheter of 14 Fr size followed by subsequent collections using sterile stomach tube with vacuum pump. Samples were immediately transferred to three centrifuge tubes of 50 ml capacity and immediately processed as described previously (Jami et al. 2013). Two millilitres of sample having both liquid and solid particles was processed for DNA extraction and remaining samples were stored at − 80 °C.
DNA extraction and 16S rRNA gene amplicon sequencing using MiSeq
Metagenomic DNA was isolated from 2 ml of rumen liquor having solid particles within it. In the first step, samples were shaken vigorously to dissociate bacteria attached to solid ruminal particles followed by centrifugation at 250 rpm to sediment out debris. The supernatant was collected carefully and further used for mechanical lysis. The same material was subjected to genomic DNA extraction using the QIAamp DNA Stool Mini Kit (QIAGEN GmbH, Germany) as per the manufacturer’s instructions with slight modifications as suggested by (Zened et al. 2013). The DNA was eluted in 50 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA) and stored at − 80 °C for further use. The quantitative and qualitative evaluation of DNA was done using Nanodrop-1000 spectrophotometer (Thermo Fisher Scientific, MA). The same procedure was repeated separately for the rest of the samples on the day of sample collection itself. All ten samples belonging to the same age group were pooled in equimolar concentration before processing further. This was done to minimise cost as well as to minimise bias of sampling procedure. Thus, a total of 19 samples were processed in downstream sequencing protocols. The primer pair covering V3–V4 regions of 16 s rRNA gene was used for amplification (Supplementary Table 1). The PCR reaction mixture, conditions and PCR purification were done using procedure described by (Rey et al. 2014) in 25 μl reaction mixture with 10 ng of metagenomic DNA. PCR products were subjected to 2.0% agarose gel electrophoresis. Bands of expected size were excised and purified using MinElute PCR Purification Kit (Qiagen GmbH, Germany). Further, libraries were prepared using Nextera XT DNA Library Preparation Kit (Illumina Inc., CA) for sequencing as described by the manufacturer’s library preparation protocols. The final purified libraries were diluted to 4 nm, denatured and mixed with PhiX (about 30% of final DNA amount) and sequenced on the MiSeq desktop sequencer (Illumina) (2 × 300 run) at the Centre of Excellence in Biotechnology, AAU, Anand, Gujarat, India.
Metagenomics data analysis
Illumina-generated sequence reads were analysed through open source online server Metagenome Rapid Annotation using Subsystem Technology (MG-RAST) version 4.0 (Meyer et al. 2008). Briefly, low-quality regions were trimmed by SolexaQA (Cox et al. 2010). Good quality sequences were uploaded to MG-RAST (http://metagenomics.anl.gov/) under deposition numbers 4569530.3 to 4569537.3 for further analysis. A range of alpha diversity parameters were estimated using statistical software Paleontological Statistics (PAST) using Bray-Curtis distance measure method. We calculated alpha-diversity (observed species, Chao1 estimator of richness and Shannon’s diversity index) and beta-diversity (PCoA, UniFrac) matrices and the rarefaction curve was generated from a number of reads for each sample against the observed species. Shannon’s index, which is a measure of microbial richness, was estimated along with Berger-Parker and Chao1 indices, to determine the taxonomic diversification. Simpson’s index was used to ascertain the uniform distribution of taxon within the sample. Taxonomic profiling for community structure was done based on 16S rRNA gene sequences from the metagenomic sequence using the Best Hit Classification function and the RDP database (Ribosomal Database Project classifier) available on MG-RAST server and TSV table generated was used for further analysis. The default parameter of min. e-val. of 1e-5; min. ident. of 97%; and a min. align. len. of 15 bp were used as cut-off for metagenomic analysis. Partial least square-discriminant analysis (PLS-DA) was performed using METAGENassist in order to undermine the variation in distribution pattern among samples (Arndt et al. 2012) after normalisation of data between all the samples. Further, comparative analysis for taxa for each sample was performed using Statistical Analysis of Metagenomic Profiles (STAMP) software package (Parks et al. 2014). PCoA analysis was also performed through STAMP for phylum, genera and species in order to determine the variation between taxonomic profiles among different samples.
Results
The aim of this study was to unveil establishment pattern of bacterial community in rumen of Indian buffalo (Bubalus bubalis). For the first time in any scientific study, we analysed 19 samples of rumen liquor from calves from birth to 365 days of age using culture-independent V3–V4 hypervariable regions of 16S rRNA analysis. A repeated sampling of same calves was done till 1 year of age to avoid individual variation affecting the microbial composition. A total of 2,524,745 sequences (average 132,882 reads per sample) comprising of 683,174,542 bp were obtained after sequencing all 19 pooled samples using MiSeq 2 × 300 bp chemistry (Supplementary Table 2).
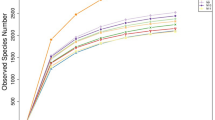
The overall number of OTUs identified based on V3–V4 regions of 16S rRNA analysis reached 7451 with 97% nucleotide sequence identity between reads as a cut-off value. The highest number of OTUs observed is 553 (15 days) and the lowest is 228 (90 days). The adequacy of reads generated for each sample was estimated through rarefaction curve at MG-RAST server (Supplementary Fig. 1). All of the samples revealed an exhaustive sampling suggesting availability of sufficient reads for identification of all the bacteria present in these samples (Table 1). High values of Simpson’s index indicate an unequal distribution of phenotypes within the samples without predominance of any particular taxa. However, evenness of taxonomic units was comparatively lower in few samples as revealed by Simpson’s index (Table 1). The higher value of Berger-Parker index in almost all samples further confirms the dominance of a few genera over the others. Shannon’s diversity index and Fisher diversity revealed a higher alpha diversity in all 19 samples. In addition, higher number of singletons and doubletons indicating rare OTUs were observed in all the samples as explained by Chao-1 index [minimum 266.6 (90 days) and maximum 767 (45 days)].
A total of 11 phyla with more than 1% abundance were identified in the samples. Among all these, Bacteroidetes, Firmicutes and Proteobacteria and unclassified bacteria were detected as dominant phyla irrespective of age group, though their ratio and abundance among all samples varied substantially (Supplementary Fig. 2). To estimate the beta diversity between the samples collected at different days of age, the principal coordinates analysis was performed. The score plot of all the 19 samples is shown (Fig. 1). The first three components (PCs) accounted for 83.5%, 7.4% and 5.1% of the variance, respectively. The closer positions of the samples indicate similar microbial composition between them. Based on distribution of variance in PCoA, it is evident that seven groups consisting of 0 D, 15 D, 30 D, 45 D, 60–165 D, 180–240 D and 270–360 D can be differentiated. During initial life up to 90 days, Bacteroidetes is the most predominant phylum followed by Firmicutes except the 0 D, where Proteobacteria is next to Bacteroidetes in terms of abundance, whose abundance significantly reduces from 15 D sample till the end of a year. At the 15 days of age Bacteroidetes:Firmicutes ratio is highest (10.06) which almost gets stabilised close to 1.0 after 60 days of age. Abundance of phylum Fibrobacter gradually increased and reached at the highest level at approximately 300–360 days of age. Relative abundance of other phyla like Planctomycetes and Cyanobacteria remained below 1% abundance except for few samples like 165 D, 45 D and 105 D. Phylum Tenericutes remained at constant abundance, around 2%, except for the first two stages of 0 D and 15 D. Verrucomicrobia phylum remained fairly constant after 90 days of age (Supplementary Table 3).
PCoA analysis of phylum level bacterial composition of ruminal samples taken at different ages. PC1, PC2 and PC3 together explain 96% of total variation at phylum level composition. It is clearly defined to make seven different groups showing different bacterial composition from all 19 samples collected at different ages
A total of 43 and 17 genera were observed with > 1% and > 5% relative abundance, respectively. The genera predominantly present among samples include unclassified bacteria, Bacteroides, Porphyromonas, Prevotella, Fibrobacter, Butyrivibrio and Ruminococcus. The genera which were observed at > 1% relative abundance mainly include Ruminobacter, Selenomonas, Treponema, Proteobacteria, Enterococcus, Acinetobacter and Alistipes (Supplementary Fig. 3 and Supplementary Table 4). The genus Porphyromonas was highest in 0 D samples (40.12% relative abundance) and decreased gradually up to 45 D, followed by rapid decrease in relative abundance among the rest of the samples. The higher proportion of this genus in initial life of the calf is mainly due to dominating presence of Porphyromonas cansulci, P. catoniae and P. gingivalis. At 0 D of age, Mannheimia was the second highest relative abundant (14.49%) genus which was significantly reduced in the next stage of 15 D and gradually disappeared as calf became older. The genus Prevotella was found least in 0 D with 0.93% relative abundance, increased gradually reached to maximum relative abundance of 33.25% at 135 D. It remained the most dominating genus across all the age groups except in the initial 15 days of age. This genus marked its presence mainly because of higher abundance of Prevotella ruminicola. The genus Clostridium was observed significantly only at 45 D of age with relative abundance of 8.1%. Proportion of the genus Ruminococcus increased gradually with age of calves and found to be constant among all age groups. The marked presence of genus Butyrivibrio was observed at 90 D and remained constant till 1 year of age. The genus Streptococcus was significantly present only in 0 D and suddenly disappeared to non-significant level of relative abundance in other age groups. The genus Bacteroides and Parabacteroides were significantly increasing from 0 D to 15 D of sampling followed by decrease to non-significant level among the rest of the age groups (Fig. 2).
Depending upon the different types of genera present in the samples, 54.8%, 18.0% and 9.2% variances are explained by PC1, PC2 and PC3, respectively in principal component analysis. Based on this, seven distinct groups can be illustrated following the same pattern as described for the phyla (Supplementary Fig. 4). Fold change of different genera indicated continuous increase in genus Butyrivibrio and Lachnospiraceae whereas decrease in Prevotella and Ruminococcus with development of rumen in calves (Fig. 3). Based on genus composition of samples, it can be very well defined that bacterial population takes significant shift at 15 D, 30 D and 45 D. After 60 days it is more or less stable at later age. Only 6.3% genera are common between the first four groups, whereas 47.1% genera are common in the last three groups comprising of 15 samples of different ages (Fig. 4).
Shared genera across different age groups. Venn plot showing the shared and unique genera found in two divisions (i) 0 D, 15 D, 30 D and 45 D and (ii) 60–165 D, 180–240 D and 270–360 D plotted groups. Only taxa at > 1% relative abundance at genus level are plotted. The parenthesis indicates percent value for respective divisions. (For details of genus please see Supplementary Table 5). Figure at bottom shows three leading genera of each group of sampling
A total of 76, 43 and 23 bacterial species found at > 1%, > 3% and > 5% relative abundance were observed across all the age groups. The predominant species found at first sampling includes, Mannheimia glucosidal, uncultured delta proteobacterium, Acinetobacter spp., Streptococcus suis, Streptococcus mutans, Clostridium ultunense, Porphyromonas cansulci and Porphyromonas levii. Most of them except Clostridium ultunense decreased significantly during 15 D sample. The day 15 samples were dominated by Parabacteroides goldsteinii, Prevotella nigrescens, Bacteroides spp., Bacteroides thetaiotaomicron, Bacteroides barnesiae, Bacteroides tectus and Bacteroides fragilis. A significant species level change was again witnessed during 30 D where bacterial population shifted to Riemerella spp. IPDH 9890, Akkermansia muciniphila, Prevotella ruminicola, Prevotella buccae, Prevotella veroralis, Prevotella melaninogenica, Prevotella loescheii, Prevotella stercorea and Prevotella baroniae. One more bacterial shift was observed in 45 D with predominant bacterial species like Prevotella multisaccharivorax, butyrate producing bacterium SS34, Paraprevotella clara, Pedobacter heparinus and Acetivibrio cellulolyticus. After 2 months of age, the bacterial population got more stabilised till 1 year of change with marked presence of species like Fibrobacter succinogenes, Prevotella ruminicola, Prevotella buccae, uncultured rumen bacterium, Acidaminococcus fermentans, Acetivibrio cellulolyticus, Anaeroplasma abactoclasticum, Alkaliphilus transvaalensis, Butyrivibrio fibrisolvens, Ruminococcus gnavus, Bacteroides cellulosolvens, Ruminococcus flavefaciens, Butyrivibrio hungatei, Gloeobacter violaceus, Prevotella bergensis, Ruminobacter amylophilus, Prevotella brevis, Ruminococcus bromii, Lachnospiraceae bacterium A4, Ruminococcus albus and uncultured proteobacterium (Fig. 5, Supplementary Table 6). It was observed that initial samples were having presence of aerobic to facultative anaerobic organism which rapidly turned in to marked presence of predominant obligate anaerobic species.
Species level composition. Colour-coded bar plot showing the relative abundance of each species at different age groups sampled at > 1% relative abundance. X axis represents the species level bacterial composition and Y axis represents relative abundance of particular species at different age group sampled
Discussion
The main objective of this experiment was to study the establishment pattern of rumen microbiota by analysing the microbial population in relation to different age groups in buffaloes. To understand this, we have collected rumen samples at every fortnight interval till 6 months of age and at monthly interval till 1 year of age. Based on V3–V4 amplicon analysis of 16S rRNA for accessing presence of bacterial species, initial days of sampling showed dynamic changes in community structure till 60 days of age (Mackie 2002; McCann et al. 2014). These may be due to dietary transitions during initial age of life happening with development of functional rumen from pre-ruminant stomach. Even though diet remained the same after 4 months of age, the less profound bacterial community shift was observed at 180 D and 270 D indicating that, microbial population in rumen was also dependent on age and other physiological factors and independent of diet, which are in line with similar observations reported in cattle by many group of researchers (Jami et al. 2013; Koenig et al. 2011). We observed greater density and diversity of microbes in age groups above 60 days which mainly includes Prevotella multisaccharivorax, Butyrivibrio spp., Ruminococcus spp., Bacteroides cellulosolvens, and Lachnospiraceae bacterium, (Malmuthuge et al. 2015) as compared to early age groups which were dominated by restricted niche which mainly includes initial colonisers like Streptococcus spp., Porphyromonas spp., as well as Bacteroides spp., Prevotella spp. and Akkermansia mucinphila. The similar type of establishment pattern was also observed in pre-ruminant stomach of other bovines (Fanaro et al. 2003; Jami et al. 2013) as well as human infants (Conroy et al. 2009; Jost et al. 2012).
We observed Bacteroidetes, Firmicutes and Proteobacteria as dominant phyla with considerable variations among all age groups. The phylum Bacteroidetes which was highest in 15 D age, mainly due to genus Bacteroides and Porphyromonas, significantly reduced thereafter and replaced by exclusive presence of Prevotella genus. This change is possibly related to change in diets from low-fibre, high-calorie which mainly includes colostrum (high in protein, sugar and fat) to high-fibre, low-calorie diet as calves advance in age; this is in accordance with other research groups (Koenig et al. 2011; Li et al. 2012; Rey et al. 2014). Reduction in proportion of phylum Bacteriodetes to Firmicutes as well as notable increase in genus Firmicutes were not observed till 30 D age, which was followed by sudden increase in proportion thereafter mainly due to increase in genus Butyrivibrio and Ruminococcus. This initial presence of genus can be attributed to Streptococcus bovis which is mainly responsible for digestion of milk-rich diet and lactate production favouring colonisation of other lactate-utilizers with increase in rapidly fermentable carbohydrates in diet. The similar observations were also reported in cattle (Drackley 1999; Hernandez-Sanabria et al. 2012) and human infants (Bergstrom et al. 2014; Ley et al. 2008a) during initial colonisation of gut microbiome. The phylum Proteobacteria was found highest in 0 D sample with highest proportion of Mannheimia spp., Acinetobacter spp. (Li et al. 2012), and Deltaproteobacteria spp., which almost get obsolete after a month of age.
At the very first day of age, the bacterial community was mainly composed of Porphyromonas gingivalis, P. cansulci and other members of this genus as well as Mannheimia glucosidal, uncultured delta proteobacterium, Acinetobacter spp., Streptococcus bovis and S. mutans. Most of them are natural habitat of mammalian gingiva, oral cavity or skin. These observations again showed concordance with hypothesis suggested by many groups of researchers that, initial microbes colonising mammalian gut are mainly derived from mother’s tongue, colostrum, skin and surrounding environment (Curtis and Sloan 2004; Ley et al. 2008b; Naeem et al. 2014; van Nimwegen et al. 2011; Zilber-Rosenberg and Rosenberg 2008). Bacterial community shifted from aerobic and facultative anaerobic initial gut coloniser to strict anaerobic organisms like Fibrobacter succinogenes, Prevotella ruminicola, Clostridium aldrichii, Butyrivibrio fibrisolvens and others with increased age (Jami et al. 2013; Rey et al. 2014).
Once again, the culture-independent method proven to be superior to culture-dependent method enabling depiction of true diversity of organisms. In our study, we identified many well-described species based on cultural studies belonging to genera Ruminoccocus, Escherichia, Fusobacterium, Streprococcus, and Lactobacillus (McCann et al. 2014; Stevenson and Weimer 2007) although with very low level of abundance suggesting important role of ‘microbial niche of both cultivable and non-cultivable organisms’ in rumen digestion. Thus, culture-independent method is an indispensable tool to estimate the actual diversity of rumen microbiome.
As reported in many companion experiments, Prevotella has emerged out as dominant bacterial species in the rumen under all dietary variations (Koenig et al. 2011; McCann et al. 2014; Rey et al. 2014). The maximum abundance of Prevotella was found at around 30 days of age where forage and concentrate gradually got incorporated in the diet. As Prevotella genus has been shown to produce propionates (O’Neill et al. 2011; Strobel 1992), we can assume that, shift in diet form milk to forage and grain around 30 to 45 days of age resulted in increased production of butyric and propionic acids with increased blood flow to the ruminal papillae and epithelium, in turn stimulation of vascular budding and epithelial cell proliferation (Hoover and Miller 1991; Matsui et al. 2000). Thus, increase in Prevotella can be correlated with rumen maturation where it gradually becomes capable of digesting grasses and grains after 60–90 days of age. We reported Prevotella ruminicola, Prevotella brevis, Prevotella loescheii, Prevotella stercorea, Prevotella baroniae and Prevotella buccae in our study but we could not detect two culturally well-defined species i.e. Prevotella bryantii and Prevotella albus (Avgustin et al. 1997; Bekele et al. 2010) in our study. This could be due to highly varied genetic divergence of Prevotella strains, low abundance in samples or database selected for analysing our data.
In accordance with the other experimental evidences, we also observed a negative correlation between Bacteroides spp. and Prevotella spp. from 15 D to 360 D of age but not at 0 D age. The relative presence of both was negligible at the time of birth, and it could be because of the absence of these bacteria in aerobic surrounding environment or at the prenatal anaerobic environment of the buffalo uterus (Scholtens et al. 2012). The similar age-related difference in bacterial genus colonisation was also reported in cattle as well as other mammals (Malmuthuge et al. 2015). Considering all these, we can conclude that Bacteroides spp. colonise faster and better after 60 days, once ruminal papillae become mature under effect of propionate produced by Prevotella spp.
In contrast to the report (Jami et al. 2013) of early presence of cellulolytic bacteria like Fibrobacter succinogenes and Ruminoccous albus at first day of life in bovines, we could not find significant proportion of these organisms in our experiment till 45 days of age i.e. in pre-ruminant stomach non-capable of digesting cellulose. Instead of that, we reported noticeable presence of Mannheimia glucosida, a facultative anaerobic bacterium, predominantly encountered in upper respiratory tract and oral cavity of ruminants (Lau et al. 2015; Poulsen et al. 2006) at 0 D with other bacterium like Porphyromonas spp. and Streptococcus bovis. Jami et al. (2013) also reported presence of a lactose fermenter and starch utilizer at 1 to 3 days of age similar in line with our report.
Conclusions
The present study constitutes the first ever report on establishment of rumen microbiota in buffalo calves from day old to 1 year of age, analysed in context with available dietary and physiological parameters. In addition, this is the first experiment of its kind, where same calves were reared from birth to 1 year of age. Based on our study, we are in agreement that, a combination of both cultural dependent and independent approach is required to identify less abundant microbes from rumen to aid our understanding of rumen microbiome establishment. Greater resolution achieved through next-generation sequencing of 16S rRNA amplicon revealed dynamic microbial composition up to 60 days of age which was gradually stabilised with increasing age. The initial colonisation of aerobic and facultative anaerobic microflora from mother’s oral cavity and surroundings of calves were gradually replaced by strict anaerobic microbiota after 90 days of age. The bacterial composition was most significantly influenced by the time of cessation of milk feeding. We found correlations between type of feed offered to animals and microbial composition available in our analysis; which mainly includes lactose fermenters and butyrate-producing genera in early life followed by cellulolytic bacterial genera in well-matured rumen after 90 days of age. Further studies defining interaction between microbiota and host in relation to other physiological parameters like VFA production, digestion efficiency, growth rate, and measurement of emission gas will improve understanding of this dynamic community, its pattern of establishment and a scope of possible modification for efficient utilisation of feed in the future.
Data availability
All data generated or analysed during this study are included in this published article as supplementary information files. In addition, the datasets generated and analysed during the current study are available in the MG-RAST. The following link can be used by the reviewer to access data as a part of data share at MG-RAST. The data will be made publically available once paper will be accepted.
Abbreviations
- NGS:
-
next-generation sequencing
- VFA:
-
volatile fatty acid
- CP:
-
crude protein
- EE:
-
ether extract
- 0D:
-
zero day
- MG-RAST:
-
metagenome rapid annotation using subsystem technology
- PAST:
-
paleontological statistics
- PCoA:
-
principal coordinates analysis
- PLS-DA:
-
partial least squares-discriminant analysis
- TSV:
-
tab separated value
- STAMP:
-
statistical analysis of metagenomic profiles
- OUT:
-
operational taxonomic units
- PC:
-
principal component
- Fr:
-
French gauge
References
Arndt D, Xia J, Liu Y, Zhou Y, Guo AC, Cruz JA, Sinelnikov I, Budwill K, Nesbo CL, Wishart DS (2012) METAGENassist: a comprehensive web server for comparative metagenomics. Nucleic Acids Res 40:W88–W95
Avgustin G, Wallace RJ, Flint HJ (1997) Phenotypic diversity among ruminal isolates of Prevotella ruminicola: proposal of Prevotella brevis sp. nov., Prevotella bryantii sp. nov., and Prevotella albensis sp. nov. and redefinition of Prevotella ruminicola. Int J Syst Bacteriol 47:284–288
Bekele AZ, Koike S, Kobayashi Y (2010) Genetic diversity and diet specificity of ruminal Prevotella revealed by 16S rRNA gene-based analysis. FEMS Microbiol Lett 305:49–57
Bergstrom A, Skov TH, Bahl MI, Roager HM, Christensen LB, Ejlerskov KT, Molgaard C, Michaelsen KF, Licht TR (2014) Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl Environ Microbiol 80:2889–2900
Connor EE, Baldwin RL, Li CJ, Li RW, Chung H (2013) Gene expression in bovine rumen epithelium during weaning identifies molecular regulators of rumen development and growth. Funct Integr Genomics 13:133–142
Conroy ME, Shi HN, Walker WA (2009) The long-term health effects of neonatal microbial flora. Curr Opin Allergy Clin Immunol 9:197–201
Cox MP, Peterson DA, Biggs PJ (2010) SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11:485
Curtis TP, Sloan WT (2004) Prokaryotic diversity and its limits: microbial community structure in nature and implications for microbial ecology. Curr Opin Microbiol 7:221–226
Dehority BA (1991) Effects of microbial synergism on fibre digestion in the rumen. Proc Nutr Soc 50:149–159
Drackley JK (1999) ADSA Foundation Scholar Award. Biology of dairy cows during the transition period: the final frontier? J Dairy Sci 82:2259–2273
Fanaro S, Chierici R, Guerrini P, Vigi V (2003) Intestinal microflora in early infancy: composition and development. Acta Paediatr 91:48–55
Fonty G, Gouet P, Jouany JP, Senaud J (1983) Ecological factors determining establishment of cellulolytic bacteria and protozoa in the rumens of meroxenic lambs. J Gen Microbiol 129:213–223
Ghattargi VC, Nimonkar YS, Burse SA, Davray D, Kumbhare SV, Shetty SA, Gaikwad MA, Suryavanshi MV, Doijad SP, Utage B, Sharma OP, Shouche YS, Meti BS, Pawar SP (2018) Genomic and physiological analyses of an indigenous strain, Enterococcus faecium 17OM39. Funct Integr Genomics 18:385–399
Hernandez-Sanabria E, Goonewardene LA, Wang Z, Durunna ON, Moore SS, Guan LL (2012) Impact of feed efficiency and diet on adaptive variations in the bacterial community in the rumen fluid of cattle. Appl Environ Microbiol 78:1203–1214
Hoover WH, Miller TK (1991) Rumen digestive physiology and microbial ecology. Vet Clin N Am Food Anim Pract 7:311–325
Hungate RE, Bryant MP, Mah RA (1964) The rumen bacteria and protozoa. Annu Rev Microbiol 18:131–166
Jami E, Israel A, Kotser A, Mizrahi I (2013) Exploring the bovine rumen bacterial community from birth to adulthood. ISME J 7:1069–1079
Jami E, White BA, Mizrahi I (2014) Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS One 9:e85423
Jost T, Lacroix C, Braegger CP, Chassard C (2012) New insights in gut microbiota establishment in healthy breast fed neonates. PLoS One 7:e44595
Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE (2011) Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 108(Suppl 1):4578–4585
Land M, Hauser L, Jun SR, Nookaew I, Leuze MR, Ahn TH, Karpinets T, Lund O, Kora G, Wassenaar T, Poudel S, Ussery DW (2015) Insights from 20 years of bacterial genome sequencing. Funct Integr Genomics 15:141–161
Lau JS, Omaleki L, Turni C, Barber SR, Browning GF, Francis MJ, Graham M, Korman TM (2015) Human wound infection with Mannheimia glucosida following lamb bite. J Clin Microbiol 53:3374–3376
Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI (2008a) Evolution of mammals and their gut microbes. Science 320:1647–1651
Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI (2008b) Worlds within worlds: evolution of the vertebrate gut microbiota nature reviews. Microbiology 6:776–788
Li RW, Connor EE, Li C, Baldwin Vi RL, Sparks ME (2012) Characterization of the rumen microbiota of pre-ruminant calves using metagenomic tools. Environ Microbiol 14:129–139
Mackie RI (2002) Mutualistic fermentative digestion in the gastrointestinal tract: diversity and evolution. Integr Comp Biol 42:319–326
Malmuthuge N, Griebel PJ, Guan le L (2015) The gut microbiome and its potential role in the development and function of newborn calf gastrointestinal tract. Front Vet Sci 2:36
Matsui H, Ogata K, Tajima K, Nakamura M, Nagamine T, Aminov RI, Benno Y (2000) Phenotypic characterization of polysaccharidases produced by four Prevotella type strains. Curr Microbiol 41:45–49
McCann JC, Wickersham TA, Loor JJ (2014) High-throughput methods redefine the rumen microbiome and its relationship with nutrition and metabolism. Bioinf Biol Insights 8:109–125
Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA (2008) The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386
Naeem A, Drackley JK, Lanier JS, Everts RE, Rodriguez-Zas SL, Loor JJ (2014) Ruminal epithelium transcriptome dynamics in response to plane of nutrition and age in young Holstein calves. Funct Integr Genomics 14:261–273
O'Neill BF, Deighton MH, O'Loughlin BM, Mulligan FJ, Boland TM, O'Donovan M, Lewis E (2011) Effects of a perennial ryegrass diet or total mixed ration diet offered to spring-calving Holstein-Friesian dairy cows on methane emissions, dry matter intake, and milk production. J Dairy Sci 94:1941–1951
Parks DH, Tyson GW, Hugenholtz P, Beiko RG (2014) STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30:3123–3124
Poulsen LL, Reinert TM, Sand RL, Bisgaard M, Christensen H, Olsen JE, Stuen S, Bojesen AM (2006) Occurrence of haemolytic Mannheimia spp. in apparently healthy sheep in Norway. Acta Vet Scand 48(19):19
Rey M, Enjalbert F, Combes S, Cauquil L, Bouchez O, Monteils V (2014) Establishment of ruminal bacterial community in dairy calves from birth to weaning is sequential. J Appl Microbiol 116:245–257
Scholtens PA, Oozeer R, Martin R, Amor KB, Knol J (2012) The early settlers: intestinal microbiology in early life. Annu Rev Food Sci Technol 3:425–447
Stevenson DM, Weimer PJ (2007) Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl Microbiol Biotechnol 75:165–174
Strobel HJ (1992) Vitamin B12-dependent propionate production by the ruminal bacterium Prevotella ruminicola 23. Appl Environ Microbiol 58:2331–2333
Tissier H (1900) Recherches sur la flore intestinale des nourrissons:(état normal et pathologique)
van Nimwegen FA, Penders J, Stobberingh EE, Postma DS, Koppelman GH, Kerkhof M, Reijmerink NE, Dompeling E, van den Brandt PA, Ferreira I, Mommers M, Thijs C (2011) Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol 128(948–955):e941–e943
Van Soest PJ (1994) Nutritional ecology of the ruminant. Cornell University Press
Zened A, Combes S, Cauquil L, Mariette J, Klopp C, Bouchez O, Troegeler-Meynadier A, Enjalbert F (2013) Microbial ecology of the rumen evaluated by 454 GS FLX pyrosequencing is affected by starch and oil supplementation of diets. FEMS Microbiol Ecol 83:504–514
Zilber-Rosenberg I, Rosenberg E (2008) Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev 32:723–735
Acknowledgements
We are thankful to the Indian Council of Agricultural Research for providing financial support under Niche Area of Excellence Programme (Grant Letter F. No. 10 (2)/2011-EPD). We thank Dr. Ghanshyambhai Patel for providing excellent technical assistance in sample collection, maintaining animals and collecting nutritional data at Animal Nutrition Research Station, Anand Agricultural University, Anand, Gujarat, India.
Funding
All the funding for the current research work was provided by Indian Council of Agricultural Research under Niche Area of Excellence programme wide NO. ICAR letter No. 10(2)/2011-EPD dated 15-07-2014 under research project entitled “Metagenomic analysis of ruminal microbes”.
Author information
Authors and Affiliations
Contributions
PGK conceptualised the project, participated in sample collection and drafted the manuscript; JRT performed DNA extraction, amplicon generation and data analysis; RJP assisted in data analysis; ATH assisted technically during experimental study; MJP performed sequencing and generated data on MiSeq; RKS maintained animals and did sample collection; SJJ drafted actual research project and improved manuscript; CGJ conceptualised research idea and provided all facilities to carryout research. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All animal ethics guidelines were followed and complied as per permission from Ethical Committee norms and letter No. IAEC 525-2015.
Conflict of interest
None of the authors has any financial or personal relationships that could inappropriately influence or bias the content of the paper. The authors declare no conflict of interest.
Electronic supplementary material
Supplementary Table 1
Details PCR primers used for detection of rumen bacterial species in this study by amplicon generation followed by NGS. (XLSX 8 kb)
Supplementary Table 2
Details of numbers of reads and bases generated for all 19 samples. (XLSX 10 kb)
Supplementary Table 3
Relative abundance of each bacterial taxa at phylum level (> 1% level of abundance) as well as ratio of Bacteriodetes:Firmicutes (XLSX 19 kb)
Supplementary Table 4
Relative abundance of each bacterial taxa at genus level (> 1% level of abundance) among seven groups consist of a total 19 samples collected at different age. (XLSX 72 kb)
Supplementary Table 5
Details of individually shared genera across different age groups. (XLSX 18 kb)
Supplementary Table 6
Relative abundance of each bacterial taxa at species level (> 1% level of abundance) among seven groups consist of a total 19 samples collected at different age. (XLSX 274 kb)
Supplementary Figure 1
Rarefaction analysis for bacterial operational taxonomic units (OTUs) in each sampling. Individual rarefaction curves for each rumen sample taken to evaluate the depth of sequencing for each sample. Each age group is distinguished by different colour of trend lines. (DOCX 1583 kb)
Supplementary Figure 2
This figure shows the relative abundance of each bacterial taxa at phylum level. X-axis, samples from different age; Y-axis, the relative abundance of bacterial taxa at each phylum level (%). (DOCX 1197 kb)
Supplementary Figure 3
This figures shows the relative abundance of each bacterial taxa at genus level. X-axis, seven age groups consist of a total 19 sampling at different age: Y-axis, the relative abundance of bacterial taxa at each genus level (%). (DOCX 737 kb)
Supplementary Figure 4
PCoA analysis of genus level bacterial composition of ruminal samples taken at different age. PC1, PC2 and PC3 together explains 82% of total variation at genus level composition. It is clearly defined to make seven different groups showing different bacterial composition from all 19 samples collected at different age. (DOCX 420 kb)
Rights and permissions
About this article
Cite this article
Koringa, P.G., Thakkar, J.R., Pandit, R.J. et al. Metagenomic characterisation of ruminal bacterial diversity in buffaloes from birth to adulthood using 16S rRNA gene amplicon sequencing. Funct Integr Genomics 19, 237–247 (2019). https://doi.org/10.1007/s10142-018-0640-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-018-0640-x