Abstract
Drought is a major abiotic stress affecting crop productivity and quality. As a class of noncoding RNA, microRNA (miRNA) plays important roles in plant growth, development, and stress response. However, their response and roles in tomato drought stress is largely unknown. Here, by using high-throughput sequencing, we compared the miRNA profiles before and after drought treatment in two tomato genotypes: M82, a drought-sensitive cultivated tomato (Solanum lycopersicum), and IL2-5, a drought-tolerant introgression line derived from M82 and the tomato wild species S. pennellii (LA0716). A total of 108 conserved and 208 novel miRNAs were identified, among them, 32 and 68 were significantly changed in expression after stress. Further, 1936 putative target genes were predicted for those differentially-expressed miRNAs. Gene ontology and pathway analysis showed that many of the target genes were involved in stress resistance, such as genes in GO terms including response to stress, defense response, response to stimulus, phosphorylation, and signal transduction. Our results suggested that miRNAs play an essential role in the drought response of tomato. This work will help to further characterize specific miRNAs functioning in drought tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As sessile organisms, plants suffer from various environmental stresses. Among all the abiotic stresses, drought is the most damaging natural disaster (Seneviratne 2012). With the change of global climate, drought occurred more frequently across the world, and it seriously affects the productivity and quality of crop (Ivits et al. 2014). Therefore, it is urgent to understand the response and underlying mechanisms of plants that challenged with drought stress (Shinozaki and Yamaguchi-Shinozaki 2007). However, the drought-responsive mechanisms of plants are extremely versatile and complex. Plants can adapt to drought by reducing size, developing a more vigorous root system, closing stomata, or enhancing wax formation on leaf surface. Plants can also be tolerant to drought by changing the physiological and biochemical status in cells, typically including increase of osmotic protectants and antioxidant compounds (Valliyodan and Nguyen 2006; Lawlor 2013). These adaptive processes are coordinated by the expression of a huge number of protein-coding RNA and also noncoding RNA.
MicroRNAs (miRNAs) are small noncoding regulatory RNAs with 20–24 nt in length, which has led to intense interest recent years (Chuck et al. 2009; Xie et al. 2015b). Plant miRNAs are synthesized through both canonical and noncanonical pathways (Budak and Akpinar 2015). In the canonical pathway, primary miRNA transcripts are transcribed by RNA polymerase II, and then processed by DCL1 (Dicer-like 1) enzyme to yield miRNA/miRNA* duplex. The duplex is methylated at the 3’ ends by HEN1 (Hua Enhancer 1), exported into the cytoplasm by HASTY, and then incorporated into AGO (Argonaute) to form RISC (RNA-induced silencing complex). In the noncanonical pathway, imprecisely processed miRNA species are produced by DLC2, DCL3, and DCL4; however, the underlying mechanism remains to be elucidated. miRNAs function in eukaryotic cells by regulating the expression of target genes at the post-transcriptional level (Jeong et al. 2011). miRNAs are involved in many biological processes, including plant development, hormone regulation, nutrition balance, and the biogenesis of their selves (Chuck et al. 2009; Liu et al. 2016). More and more evidence has shown that miRNAs play important roles in plant response to abiotic stresses (Sunkar et al. 2012; Budak et al. 2015; Ferdous et al. 2015). miRNAs are involved in plant drought response through regulating its downstream targets in ABA response, auxin signaling, osmotic protection, and antioxidant system (Ding et al. 2013). With the rapid development of sequencing technology, transcriptomic profiling has been successfully applied to identify miRNAs from different crop species, especially the lowly expressed miRNAs. Drought-related miRNAs have been identified in many plant species, such as Arabidopsis (Sunkar and Zhu 2004; Li et al. 2008; Liu et al. 2008), rice (Zhao et al. 2007; Zhou et al. 2010; Cheah et al. 2015), maize (Xu et al. 2014), barley (Kantar et al. 2010), cotton (Xie et al. 2015a), and Triticeae (Alptekin et al. 2017). However, information is very limited for the miRNAs involved in the drought response of tomato.
Tomato (Solanum lycopersicum) is one of the most important vegetable species grown worldwide (Klee and Giovannoni 2011). Tomato fruits have low calories and are rich in lycopene, vitamins, and ions (Ilahy et al. 2016). However, abiotic stresses, such as drought, are major factors limiting the sustainable production of tomato. Stresses not only reduce tomato yield, but also affect the fruit quality (Capel et al. 2015; Iovieno et al. 2016). Therefore, it would be very important to exploit drought-tolerant genes and to elucidate their mechanisms. Modern tomato cultivars have very narrow genetic bases after long-term and high-pressure of selection, and the vast majority of them are sensitive to drought stress (Shirasawa et al. 2010). However, elite genes for stress tolerance are well reserved in some wild tomato species. For instance, S. pennellii grows in Andean regions in South America where it is extremely drought, and accessions from this species show high level of drought tolerance (Bolger et al. 2014). Using the processing tomato cultivar M82 as recurrent parent, Eshed and Zamir (1994) have developed an introgression line population which covers the whole genome of S. pennellii LA0716, and it serves as a classic population for addressing complex quantitative trait loci (QTLs), including drought-tolerant QTLs (Alseekh et al. 2013).
With the decoding of the genome, tomato appears to be a new model to study the interaction of miRNAs and their targets (The Tomato Genome Consortium 2012; Bolger et al. 2014; Cao et al. 2014; Lin et al. 2014). Using next-generation sequencing, miRNAs such as miR156 and miR172 have been detected to be involved in fruit development (Karlova et al. 2013). Also, Jin and Wu (2015) and Pradhan et al. (2015) have investigated the change of miRNAs in tomato before and after the inoculation of Botrytis cinerea and tomato leaf curl New Delhi virus, respectively. Cao et al. (2014) have identified 161 conserved and 236 novel miRNAs in the two libraries involved in chilling stress from S. habrochaites. Nevertheless, only a couple of reports have been published so far on miRNAs involved in tomato drought response (Candar-Cakir et al. 2016; Liu et al. 2017).
In this study, by employing high-throughput sequencing, we profiled drought-responsive miRNAs in a drought-tolerant line (IL2-5) from the introgression line population of LA0716 (Gong et al. 2010), and its drought-sensitive recurrent parent (M82). A total of 108 conserved and 208 novel miRNAs were detected in the small RNA libraries constructed from RNA samples with and without drought stress, and 32 and 68 out of them changed significantly in expression after stress, respectively. Many putative targets of those miRNAs are involved in stress tolerance, which indicated that these miRNAs and their corresponding targets may play important roles in tomato drought response.
Materials and methods
Plant materials and drought treatment
Seeds of M82 and IL2-5 were obtained from the Tomato Genetic Resource Center (TGRC: https://tgrc.ucdavis.edu). M82 (S. lycopersicum) is a drought-sensitive tomato cultivar (Gong et al. 2010). IL2-5 is a drought-tolerant introgression line with S. pennellii LA0716 and M82 as the donor and recurrent parent, respectively (Eshed and Zamir 1994; Gong et al. 2010). This line carries a single chromosomal fragment from 44,867,267 to 53,854,595 bp of chromosome 2 of S. pennellii LA0716 (https://solgenomics.net, Ver. SL2.5).
Seed germination, soil mixture, and seedling management were the same as described in our recent publication (Liu et al. 2017). The major difference is the growth facility. The seedlings in this study were placed in a glasshouse (belonging to the National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University). A photoperiod of 16/8 h light/dark cycle was achieved by using artificial light to supplement natural light, and favorable temperature conditions (25/20 °C day/night) were maintained through a hot-water pipe heating system. At the five-leaf stage, the tomato seedlings were challenged with drought stress by withdrawing water, and the control plants were irrigated as usual. After 10 days of stress, the third leaf from the bottom of each seedling was removed, quick-frozen in liquid nitrogen, and stored at − 70 °C until use.
Small RNA library construction and sequencing
Total RNA was extracted from the leaf samples using TRIzol reagent (Invitrogen, CA, USA) according to the supplier’s manual. RNA quality and concentration were determined by a Nanodrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE), and the RNA integrity was further checked by 1% agarose gel analysis. All the RNA samples were sent to Novogene (Beijing, China) to construct libraries for small RNA sequencing. Single-end reads (SE50) were sequenced on an Illumina HiSeq 2500 platform (Illumina Inc., San Diego, CA, USA).
Identification of conserved and novel miRNAs
As described in our recent publication (Liu et al. 2017), raw reads were filtered to obtain clean reads, and potential small RNAs were obtained by mapping the clean reads to the tomato reference genome (https://solgenomics.net, Ver. SL2.5) with bowtie software (Langmead and Salzberg 2012). Rfam database (https://rfam.xfam.org, Ver. 11.0) was employed to identify rRNA, tRNA, snRNA, and snoRNAs (Burge et al. 2013). After removing the house-keeping noncoding RNAs mentioned above, the remaining reads were blasted (E value ≤ 1e-6) against the tomato miRNAs in miRBase 21 to obtain conserved miRNA (Griffiths-Jones et al. 2008). After removing the reads corresponding to the newly identified conserved RNAs, the remaining reads were predicted by miR-PREFeR (https://github.com/hangelwen/miR-PREFeR) to obtain potential novel miRNAs. During the prediction, putative miRNAs with less than 10 reads and no expression of the STAR sequence were removed to increase the accuracy (Lei and Sun 2014).
Identification of differentially-expressed miRNAs
Differentially-expressed miRNAs were calculated using the expression of transcript per million (TPM) value. TPM is a standardized method used to calculate miRNA expression levels. TPM values were calculated as follows: (actual miRNA count/total count of mapped reads) × 1,000,000. Then, the fold-change, p value and q value were calculated using perl scripts. Differentially-expressed miRNAs were identified according to the criteria of |log2 (fold-change) | ≥ 1 and FDR-adjusted q value ≤ 0.05.
Prediction of miRNA targets, GO, and KEGG pathway analysis
Potential target genes of miRNA were predicted using the psRNATarget server (https://plantgrn.noble.org/psRNATarget/) (Dai and Zhao 2011). The parameters in prediction were set as follows: maximum expectation score (3.0), length for complementarity (17 bp), and range of central mismatch (10–11 nt). Gene ontology (GO) enrichment was conducted to analyze the functions of target genes for the miRNAs, using the GO Enrichment tool in PlantRegMap (https://plantregmap.cbi.pku.edu.cn/) (Jin et al. 2017). Pathway analysis was performed using KEGG (The Kyoto Encyclopedia of Genes and Genomes) database (https://www.genome.jp/kegg/) (Kanehisa et al. 2014).
Validation of miRNA and target gene expression with qRT-PCR
The expression of some miRNAs and target genes were validated by quantitative real-time PCR (qRT-PCR). Total RNA sample was treated by DNase I (Takara, Dalian, China) to remove residual genomic DNA. RNA was reverse transcribed to cDNA using the Mir-X miRNA qRT-PCR SYBR® Kits (Takara), according to the supplier’s instruction. The cDNA was diluted to a final volume of 100 μl, and qRT-PCR was performed to detect the expression of the selected miRNAs and target genes. The primers were listed in Table S6. PCR cycling conditions were: 95 °C 10 s, 40 cycles of 95 °C 5 s, and 60 °C 20 s. Specificity of the amplified product was confirmed by performing a standard melting curve analysis (95 °C 55 s, 55 °C 30 s, 95 °C 30 s). All reactions were repeated three times, and the U6 and Actin4 (Solyc04g011500) were served as internal control. The relative expression level of miRNAs and the predicted target genes was calculated using the 2−ΔΔCT method (Schmittgen and Livak 2008).
Availability of data and materials
The sequencing data of this study are available in the Sequence Read Archive (SRA) at the National Center for Biotechnology Information (NCBI) (Accession number: SRX2820300, SRX2820301, SRX2820302 and SRX2820303). The other supporting data are included as supplementary files.
Results
Basic statistics of the small RNA data from tomato libraries
To identify drought stress-related miRNAs in tomato, small RNA libraries were constructed for the drought-sensitive tomato cultivar M82 with (SD) and without (SCK) drought stress, and also for the drought-tolerant introgression line IL2-5 before (TCK) and post (TD) stress. After sequencing, 48,776,425 raw reads were acquired (Table 1). A total of 32,017,598 clean reads were obtained after removing the adaptors and junk reads. Sequences within the range of 16–28 nt were further analyzed. The length distributions from the libraries tested were overall similar, most of the small RNAs are between 21 and 24 nt (Fig. 1). Among them, small RNAs with 24 nt were the most abundant, with an average percentage of 38.83% (Fig. 1).
Conserved miRNAs from tomato
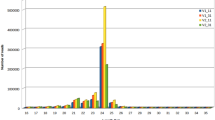
To identify conserved miRNAs from the small RNA libraries tested, reads with the length of 16–28 nt were mapped to tomato conserved miRNAs in miRBase 21. In total, 108 conserved miRNAs were obtained, which were belonging to 46 miRNA families (Table S1). Different families had different numbers of miRNAs (Table S1). The families of sly-miR156 and sly-miR482 had the most members, each with seven miRNAs. The expression level of different miRNAs was widely different, with reads number from a few to millions (Fig. 2, Table S1). The highly-expressed ten miRNAs were sly-miR159, sly-miR162, sly-miR166a, sly-miR166b, sly-miR166c-3p, sly-miR396a-5p, sly-miR396b, sly-miR482e-3p, sly-miR9471a-3p, and sly-miR9471b-3p, and their accumulative reads accounted for 83.57% of the total conserved reads. Among them, sly-miR159 had the highest number of reads, with an average of 142,249 reads in each library, which accounted for approximate 30% of the total conserved reads (Fig. 2).
Abundance of the top 10 highly-expressed conserved miRNAs in the four tomato libraries. SCK: miRNA library from the drought-sensitive genotype M82 without drought stress, SD: from M82 with drought stress, TCK: from the drought-tolerant genotype IL2-5 without drought stress, TD: from IL2-5 with drought stress
It was found that 98 conserved miRNAs were detected in all the libraries tested (Fig. 3a, Table S1). For the drought-sensitive M82, 100 miRNAs were detected both before and after drought treatment. Only two miRNAs (sly-miR169b and sly-miR169c) were specifically expressed before stress, while four miRNAs (sly-miR5302a, sly-miR9469-5p, sly-miR9472-3p, and sly-miR9473-3p) were only detected after stress (Table S1). As for the drought-tolerant line IL2-5, 99 miRNAs were detected both before and after drought stress. Five miRNAs (sly-miR169c, sly-miR169e-5p, sly-miR9469-3p, sly-miR9469-5p, and sly-miR9478-5p) were detected specifically before stress; however, no specific miRNA was detected only after drought treatment (Table S1). Regarding genotype-specific miRNAs, four (sly-miR169b, sly-miR5302b-3p sly-miR9472-3p, and sly-miR9473-3p) and two (sly-miR169e-5p and sly-miR9469-3p) were found to be specific to M82 and IL2-5, respectively (Fig. 3a, Table S2). In regard to treatment, four (sly-miR169b, sly-miR169e-5p, sly-miR169c, and sly-miR9469-3p) and two (sly-miR9472-3p and sly-miR9473-3p) miRNAs were detected in the small RNA libraries from controland drought-treated plants, respectively (Fig. 3a, Table S1). In addition, three of the 108 conserved miRNAs (sly-miR156b, sly-miR156c, sly-miR171a) identified were located in the introgression region of IL2-5 (Table S1).
Venn diagrams of tomato miRNAs identified in the four libraries. a Conserved miRNAs. b Novel miRNAs. SCK: miRNA library from the drought-sensitive genotype M82 without drought stress, SD: from M82 with drought stress, TCK: from the drought-tolerant genotype IL2-5 without drought stress, TD: from IL2-5 with drought stress
Novel miRNAs from tomato
To identify novel miRNAs in the clean reads, miR-PREFeR was used to predict putative novel miRNAs after removing the identified conserved ones. After prediction, a total of 208 novel miRNAs were obtained. There were 119, 114, 113, and 113 novel miRNAs detected in the library of SCK, SD, TCK, and TD, respectively (Table S2). Forty-nine novel miRNAs were common for all the libraries tested (Fig. 3b). Regarding genotype-specific novel miRNAs, each genotype had 49 specific ones (Fig. 3b). There were 18, 15, 11, and 13 novel miRNAs detected specifically in the library of SCK, SD, TCK, and TD, respectively. As expected to be different from conserved miRNAs, most novel miRNAs had a low reads number (Fig. 3b, Table S2). Only two novel miRNAs (sly_miN_545 and sly_miN_945) had more than 1000 reads across the libraries (Table S2). After chromosome mapping analysis, three novel miRNAs (sly_miN_209, sly_miN_211, and sly_miN_213) were found to be located in the introgression region of IL2-5 (Table S2).
Differential expression of conserved and novel miRNAs under drought stress
To identify differentially-expressed miRNA, the expression levels of conserved and novel miRNAs after drought treatment were compared with those before stress. Thirty-two conserved miRNAs, belonging to 16 families, were detected to be differentially expressed (Fig. 4a, Table S1). Among them, 19 were detected in M82, with seven upregulated and 12 downregulated, respectively. There were 23 differentially-expressed miRNAs detected in IL2-5, in which four were induced and 19 were downregulated (Fig. 4a, Table S1). Ten differentially-expressed conserved miRNAs were shared by both genotypes; among them, three (sly-miR156c, sly-miR164a-5p, and sly-miR172a) were induced by drought, while seven (sly-miR156a, sly-miR164b-5p, sly-miR167b-3p, sly-miR168a-3p, sly-miR169e-3p, sly-miR172b, and sly-miR9474-5p) were repressed under stress. A few conserved miRNAs showed opposite expression pattern between the drought-sensitive and drought-tolerant genotype. For instance, sly-miR156d-3p, sly-miR394-3p, and sly-miR477-5p were induced in M82, but they were downregulated in IL2-5 (Fig. 4a, Table S1).
Heatmap showing differentially-expressed miRNAs upon drought stress in tomato. a Conserved miRNAs. b Novel miRNAs. T: drought-tolerant genotype IL2-5, S: drought-sensitive genotype M82, D: drought treatment, CK: non-stress condition. The color scale indicates log2-transformed values of the fold-change
Sixty-eight novel miRNAs were differentially expressed under drought stress (Fig. 4b, Table S2). Among them, 40 were detected in the drought-sensitive M82, with 16 upregulated and 24 downregulated, respectively (Fig. 4b, Table S2). There were 30 differentially-expressed miRNAs detected in IL2-5, in which 16 were induced and 14 were downregulated. Only two (sly_miN_301 and sly_miN_602) differentially-expressed novel miRNAs were shared by both genotypes (Fig. 4b, Table S2). Sly_miN_602 was downregulated in both genotypes after drought stress. However, sly_miN_301 showed a contrary expression pattern in the two genotypes, which was induced in M82 but downregulated in IL2-5.
Target gene prediction and functional classification of the drought-responsive miRNAs
The psRNATarget server was used to predict the target genes of drought-responsive miRNAs. A total of 1936 putative targets were retrieved for the 32 conserved and 68 novel differentially-expressed miRNAs (Table S3). Many of the target genes encode proteins functioning as transcription factors, stress-related proteins, protein kinase, phosphatase, and signal and metabolic pathway-related proteinase. For instance, the targets of sly-miR156a, sly-miR164a-3p, sly-miR166a, sly-miR477-3p, sly_miN_549, and sly_miN_746 encode SPL, bHLH, BZIP, GRAS, WRKY or MYB, whose homologues in other plant species are well-known transcription factors involved in plant development and stress response. The majority targets of the 21 conserved and 32 novel miRNAs encode stress-related proteins and protein kinase.
To further understand the potential roles of miRNAs in tomato drought response, gene ontology (GO) and KEGG pathway analyses were carried out on the 1936 target genes. Among them, 1592 genes have GO annotation and 451 GO terms were enriched (Threshold p value ≤ 0.05), including 295 biological processes, 109 molecular function, and 47 cellular components. Many enriched GO terms were related to plant stress tolerance, such as response to stress (GO:0006950), defense response (GO:0006952), response to stimulus (GO:0050896), phosphorylation (GO:0016310), and cellular lipid metabolic process (GO:0044255). GO terms involved in signaling transduction were also enriched, including signal transduction (GO:0007165), response to ethylene (GO:0009723), regulation of ethylene-activated signaling pathway (GO:0010104), and gibberellin metabolic process (GO:0009685) (Table S4).
A total of 111 KEGG pathways were retrieved for the miRNA target genes. Many of the pathways were related to stress resistance, including plant hormone signal transduction, peroxisome, plant-pathogen interaction, ascorbate and aldarate metabolism, phosphatidylinositol signaling system, cutin, suberine and wax biosynthesis, arginine and proline metabolism, and oxidative phosphorylation (Table S5). It was found that 30 differentially-expressed miRNAs were involved in the regulation of plant hormone signal transduction, including auxin, cytokinine, abscisic acid, ethylene, brassinosteroid, jasmonic acid, and salicylic acid (Fig. 5).
Validation of the expression of miRNAs and their targets
To verify the expression of miRNAs and their targets, nine miRNAs and some of their putative targets were picked up for qRT-PCR validation. The selected miRNAs were sly-miR156d-3p, sly-miR164a-3p, sly-miR166c-5p, sly-miR168a-3p, sly-miR169e-3p, sly-miR171c, sly-miR390a-5p, sly-miR390b-3p, and sly-miR9472-5p. The qRT-PCR results were consistent with those of RNA sequencing (Fig. 6). The expression patterns of the nine target genes picked were also in agreement with expectation, which were opposite to the expression of their corresponding miRNAs. For instance, RNA sequencing results showed that sly-miR166c-5p and sly_miN_429 were significantly downregulated in IL2-5, and their corresponding target gene Solyc03g026340 and Solyc01g008980 showed increased expression after stress (Fig. 7). The expression of sly-miR9474-5p was reduced after drought stress both in M82 and IL2-5, while its target gene, Solyc01g107240, was upregulated significantly in both genotypes after drought stress (Fig. 7). These indicated that the expression profile based on RNA sequencing can reflect the true scenario of tomato miRNAs under drought stress condition.
qRT-PCR validation of miRNA target genes in tomato. S-miRNA: miRNA in drought-sensitive tomato M82, deep sequencing; S-Target: target of miRNA in drought-sensitive tomato M82, qRT-PCR; T-miRNA: miRNA in drought-tolerant tomato IL2-5, deep sequencing; T-Target: target of miRNA in drought-tolerant tomato IL2-5, qRT-PCR; D: drought treatment, CK: non-stress condition
Discussion
Drought is a major abiotic stress which limits crop productivity and quality (Seneviratne 2012). Drought tolerance of plant is a complex trait, which involves miRNAs and many regulatory networks (Shinozaki and Yamaguchi-Shinozaki 2007; Ferdous et al. 2015). As noncoding small RNAs, miRNAs have been found to play important roles in plant growth and development, and stress response (Kidner and Martienssen 2005; Budak et al. 2015; Ferdous et al. 2015; Xie et al. 2015b). Previous studies have demonstrated that miRNAs are involved in the regulation of many stress-related genes, such as those encode dehydrins, late embryo abundant proteins (LEA), glutathione S-transferase (GST), transcription factors (GRAS, ARF and MYB), and hormonal pathway components (Valliyodan and Nguyen 2006; Xie et al. 2015b; Zhang 2015). Recent years, with the booming of the next-generation sequencing, high-throughput sequencing has been widely used to identify conserved and novel miRNAs in plants (Sunkar et al. 2012; Zhuang et al. 2014; Cheah et al. 2015). However, less effort has been made to disclose tomato miRNAs involved in drought response. Therefore, we investigated here the miRNA profiles of two tomato genotypes under drought stress, using RNA sequencing technology.
In our results, 108 conserved miRNAs, belonging to 46 families, were detected in the small RNA libraries constructed. Among them, 98 were detected in all the libraries, whereas only a few conserved miRNAs were specifically expressed. Similar to our previous report (Liu et al. 2017), most of the conserved miRNAs were detected across all the libraries tested. The reads number of different miRNAs varied from a few to millions, in which sly-miR159 accounted for about 30% of the conserved reads. This result is consistent with that obtained from Arabidopsis (Fahlgren et al. 2007). Simultaneously, we predicted novel miRNAs from the small RNA libraries tested; among them, 49 were detected across all the libraries. It was not surprise that most novel miRNAs showed a low level of expression, which agreed with previous reports on tomato (Cao et al. 2014; Liu et al. 2017), Arabidopsis (Rajagopalan et al. 2006), and wheat (Yao et al. 2007).
Thirty-two conserved miRNAs, belonging to 16 families, were differentially expressed upon drought stress, with 19 for M82 and 23 for IL2-5, respectively. Ten differentially-expressed miRNAs were shared by the two genotypes, and a few miRNAs showed opposite expression pattern between the two genotypes. For instance, sly-miR156d-3p, sly-miR394-3p, and sly-miR477-5p were induced in M82 by drought stress, but they were repressed in IL2-5 after drought treatment. Similar results were documented in other plant species, such as Oryza sativa, Triticum aestivum, and Triticum turgidum (Ferdous et al. 2015; Liu et al. 2015, 2016). Candar-Cakir et al. (2016) have investigated the miRNA profiles in tomato under polyethylene glycol (PEG) treatment. miRNAs such as sly-miR396a-3p and sly-miR9474-5p are induced upon PEG treatment, which were in accordance with our results. Meanwhile, some of our results were different from those obtained under PEG treatments. For instance, sly-miR477-3p was induced by PEG treatment (Candar-Cakir et al. 2016), while in our analysis, it was downregulated under drought treatment. In addition, we have detected some extra conserved miRNAs which were differentially expressed under drought stress, including sly-miR166c-5p, sly-miR390a-3p, and sly-miR1919c-5p. We also compared the result from IL2-5 here with that from IL9-1 (Liu et al. 2017). Some drought-responsive conserved miRNAs were detected in both introgression lines, such as sly-miR168b-5p and sly-miR9474-5p. Nevertheless, a number of conserved miRNAs showed different expression pattern in the two studies. Cases were sly-miR156a, sly-miR164a-3p, and sly-miR477-3p, which were induced in IL9-1 under stress (Liu et al. 2017) while downregulated in IL2-5 in this study. In addition, we also detected 68 differentially-expressed novel miRNAs, with 40 and 30 from M82 and IL2-5, respectively. Among them, sly_miN_602 and sly_miN_611 were induced in IL2-5 upon drought treatment, and the result was consistent with that from IL9-1 (Liu et al. 2017). Besides, we also detected that some novel miRNAs were specifically downregulated in IL2-5, such as sly_miN_716 and sly_miN_835. Potential reasons for the differences of results could be (1) different treatment methods (PEG versus natural drought), (2) different seedling ages, (3) different genotypes, and (4) different growth environment. Genetically, the introgression line IL2-5 used in this study carries a segment of chromosome 2 from the drought-tolerant donor parent S. pennellii LA0716, while IL9-1 in our previous study harbors a large fragment of chromosome 9 from LA0716 (Eshed and Zamir 1994). Although both introgression lines are tolerant to drought stress (Gong et al. 2010), the underlying mechanisms could be very different, which can further result in the difference on drought-responsive miRNAs from these two lines (Reinhart et al. 2002; Alseekh et al. 2013; Ferdous et al. 2015). Furthermore, many studies have shown that even in the same plant species, an miRNA can response differently to drought, according to different degrees of drought, different tissue types, and different developmental stages (Reinhart et al. 2002; Wang et al. 2011; Ferdous et al. 2015).
A total of 1936 putative target genes were predicted for the 32 conserved and 68 novel miRNAs that expressed differentially under drought stress. Many of these targets are potentially involved in plant stress tolerance, as they encode transcription factors, stress-related proteinase, protein kinase, phosphatase, and signal transduction components. For instance, miR156 can modulate plant stress tolerance by regulating its target, SPL (Squamosa promoter binding-like protein), which functions in anthocyanin metabolism (Cui et al. 2014). miR156 is involved in various stress response, including drought, salt, and cold stress (Sunkar and Zhu 2004; Liu et al. 2008; Zhou et al. 2008). Kantar et al. (2010) found that miRNA156a in barley leaf is upregulated under drought stress, and this would modulate the plant response to drought by regulating the downstream targets of miRNA156a, the SPL genes. Cui et al. (2014) have found that the miR156c plays dominant roles in plant drought stress response. We also detected that sly-miR156c was induced significantly by drought stress both in M82 and IL2-5, and its targets included SPL gene. This indicated that sly-miR156c plays a conserved role on the drought tolerance of tomato. Besides, many differentially-expressed novel miRNAs may also be enrolled in stress response. For instance, sly_miN_716 was downregulated in the drought-tolerant line, IL2-5, whose targets included annexin. Annexin family protein is also called Ca2+-dependent phospholipid binding protein, which plays important roles in plant stress tolerance and development (Gerke et al. 2005; Mortimer et al. 2008; Laohavisit and Davies 2011). Overexpression of annexin in Arabidopsis improves drought tolerance through alleviating oxidative damage (Konopka-Postupolska et al. 2009). In cotton, overexpression of annexin improves drought tolerance, as it increases the contents of chlorophyll, proline, and soluble sugars, enhances the activity of peroxidase, and reduces the level of membrane lipid oxidation (Zhang et al. 2015). In tomato, annexin is also induced by drought treatment (Lu et al. 2012). These results indicated that sly_miN_716 may serve as an important regulator in tomato drought tolerance.
Further analysis on target genes using gene ontology and KEGG pathway tools enriched many stress-related GO terms and pathways. Among them, 182 target genes were classified into GO category response to stress (GO:0006950), and their upstream miRNAs were sly-miR169e-3p, sly-miR395a, sly_miN_92 and sly_miN_586, etc. (Table S3, Table S4). As compared to our previous investigation on IL9-1 (Liu et al. 2017), GO terms response to stress (GO:0006950) and defense response (GO:0006952) were also enriched in IL2-5 under drought stress. Besides, other GO terms related to stress response were enriched only in IL2-5, including response to external stimulus (GO:0009605) and signal transduction (GO:0007165) (Table S3, Table S4). Seventy-one genes were enriched in GO term response to external stimulus (GO:0009605), and the involved miRNAs included sly-miR156e-5p, sly-miR164a-5p, sly-miR477-5p and sly_miN_272, etc. (Table S3, Table S4). Previous studies have shown that wax, ascorbic acid, and proline play important roles in stress response (Ashraf and Foolad 2007; Gallie 2013; Zhu and Xiong 2013). Similar to our previous study (Liu et al. 2017), many target genes of the drought-responsive miRNAs identified here were involved in drought tolerance-related pathways, such as the pathway of ascorbate and proline metabolism. Besides, we also identified different pathways, typically, the pathway of wax biosynthesis, which has been mentioned to be important to the drought tolerance of IL2-5 (Bolger et al. 2014).
Finally, phytohormones are important regulators for plant development and stress response (Nemhauser et al. 2006; Peleg and Blumwald 2011). GO enrichment and pathway analysis showed that many drought-responsive miRNA targets were involved in hormone pathways, including response to ethylene, regulation of ethylene-activated signaling, gibberellin metabolic process, and plant hormone signal transduction. A total of 30 differentially-expressed miRNAs were involved in plant hormone signal transduction. In Arabidopsis and rice, it has been found that miRNAs control plant hormonal pathways via regulating their target genes, thus to modulate plant drought tolerance (Sunkar and Zhu 2004; Zhao et al. 2007; Wei et al. 2009). In our previous study on IL9-1, drought-responsive microRNAs have been found to be involved in three hormone-related pathways (ABA, CTK and ETH) (Liu et al. 2017); however, in this investigation, genes from seven signal transduction pathways of different hormones were detected to be target of the drought-responsive miRNAs identified from IL2-5 (Fig. 5). ABA acts as a stress hormone, which plays key roles in regulating drought-responsive genes and controlling stomatal movement (Wilkinson and Davies 2002; Nambara and Marion-Poll 2005). In our results, there were nine (Fig. 5) drought-responsive miRNAs involving in ABA signaling pathway. Besides, auxin also functions importantly in plant growth and development, especially when a plant is suffered from drought stress (Bari and Jones 2009; Lawlor 2013). A number of reports have shown that, under drought stress, miRNAs modulate plant growth and development via regulating auxin signaling (Ding et al. 2013; Ferdous et al. 2015). We also detected five auxin signaling-related miRNAs (Fig. 5) that were drought responsive. All these results strongly suggested that many of the drought-responsive miRNAs would modulate tomato drought tolerance through regulating hormonal pathways.
Conclusions
In summary, we have identified 108 conserved and 208 novel miRNAs in tomato using high-throughput RNA sequencing. Among them, there were 32 conserved and 68 novel miRNAs were expressed differentially after drought treatment. GO enrichment and KEGG pathway analysis revealed that many miRNAs and their targets were involved in stress response. Our results indicated that miRNAs play essential roles in tomato drought tolerance. This would also further help us to functionally characterize particular stress-related miRNAs in tomato.
Abbreviations
- miRNA:
-
microRNA
- nt:
-
Nucleotide
- qRT-PCR:
-
Quantitative real-time PCR
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
References
Alptekin B, Langridge P, Budak H (2017) Abiotic stress miRNomes in the Triticeae. Funct Integr Genomics 17:145–170
Alseekh S, Ofner I, Pleban T, Tripodi P, Dato FD, Cammareri M, Mohammad A, Grandillo S, Fernie AR, Zamir D (2013) Resolution by recombination: breaking up Solanum pennellii introgressions. Trends Plant Sci 18:536–538
Ashraf M, Foolad MR (2007) Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ Exp Bot 59:206–216
Bari R, Jones JDG (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69:473–488
Bolger A, Scossa F, Bolger ME et al (2014) The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat Genet 46:1034–1038
Budak H, Akpinar BA (2015) Plant miRNAs: biogenesis, organization and origins. Funct Integr Genomics 15:523–531
Budak H, Kantar M, Bulut R, Akpinar BA (2015) Stress responsive miRNAs and isomiRs in cereals. Plant Sci 235:1–13
Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, Eddy SR, Gardner PP, Bateman A (2013) Rfam 11.0: 10 years of RNA families. Nucleic Acids Res 41:226–232
Candar-Cakir B, Arican E, Zhang B (2016) Small RNA and degradome deep sequencing reveals drought-and tissue-specific micrornas and their important roles in drought-sensitive and drought-tolerant tomato genotypes. Plant Biotechnol J 14:1727–1746
Cao X, Wu Z, Jiang F, Zhou R, Yang Z (2014) Identification of chilling stress-responsive tomato microRNAs and their target genes by high-throughput sequencing and degradome analysis. BMC Genomics 15:1130
Capel C, Fernandez del Carmen A, Alba JM, Lima-Silva V, Hernandez-Gras F, Salinas M, Boronat A, Angosto T, Botella MA, Fernandez-Munoz R, Granell A, Capel J, Lozano R (2015) Wide-genome QTL mapping of fruit quality traits in a tomato RIL population derived from the wild-relative species Solanum pimpinellifolium L. Theor Appl Genet 128:2019–2035
Cheah BH, Nadarajah K, Divate MD, Wickneswari R (2015) Identification of four functionally important microRNA families with contrasting differential expression profiles between drought-tolerant and susceptible rice leaf at vegetative stage. BMC Genomics 16:692
Chuck G, Candela H, Hake S (2009) Big impacts by small RNAs in plant development. Curr Opin Plant Biol 12:81–86
The Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485:635–641
Cui LG, Shan JX, Shi M, Gao JP, Lin HX (2014) The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J 80:1108–1117
Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39:W155–W159
Ding Y, Tao Y, Zhu C (2013) Emerging roles of microRNAs in the mediation of drought stress response in plants. J Exp Bot 64:3077–3086
Eshed Y, Zamir D (1994) A genomic library of Lycopersicon pennellii in L. esculentum: A tool for fine mapping of genes. Euphytica 79:175–179
Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, Carrington JC (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One 2:e219
Ferdous J, Hussain SS, Shi BJ (2015) Role of microRNAs in plant drought tolerance. Plant Biotechnol J 13:293–305
Gallie DR (2013) The role of L-ascorbic acid recycling in responding to environmental stress and in promoting plant growth. J Exp Bot 64:433–443
Gerke V, Creutz CE, Moss SE (2005) Annexins: linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol 6:449–461
Gong P, Zhang J, Li H, Yang C, Zhang C, Zhang X, Khurram Z, Zhang Y, Wang T, Fei Z, Ye Z (2010) Transcriptional profiles of drought-responsive genes in modulating transcription signal transduction, and biochemical pathways in tomato. J Exp Bot 61:3563–3575
Griffiths-Jones S, Saini HK, Van DS, Enright AJ (2008) MiRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–D158
Ilahy R, Piro G, Tlili I, Riahi A, Sihem R, Ouerghi I, Hdider C, Lenucci MS (2016) Fractionate analysis of the phytochemical composition and antioxidant activities in advanced breeding lines of high-lycopene tomatoes. Food Funct 7:574–583
Iovieno P, Punzo P, Guida G, Mistretta C, Van Oosten MJ, Nurcato R, Bostan H, Colantuono C, Costa A, Bagnaresi P, Chiusano ML, Albrizio R, Giorio P, Batelli G, Grillo S (2016) Transcriptomic changes drive physiological responses to progressive drought stress and rehydration in tomato. Front Plant Sci 7:371
Ivits E, Horion S, Fensholt R, Cherlet M (2014) Drought footprint on European ecosystems between 1999 and 2010 assessed by remotely sensed vegetation phenology and productivity. Globe Change Biol 20:581–593
Jeong DH, Park S, Zhai J, Gurazada SG, De Paoli E, Meyers BC, Green PJ (2011) Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23:4185–4207
Jin W, Wu F (2015) Characterization of miRNAs associated with Botrytis cinerea infection of tomato leaves. BMC Plant Biol 15:1
Jin J, Tian F, Yang DC, Meng YQ, Kong L, Luo J, Gao G (2017) PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res 45:D1040–D1045
Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M (2014) Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42:D199–D205
Kantar M, Unver T, Budak H (2010) Regulation of barley miRNAs upon dehydration stress correlated with target gene expression. Funct Integr Genomics 10:493–507
Karlova R, van Haarst JC, Maliepaard C, van de Geest H, Bovy AG, Lammers M, Angenent GC, de Maagd RA (2013) Identification of microRNA targets in tomato fruit development using high-throughput sequencing and degradome analysis. J Exp Bot 64:1863–1878
Kidner CA, Martienssen RA (2005) The developmental role of microRNA in plants. Curr Opin Plant Biol 8:38–44
Klee HJ, Giovannoni JJ (2011) Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet 45:41–59
Konopka-Postupolska D, Clark G, Goch G, Debski J, Floras K, Cantero A, Fijolek B, Roux S, Hennig J (2009) The role of annexin 1 in drought stress in Arabidopsis. Plant Physiol 150:1394–1410
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Laohavisit A, Davies JM (2011) Annexins. New Phytol 189:40–53
Lawlor DW (2013) Genetic engineering to improve plant performance under drought: physiological evaluation of achievements, limitations, and possibilities. J Exp Bot 64:83–108
Lei J, Sun Y (2014) MiR-PREFeR: an accurate, fast and easy-to-use plant miRNA prediction tool using small RNA-Seq data. Bioinformatics 30:2837–2839
Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20:2238–2251
Lin T, Zhu G, Zhang J, Xu X, Yu Q, Zheng Z, Zhang Z, Lun Y, Li S, Wang X (2014) Genomic analyses provide insights into the history of tomato breeding. Nat Genet 46:1220–1226
Liu HH, Tian X, Li YJ, Wu CA, Zheng CC (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14:836–843
Liu H, Searle IR, Watson-Haigh NS, Baumann U, Mather DE, Able AJ, Able JA (2015) Genome-wide identification of microRNAs in leaves and the developing head of four durum genotypes during water deficit stress. PLoS One 10:e0142799
Liu H, Able AJ, Able JA (2016) SMARTER de-stressed cereal breeding. Trends Plant Sci 21:909–925
Liu M, Yu H, Zhao G, Huang Q, Lu Y, Ouyang B (2017) Profiling of drought-responsive microRNA and mRNA in tomato using high-throughput sequencing. BMC Genomics 18:481
Lu Y, Ouyang B, Zhang J, Wang T, Lu C, Han Q, Zhao S, Ye Z, Li H (2012) Genomic organization, phylogenetic comparison and expression profiles of annexin gene family in tomato (Solanum lycopersicum). Gene 499:14–24
Mortimer JC, Laohavisit A, Macpherson N, Webb A, Brownlee C, Battey NH, Davies JM (2008) Annexins: multifunctional components of growth and adaptation. J Exp Bot 59:533–544
Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56:165–185
Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126:467–475
Peleg Z, Blumwald E (2011) Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol 14:290–295
Pradhan B, Naqvi AR, Saraf S, Mukherjee SK, Dey N (2015) Prediction and characterization of Tomato leaf curl New Delhi virus (ToLCNDV) responsive novel microRNAs in Solanum lycopersicum. Virus Res 195:183–195
Rajagopalan R, Vaucheret H, Trejo J, Bartel DP (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20:3407–3425
Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16:1616–1626
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108
Seneviratne SI (2012) Climate science: historical drought trends revisited. Nature 491:338–339
Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58:221–227
Shirasawa K, Isobe S, Hirakawa H, Asamizu E, Fukuoka H, Just D, Rothan C, Sasamoto S, Fujishiro T, Kishida Y, Kohara M, Tsuruoka H, Wada T, Nakamura Y, Sato S, Tabata S (2010) SNP discovery and linkage map construction in cultivated tomato. DNA Res 17:381–391
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019
Sunkar R, Li YF, Jagadeeswaran G (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17:196–203
Valliyodan B, Nguyen HT (2006) Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr Opin Plant Biol 9:189–195
Wang T, Chen L, Zhao M, Tian Q, Zhang WH (2011) Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genomics 12:367
Wei L, Zhang D, Xiang F, Zhang Z (2009) Differentially expressed miRNAs potentially involved in the regulation of defense mechanism to drought stress in maize seedlings. Int J Plant Sci 170:979–989
Wilkinson S, Davies WJ (2002) ABA-based chemical signalling: the co-ordination of responses to stress in plants. Plant Cell Environ 25:195–210
Xie F, Wang Q, Sun R, Zhang B (2015a) Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton. J Exp Bot 66:789–804
Xie M, Zhang S, Yu B (2015b) MicroRNA biogenesis, degradation and activity in plants. Cell Mol Life Sci 72:87–99
Xu J, Yuan Y, Xu Y, Zhang G, Guo X, Wu F, Wang Q, Rong T, Pan G, Cao M, Tang Q, Gao S, Liu Y, Wang J, Lan H, Lu Y (2014) Identification of candidate genes for drought tolerance by whole-genome resequencing in maize. BMC Plant Biol 14:83
Yao Y, Guo G, Ni Z, Sunkar R, Du J, Zhu JK, Sun Q (2007) Cloning and characterization of microRNAs from wheat (Triticum aestivum L.) Genome Biol 8:R96
Zhang B (2015) MicroRNA: a new target for improving plant tolerance to abiotic stress. J Exp Bot 66:1749–1761
Zhang F, Li S, Yang S, Wang L, Guo W (2015) Overexpression of a cotton annexin gene, GhAnn1, enhances drought and salt stress tolerance in transgenic cotton. Plant Mol Biol 87:47–67
Zhao B, Liang R, Ge L, Li W, Xiao H, Lin H, Ruan K, Jin Y (2007) Identification of drought-induced microRNAs in rice. Biochem Biophys Res Commun 354:585–590
Zhou X, Wang G, Sutoh K, Zhu JK, Zhang W (2008) Identification of cold-inducible microRNAs in plants by transcriptome analysis. Biochim Biophys Acta 1779:780–788
Zhou L, Liu Y, Liu Z, Kong D, Duan M, Luo L (2010) Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot 61:4157–4168
Zhu X, Xiong L (2013) Putative megaenzyme DWA1 plays essential roles in drought resistance by regulating stress-induced wax deposition in rice. PNAS 110:17790–17795
Zhuang J, Zhang J, Hou XL, Wang F, Xiong AS (2014) Transcriptomic, proteomic, metabolomic and functional genomic approaches for the study of abiotic stress in vegetable crops. Crit Rev Plant Sci 33:225–237
Funding
This work was supported by the National Natural Science Foundation of China (U1503186, 31572133) and the Applied Basic Research Program (2016020101010092) of Science and Technology Bureau of Wuhan City, Hubei, China.
Author information
Authors and Affiliations
Contributions
Bo Ouyang and Minmin Liu designed the experiments and wrote the manuscript. Minmin Liu and Huiyang Yu analyzed the data. Minmin Liu, Gangjun Zhao, Qiufeng Huang, and Yongen Lu carried out the experiments. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Supplementary Table S1
- Conserved miRNAs expressed in the drought-sensitive and -tolerant tomato genotypes. (XLSX 27 kb)
Supplementary Table S2
- Novel miRNAs expressed in drought-sensitive and -tolerant tomato genotypes. (XLSX 37 kb)
Supplementary Table S3
- Target gene prediction results of differentially-expressed miRNAs. (XLSX 456 kb)
Supplementary Table S4
- Enriched GO terms of drought-responsive miRNA targets in tomato. (XLSX 90 kb)
Supplementary Table S5
- KEGG pathway terms of drought-responsive miRNA targets in tomato. (XLSX 15 kb)
Supplementary Table S6
- The primers of miRNAs and target genes used for qRT-PCR verification. (XLSX 9 kb)
Rights and permissions
About this article
Cite this article
Liu, M., Yu, H., Zhao, G. et al. Identification of drought-responsive microRNAs in tomato using high-throughput sequencing. Funct Integr Genomics 18, 67–78 (2018). https://doi.org/10.1007/s10142-017-0575-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-017-0575-7