Abstract
Global warming is a major threat for agriculture and food security, and in many cases the negative impacts are already apparent. Wheat is one of the most important staple food crops and is highly sensitive to the heat stress (HS) during reproductive and grain-filling stages. Here, whole transcriptome analysis of thermotolerant wheat cv. HD2985 was carried out at the post-anthesis stage under control (22 ± 3 °C) and HS-treated (42 °C, 2 h) conditions using Illumina Hiseq and Roche GS-FLX 454 platforms. We assembled ~24 million (control) and ~23 million (HS-treated) high-quality trimmed reads using different assemblers with optimal parameters. De novo assembly yielded 52,567 (control) and 59,658 (HS-treated) unigenes. We observed 785 transcripts to be upregulated and 431 transcripts to be downregulated under HS; 78 transcripts showed >10-fold upregulation such as HSPs, metabolic pathway-related genes, etc. Maximum number of upregulated genes was observed to be associated with processes such as HS-response, protein-folding, oxidation-reduction and photosynthesis. We identified 2008 and 2483 simple sequence repeats (SSRs) markers from control and HS-treated samples; 243 SSRs were observed to be overlying on stress-associated genes. Polymorphic study validated four SSRs to be heat-responsive in nature. Expression analysis of identified differentially expressed transcripts (DETs) showed very high fold increase in the expression of catalytic chaperones (HSP26, HSP17, and Rca) in contrasting wheat cvs. HD2985 and HD2329 under HS. We observed positive correlation between RNA-seq and qRT-PCR expression data. The present study culminated in greater understanding of the heat-response of tolerant genotype and has provided good candidate genes for the marker development and screening of wheat germplasm for thermotolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heat stress is one of the major yield limiting factors in the crops’ growth and development. High temperature has been reported to have adverse effects on the quality and quantity of grains (Chen et al. 2006; Hatfield and Prueger 2015). Information is, however, very scarce on the thermotolerance mechanism in wheat, as compared with Arabidopsis and rice (Bokszczanin et al. 2013; Matsui et al. 2008). Wheat (Triticum aestivum L.) is one of the staple food grain crops in major parts of the world, providing the required carbohydrates and proteins in the diet (Kumar and Rai 2014; Budak et al. 2013a, b). Due to the global climate change, increase in the temperature has become one of the major factors adversely affecting the quantity and quality of wheat grains (Kumar et al. 2013a). Heat stress especially during the critical stage of wheat growth and development causes significant reduction in the individual kernel weight and number (Hays et al. 2007; Kumar et al. 2013b).
Higher plants exposed to heat stress exhibit a characteristic set of cellular and metabolic responses like decrease in the synthesis of normal proteins and an accelerated transcription and translation of stress-associated genes (SAGs) (Goswami et al. 2014; Kumar et al. 2013b; Wahid et al. 2007). High temperature also causes a reversible effect on the rate of photosynthesis (Kumar and Rai 2014). Budak et al. (2013a, b) reported several differentially expressed proteins associated with drought stress tolerance in wild emmer and modern durum wheat through the proteome approach. Besides altering the pattern of gene expression, HS also damage the cellular structure of organelles and the cytoskeleton, and impair the membrane function (Bita and Gerats 2013). Studies have demonstrated that cellular and metabolic changes are, in fact, required by the plants to survive high temperatures (Hasanuzzaman et al. 2013). Although the research on abiotic stress tolerance has advanced considerably in recent years, very limited work has been executed for characterizing the genes responsive to acclimation to high temperature in wheat, as evident from the literature available on public databases.
Several techniques like array chip, massively parallel sequencing, microarray analysis, etc., have recently been used to catalogue the effects of HS on the expression of thousands of genes in the model species (Martin and Wang 2011; Ergen and Budak 2009). Transcriptome sequencing using next-generation sequencing (NGS) technology is a powerful tool for studying global transcriptional networks and provides high-resolution data (Mutz et al. 2013). Sequence-based bioinformatic characterization reveals the expression profiles of stress-responsive genes (SAGs) and provides the functional annotation of genes associated with complex traits in different crops (Priest et al. 2014, Budak et al. 2015). Zhao et al. (2011) optimized the de novo transcriptome assembly from short-read RNA-seq data. Transcriptome profiles and digital expression of genes in the model species is generally performed by mapping reads from the NGS analysis to a reference genome sequence and further annotation of the genes using different databases (Brautigam and Gowik 2010). The strategies deployed for the model species are not feasible in wheat, as its reference genome sequence and gene annotation are still incomplete; an international project to achieve these goals is, however, under progress (IWGSC: International Wheat Genome Sequencing Consortium; http://www.wheatgenome. org/). This project may take considerable time, because of the difficulties involved in sequencing the huge (40 times larger than rice), highly-repetitive hexaploid genome of wheat (Chapman et al. 2015). Recently, the first draft of the wheat genome has, however, been reported (IWGSC 2014). The draft genome sequence is based on Illumina HiSeq sequencing and covers about 60% of the genome (Dong et al. 2015). It provides very comprehensive information about the genes, their evolution and expression. The highly fragmented nature of the sequence assemblies limits its use as reference sequence in transcriptome experiment. Next-generation sequencing technologies and de novo assembly of short sequences of transcripts enable researchers to reconstruct the sequences of the entire transcriptome, identify the expressed genes, separate isoforms and capture the transcript expression levels (Zhao et al. 2011).
The present investigation aimed at gaining greater and comprehensive insights into the novel heat-responsive genes and molecular markers in wheat using the de novo transcriptomic approach and their potential role in tolerance of photosynthetic carbon metabolism processes under terminal heat.
Material and methods
Sowing and heat stress treatment
We used HD2985 (thermotolerant)—a popular wheat cv. (T. aestivum L.)—for the present investigation. Pre-treated seeds with Bavistin (0.5%) were sown in 18 pots (having equal quantity of perlite to farmyard manure mixture) inside the regulated chamber (22 ± 3 °C, relative humidity of 75% and 8 h light with intensity of 250 μmol m−1 s−1 photosynthetic active radiation) in the National Phytotron Facility at Indian Agricultural Research Institute, New Delhi. Irrigation was done at regular intervals, and plants at the pollination, milky-ripe and mealy-ripe stages (3 pots each) were exposed to HS (42 °C for 2 h), whereas the other three pots served as control (22 ± 3 °C) (ESM_1.TIFF). The HS was given in a sinusoidal mode using a microprocessor-regulated controller. Samples (flag leaf, stem and immature endospermic tissues) collected in duplicates from control and HS-treated wheat cv. HD2985 during the milky-ripe sub-stage of grain-filling (based on the Feekes scale) were frozen in liquid nitrogen for further de novo sequencing. Validation of identified transcripts was carried out in seedlings of wheat cvs. HD2985 and HD2329 (15 pots were used for each cultivar) kept under control (22 ± 3 °C) and HS-treated conditions (42 °C for 0.5, 1, 2 and 4 h). Flow-chart of the work plan has been depicted in the doc file (online resource 2; ESM_2.doc).
Photosynthetic measurement through infra-red gas analyser
The wheat cv. HD2985 was exposed to HS (42 °C, 2 h) during pollination, milky-ripe (Feekes scale—11.1) and mealy-ripe (Feekes scale - 11.2) sub-stages of grain-filling (critical stages), and further fully expanded leaf was subjected to infra-red gas analyser (IRGA) (LCA-4, ADC, Hoddesdon, UK) for analysing the effect of HS on the performance of photosynthetic apparatus as suggested by Long and Bernacchi (2003). Photosynthetic rate, transpiration rate and stomatal conductance were measured by taking readings in triplicates between 11 am to 12 pm. Visual observations of leaf disc collected from the control and HS-treated plants were also scanned for analysing the pigment disintegration.
Whole transcriptome analysis of wheat
RNA isolation and sequencing
Two biological replicates collected from the control (22 ± 3 °C) and HS-treated (42 °C, 2 h) wheat cv. HD2985 during the milky-ripe stage were used for the de novo transcriptome sequencing. Total RNA was isolated separately from the control and HS-treated tissues (100 mg of each tissue of flag leaf, stem and immature endosperm) using the Trizol method (Invitrogen, UK). The separately isolated total RNA was re-purified with the RNeasy kit (Qiagen, Valencia, CA, USA) following the manufacturer’s protocol. The quality of RNA was analysed using Bioanalyzer (Agilent, UK) as well as electrophoretically on 1% denaturing agarose gel. Pooled total RNAs in equimolar concentration (from different tissues) were used as one sample (~60 μg) for the cDNA library construction (as mentioned in Kumar et al. 2015). The paired-end cDNA sequencing libraries were prepared for both the control and HS-treated RNA (in duplicates) using Illumina TruSeq RNA Library Preparation Kit (Illumina, San Diego, CA, USA) as per the protocol mentioned in our earlier publication (Kumar et al. 2015). DNA of each cluster was sequenced on the HiSeq 2000 platform (Illumina, San Diego, CA, USA) using TruSeq SBS v3-HS kit. For the Roche GS-FLX 454 platform, cDNA synthesis was performed using SMART II™ cDNA Synthesis kit (Clontech Laboratories, Inc. Mountain View, CA, USA), and further protocol was followed as described by Kumar et al. (2015).
Preprocessing of raw reads
Raw reads generated from the control and HS-treated wheat samples using Illumina HiSeq and Roche GS-FLX 454 were processed using the Newbler program v 2.5 to remove the adaptor contamination, and low-quality reads (reads mean Q-value 20). Low-quality (<Q20) 3′ end of reads were trimmed off by custom PERL script. The trimmed reads were evaluated with FastQC, a Java-based, quality control tool for high-throughput sequence data, and normalized with Trinity’s in silico read normalization (http://Trinityrnaseq.sourceforge.net/), with maximum coverage of 30 and k-mer size of 25.
De novo transcriptome assemblies
Wheat, being allohexaploid, creates a real challenge for any de novo transcriptome assemblies because of the presence of duplicated genes, homologues gene copies, etc., causing partially assembled contigs. Similarly, with given specific parameters, it will be difficult for any assembler to reassemble all the transcripts from the wheat transcriptome. In order to maximize the diversity and completeness of the identified contigs, four de novo transcriptome assemblers were used in the present investigation (reported to be the best de novo assemblers for the Roche 454 and Illumina Hiseq transcriptomic reads; Mundry et al. 2012)—(Cap3 (http://seq.cs.iastate.edu/cap3.html), MIRA (version 3.2.1; http://sourceforge.net/projects/miraassembler/), GS De novo Assembler (Newbler v 2.5.3; http://www.454.com/products-solutions/analysis-tools/gs-de-novoassembler.asp) and Velvet-Oases assembly program v 0.2.04 (Zerbino and Birney 2008)). CAP3 detects the overlap regions, generate consensus sequences and produces fewer errors than the other assemblers (Chopra et al. 2014). MIRA correct the ambiguous regions of an assembly. Velvet is a graph based short-read assembler which resolves the ambiguities caused by the sequencing errors. It uses paired-end information to remove repeats. Newbler 2.5 gave longer contig, better alignments to reference sequences and is fast and easy to use. The reads are more readily merged into a longer contig with less redundancy. Oases produced the overall shortest assembly. CAP3 was used to assemble k-mer sizes of 48 to 86 with step size of 2, MIRA was used to assemble a k-mer size of 31, GS De novo assembler was used to assemble a k-mer size of 20 to 44 with a step size of 4 and velvet-Oases was assembled with K-mer sizes of 25 to 75 with step size of 10. K-mer assemblies from each assembler were merged using the native merge utilities in each software package. Contigs shorter than 200 bp were discarded from all assemblies.
Reducing redundancy of assemblies
To generate a non-redundant set of reference transcripts (Unigenes, Plant Unique Transcripts), we merged the ‘raw’ assemblies using CD-HIT-EST / CD-HIT-454 (http://weizhongli-lab.org/cd-hit/) with 100% identity to remove identical fragments. Contigs that overlapped with minimum length of 50 bp and minimum identity of 99% were collapsed into a single contig using the TGI clustering tool (Pertea et al. 2003) (http://www.tigr.org/tdb/tgi/software/) to further remove redundancy. The options used for the TGI clustering was “-p 99 –l 50 –v 100”. CLC Genomics workbench (CLC bio, Aarhus, Denmark) was used for mapping the reads. For the data set used here, we selected the heaviest penalty for mismatch, insertion and deletion costs which is 3. Length fraction was set to 0.5 and similarity was set to 0.95. We set the minimum contig length to 200 nt.
Sequence annotation
Assessment and annotation of unigenes were performed by sequence similarity on five different databases, i.e. NCBI non-redundant (http://www.ncbi.nlm.nih.gov/refseq/), UniProtKB/Swiss-Prot (http://www.ebi.ac.uk/uniprot), Triticeae full-length CDS database ((TriFLDB)—http://trifldb.psc.riken.jp/v3/index.pl), PlantCyc (http://www.plantcyc.org/) and Pfam (http://pfam.xfam.org/) using Basic Local Alignment Search Tool ((BLAST)—http://blast.ncbi.nlm.nih.gov/Blast.cgi) with E values of 1e-5 unless otherwise stated. Gene function was assigned to each unigene based on the best BLAST hit (lowest E-value). To increase the computational speed, such search was limited to the first 10 significant hits for each query.
Calculation of RPKM value and clustering of DETs
The transcript level was quantified in terms of Reads Per Kilobase of Transcript Per Million Reads Mapped (RPKM) as mentioned in Kumar et al. (2015). Digital gene expression profiles of all unique transcripts mapped onto the unigenes were quantified using the edgeR package (Robinson et al. 2010). Differential expression (DE) was detected using the edgeR Bioconductor package with log2 fold change threshold of 2 and FDR value cut-off <0.01.
Gene Ontology and pathway analysis
To annotate the assembled sequences with Gene Ontology (GO) terms describing biological processes, molecular functions and cellular components, the NCBI-nr BLAST result was imported into Blast2GO suite, a software package (http://www.blast2go.com/b2ghome) that retrieves GO terms, allowing gene functions to be determined and compared (Gotz et al. 2008). The unigenes were also aligned to the Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway database using the online KEGG Automatic Annotation Server (KAAS) (http://www.genome.jp/kegg/kaas/). The bi-directional best hit (BBH) method (http://www.nmpdr.org/) was used to obtain KEGG Orthology (KO) system (http://www.genome.jp/kegg/ko.html).
Expression analysis at transcript and protein level under HS
Validation of expression of transcripts through quantitative real-time PCR (qRT-PCR)
For the validation of identified DETs, transcript-specific forward and reverse primers were designed using the Genefisher2 primer designing software (http://bibiserv.techfak.uni-bielefeld.de/genefisher2/). The list of primers used for the comparative expression analysis has been presented as Online Resource 3 (ESM_3.doc).
Total RNA were isolated from the control (22 ± 3 °C) and HS-treated (42 °C, 2 h) wheat samples (three biological and three technical replicates) using the Trizol method (Invitrogen, UK). The quality of the isolated total RNA was checked on Bio-analyser (Agilent, USA). Two prominent peaks were observed in all the samples with OD 260/280 ratio of >2.0. First strand cDNA synthesis was carried out using RevertAid™ H minus First Strand cDNA synthesis kit (Thermo Fisher Scientific, USA). The cDNA products were diluted 50-fold with deionized water before using it as a template in qRT-PCR. Quantitative real-time PCR was performed using the CFX96 platform (Bio-Rad, UK) as per the protocol mentioned in our earlier publication (Kumar et al. 2015). All the expression levels were normalized to the arithmetic mean of the selected β-actin gene (accession no AB181991.1). The relative expression was calculated using the comparative Ct method (Pfaffl 2001).
Validation of protein accumulation pattern under HS through immunoblot assay
The most abundant protein involved in carbon metabolism processes, i.e. RuBisCo, was used for characterizing the accumulation pattern under HS. The leaves of control (22 ± 3 °C) and HS-treated (42 °C, 2 h) wheat cvs. HD2985 and HD2329 collected during the pollination, milky-ripe and mealy-ripe stages (based on the Feekes scale) were subjected to Western blot analysis using the protocol as mentioned in our earlier publication (Kumar et al. 2013a). The dilution used for the incubation of membrane in primary monoclonal antibody (Anti-RuBisCo plant antibody produced in chicken, Sigma-Aldrich, USA) and secondary antibody (peroxidase-conjugated rabbit anti-chicken IgG; Sigma-Aldrich, USA) was 1:2000. Band of desired intensity was visible after incubation (2–5 min), and the membrane was immediately washed in water, followed by Tris-buffered saline (TBS). The dried membrane was used for the photography using Gel Doc Easy (Bio-Rad, UK).
Validation of biological activity of protein under HS through 14C labelling method
In order to characterize the effect of HS on the activity of enzyme, we selected one of the most important enzyme of the carbon assimilatory pathway, i.e. RuBisCo, for the activity assay. One gramme of leaves collected from control (22 ± 3 °C) and HS-treated (42 °C, 2 h) wheat cv. HD2985 during pollination, milky-ripe and mealy-ripe stages were crushed into fine powder using liquid nitrogen in 5 mL extraction buffer (50 mM Tris-HCl (pH 8.0), 1 mM ethylene diaminetetraacetic acid (EDTA), 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.1% BSA). The extract was centrifuged at 9000×g for 15 min and further supernatant was separated into fresh tubes; 1 mL supernatant was activated by incubating at room temperature (RT) with activation buffer (0.1 mL each of 10 mM sodium bicarbonate (NaHCO3), 10 mM MgCl2, 5 mM glutathione, 0.1 mM EDTA and 0.6 mL Tris-HCl (50 mM) pH 8.0). Reaction mixture was prepared by adding 0.2 mL Tris-HCl (30 μmol; pH 8.0), 0.1 mL labelled NaH14CO3 and 0.1 mL unlabeled NaHCO3, 0.01 mL MgCl2 (3.03 μmol), 0.01 mL EDTA, 0.1 mL activated enzyme extract and 0.1 mL ribulose-1,5-bisphosphate (RuBP; 0.2 μmol). Assay medium without RuBP served as blank for each assay. The reaction was started by the addition of RuBP and further terminated after 120 min with the addition of 0.2 mL acetic acid (6 N). The content of the vials were evaporated to dryness at 65 °C, and the acid stable 14C was subsequently counted in liquid scintillation counter (Packard, Tri-Carb, 1600 TR USA). The calculation was carried out as mentioned in our earlier publication (Kumar et al. 2016).
Development of expressed sequence tags—simple sequence repeat (EST-SSR) markers and their validation
The high-quality reads obtained in the present investigation were used for the SSRs identification using the MIcroSAtellite identification tool (MISA; http://pgrc.ipk-gatersleben.de/misa/misa.html). SSR identification was carried out using the self-developed pipeline based on the following criteria as reported by Haq et al. (2014)—a dinucleotide pattern should appear at least six times, trinucleotide pattern should appear at least five times, tetranucleotide pattern should appear at least five times, pentanucleotide pattern should appear at least five times and hexanucleotide pattern should appear at least five times.
We selected 243 SSRs localised on the heat-responsive DETs for the validation. Genomic DNA was isolated from the leaves of 30 genotypes of wheat (selected from the mini-core subset developed for the thermotolerance at the Indian Agricultural Research Institute, New Delhi) by the cetyl trimethylammonium bromide (CTAB) method. The isolated genomic DNA was quantified by UV-260 absorbance and adjusted to a final concentration of 30 ng/μL. All the 243 genic-SSR loci with SSR length of 18 bp or longer were first tested for the amplification in order to optimize the annealing temperature. The list of SSR primers designed for the validation has been included as Online Resource 4 (ESM_4.pdf). The PCR reactions were performed using Bio-Rad Gradient Cycler. Each PCR reaction consisted of 2 μL of 10× reaction buffer, 0.20 μL of 10 mM dNTPs, 1.5 μL of 10 μM each of forward and reverse primers, 2 μL of template genomic DNA and 0.2 μL of Taq DNA polymerase in a final reaction volume of 20 μL. The PCR reaction profile was DNA denaturation at 95 °C for 5 min followed by 35 cycles of 94 °C for 1 min, 55 °C for 1 min, 72 °C for 1 min and finally 72 °C for a final extension of 10 min. The optimized SSR primers were then used for PCR amplification in 30 lines of wheat. The PCR products were separated by electrophoresis in 4% Metaphor agarose gels (Lonza, Rockland ME, USA) containing 0.1 μg/mL ethidium bromide in 1× TBE buffer at 80 V for 1 h. After electrophoresis, PCR products were visualized and photographed using Gel Doc Easy (Bio-Rad, UK). The genotype profiles of the different wheat varieties produced by SSR markers were scored manually. Each allele of the concerned marker was scored as present (1) or absent (0) for each of the SSR loci. A total of 243 SSR markers giving consistent expected size products were used for the genotyping of 30 diverse wheat germplasm; three highly polymorphic loci of these SSRs were used for the diversity analysis. Markers that produced expected size of amplicon (100–200 bp) was scored for variation in amplicon size and the Polymorphism Information Content (PIC) was calculated using the formula-
where P i is the frequency of the i th allele in the set of genotypes analysed, calculated for each SSR locus. The genetic similarity between any two genotypes was estimated based on Jaccard’s similarity coefficient. All the 30 genotypes were clustered with the unweighted pair group method with arithmetic mean (UPGMA) analysis and SAHN procedure of the NTSYS-PC v2.10 t.
Results
Effect of HS on photosynthetic parameters of wheat under HS
Heat stress adversely affects the carbon assimilatory processes in crop plants. The synthesis, transportation and accumulation of photosynthates are altered in plants under elevated temperature. Here, we analysed the photosynthesis-related parameters of fully expanded leaves of wheat cv. HD2985 under control (22 ± 3 °C) and HS-treated (42 °C, 2 h) conditions at different stages of growth (pollination, milky-ripe and mealy-ripe). Differential behaviour of wheat cv. HD2985 was observed under HS at pollination and grain-filling stages (milky-ripe and mealy-ripe sub-stages; Fig. 1). A significant decrease in the photosynthetic rate, conductance and transpiration rate was observed during pollination, milky-ripe and mealy-ripe stages under HS, as compared with control; decrease in the photosynthetic rate was observed more pronounced during mealy-ripe, as compared with other stages of growth. Similarly, decrease in the conductance was observed in HS-treated samples compared to control at different stages of growth. WUE was observed maximum in HS-treated HD2985 during the pollination stage. A non-significant difference in the WUE was observed in the leaves of HS-treated samples collected during pollination and mealy-ripe stages.
Infra-red gas analyser (IRGA) analysis of wheat cv. HD2985 during pollination and grain-filling stages. Fully expanded leaves of control (22 ± 3 °C) and HS-treated (42 °C, 2 h) plants were selected for the reading in triplicates; plants were exposed to HS in a microprocessor-regulated closed chamber
The leaf discs collected from the flag leaves used for the IRGA analysis showed significant degradation of chlorophyll as shown in Online Resource 5 (ESM_5.TIFF). The percent yellow zone was observed maximum in the leaves exposed to HS during mealy-ripe, as compared with milky-ripe sub-stages of grain-filling.
Transcriptome sequencing and read assembly
Sequencing of control and HS-treated libraries generated a total of 59.3 million raw reads. Data filtering and quality assessment generated 47.9 million high-quality reads (control—24.8 and HS-treated—23.1). The assembly of high-quality reads yielded 52,567 (control) and 59,658 (HS-treated) contigs. The N50 value observed was 585 bp (control) and 632 bp (HS-treated). The merged contigs were subjected to cluster and assembly analysis. A total of 23,470 unigenes were observed with 4706 genes (20.0%) showing size greater than 1 kb and 16,000 unigenes (68.3%) having length greater than 500 bp (ESM_6.doc, Fig. 2). The raw data sets were deposited in the Short Read Archive (SRA) repository of NCBI under the accession number PRJNA171754.
Distribution of identified unigenes length generated by different assemblers. High-quality reads generated from the control and heat shock-treated samples of HD2985 cultivar of wheat was used; Illumina HiSeq 2000 and Roche GS-FLX 454 platform were used for the sequencing; x-axis—contig length, y-axis—number of transcript contigs
Functional annotation and classification of wheat transcriptome
Five different databases were used (NCBI non-redundant, UniProt: Swiss-Prot, TriFLDB, PlantCyc and Pfam) to maximize the annotation percentages. Of 23,470 unigenes, 16,978 (72.3%) were annotated by the NCBI-nr database. We annotated 10,320 (43.9%) unigenes using Uniprot BLAST analysis (Altschul et al. 1990). Some of the unigenes which were identified to be associated with HS-tolerance using NCBI non-redundant database are—predicted calcineurin subunit B-like (CL90Contig1), ascorbate peroxidase (CL138Contig2), HSP70 (CL152Contig1), HSP101 (CL157Contig1), etc. Most of the unigenes identified were functionally annotated to be predicted/hypothetical protein (ESM_7.xls). Similarly, unigenes identified using the TriFLDB were annotated to play an important role in thermotolerance such as stress-associated protein (CL34Contig2), cytosolic malate dehydrogenase (CL32Contig2), TaHSP70d (CL38Contig1), NAC domain transcription factor (CL64Contig2), etc. Annotations of the unigenes using other databases are provided as Online Resources 8, 9 and 10 (ESM_8.xls, ESM_9.xls and ESM_10.xls).
Prediction of enzyme coding transcripts was performed for the unigenes based on sequence similarity with the ENZYME database (http://www.brenda-enzymes.info/). We identified 1259 unigenes to have at least one enzyme hit. The Pfam database was mined for the identification of domains in unigenes using Reverse Position Specific Blast (RPSBlast). Eight thousand two hundred seventy-three8 sequences were observed to have at least one domain with domain coverage >50%. The quality of unigenes were assessed by comparing the transcript sequences to TriFLDB (wheat fl cDNA) and PlantCyc (Plant metabolic pathway database) using BLAST (E value <1e-5). Out of 23,470 unigenes, 12,992 (55.3%) and 8480 (36.1%) were aligned to the full-length wheat cDNAs and plant metabolic pathway genes. Unigenes identified from wheat cv. HD2985 were subjected to Gene Ontology (GO) analysis for their categorization under biological or molecular functions. Under biological processes, most of the unigenes were associated with cellular processes followed by metabolic and response to stimulus. Similarly, under molecular function, most of the identified unigenes were involved in binding followed by catalytic and transporter activities.
Identification of differentially expressed transcripts (DETs) in wheat under heat stress
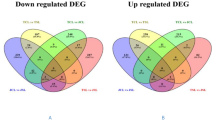
Uniquely mapped reads were used in the expression analysis of each sample using edgeR statistical package to calculate the RPKM value (Mortazavi et al. 2008), which indicates the relative transcript amount. Average RPKM values from samples were 43.9 and 44.6. The maximum RPKM values reported for the control and HS-treated samples were 76,080.48 and 24,031.95, respectively. The RPKM values of 1140 and 1559 unigenes were observed to be greater than 100 for control and HS-treated samples, suggesting higher number of modulated genes under the HS. DETs were detected with log fold value threshold 2 and false discovery rate (FDR) value cut-off <0.01 (ESM_11.xls). We observed 785 upregulated transcripts and 431 downregulated transcripts in the HS-treated sample (ESM_11.xls); 78 transcripts showed more than 10-fold upregulation such as HSPs—heat shock protein (HSP26), 101 kDa heat shock protein (HSP101), heat shock cognate 70 kDa protein (HSC70), small heat shock protein (HSP17.8) and other metabolic pathways-related genes (Table 1). Similarly, 109 transcripts showed less than −fivefold downregulation under HS like 3-oxoacyl-(acyl-carrier-protein) reductase, phosphoglucomutase-like, MADS-box transcription factor 1, kDa proline-rich protein, sucrose transporter, etc. (Table 2).
The significant alteration in the expression level of catalytic chaperons like HSPs and its regulator HSFs in response to HS showed the potential role of these DETs in thermotolerance. Downregulation of some of the transcripts under HS makes us to conclude that pathways like lipid metabolism, secondary metabolite synthesis, distribution of photosynthates, floral growth and differentiation, etc., are most affected because of HS. The variations in the expression of candidate genes in the HS-treated sample, as compared to control, are shown in the heat map with different colours which depicts the relative expression of the identified candidate genes (Fig. 3). Significant upregulation and downregulation were observed in the expression of identified transcripts in response to HS.
Functional classification
The modulated genes with hits to the GenBank nr database were grouped into 251 functional categories like ATP binding, stress-related, heat shock protein binding, photosynthesis, etc. (Fig. 4). The most altered biological processes observed was HS-response followed by protein folding, oxidation-reduction process, heat acclimation, phytohormone response and electron transport chain. The largest group of genes with known function altered due to HS was metabolism-related genes. Stress-related, chloroplast-associated and photosynthesis-related genes were observed to play an important role in plant under HS condition. Data mining in the present investigation identified a large number of transcripts showing homology with heat shock transcription factors (HSFs) and heat shock proteins (HSPs) (Table 3). Similarly, genes associated with molecular functions like ATP binding, protein unfolding, Zn-ion binding, etc., was observed to be most affected by elevation in temperature (Fig. 4).
Distribution of upregulated and downregulated transcripts based on Gene Ontology. Upregulated and downregulated transcripts were grouped based on their biological and molecular functions and involvement in different pathways using the GO database (http://www.geneontology.org/)
To comprehensively assess the biological functions of the DETs, we mapped the modulated transcripts sequences to the KEGG database using BLAST. Pathways such as metabolic and biosynthesis of secondary metabolites were highly influenced by HS (Fig. 4). Metabolic pathway characterization showed 33 transcripts to be upregulated and 14 to be downregulated under the HS. Similarly, 16 transcripts associated with the secondary metabolite biosynthesis pathway were observed to be upregulated and 10 to be downregulated under the HS.
Co-expressed genes identified through expression cluster analysis
Cluster analysis of gene expression data has been reported to be a useful tool for identifying co-expressed genes. We identified 16 expression clusters among which 6 of them have more than 100 genes each as shown in Online Resource 12 (ESM_12.xls). Subcluster_3 has maximum genes (240) showing a similar pattern of expression under HS followed by subcluster_6 with 165 genes. Subcluster_2 showed upregulated genes like HSF B-2b, HSC 70 kDa protein 4, alternative oxidase 1a- mitochondrial, etc. Ribulose bisphosphate carboxylase/oxygenase activase A, peroxisomal (S)-2-hydroxy-acid oxidase GLO1, etc., were observed to be upregulated (subcluster_16). Eleven of the 16 sub-clusters (such as 2, 3, 5, 6, 7, 8, 11, 12, 14, 15 and 16) showed upregulation, whereas five sub-clusters showed downregulation of the identified DETs as presented in Online Resource 12 (ESM_12.xls).
Spatio-temporal expression analysis at transcript level under HS
Variations in the expression of identified DETs in wheat under heat stress
The DETs identified in the present investigation were randomly selected for the expression analysis in contrasting wheat cvs. HD2985 (thermotolerant) and HD2329 (thermosusceptible) under HS (42 °C, 2 h). The transcripts selected for the validation were ferredoxin NADP reductase (transcript_contig_31), HSP26 (CL4695Contig1), HSP17.8 (CL2208Contig1), serine threonine-protein kinase (CL17268Contig1), heat shock cognate 70 kDa protein (CL358Contig1), malate dehydrogenase (CL4446Contig1), novel transcript (CL13346Contig1), novel transcript (CL1679Contig1), hypothetical protein (transcript_contig_11580), novel transcript (transcript_contig_26692), novel transcript (transcript_contig_12003) and ribulose bisphosphate carboxylase/oxygenase activase A (CL2092Contig1).
The relative expression of HSP17.8 (CL2208Contig1) was observed maximum under HS in both the cultivars followed by hypothetical protein (transcript_contig_11580) and HSP26 (CL4695Contig1). The novel transcript (CL1679Contig1) showed relatively high fold expression in both the cultivars, as compared with other transcripts selected for the validation under the HS (42 °C, 2 h). RCA-A which acts as catalytic chaperone augmenting the activity of RuBisCo under stress showed 7.4-fold (HD2985) and 4.1-fold (HD2329) increase in the expression under the HS (Fig. 5). Similarly, HSC70, malate dehydrogenase (CL4446Contig1) and ferredoxin NADP reductase (transcript_contig_31) showed increase in the expression under HS; relative expression was more pronounced in wheat cv. HD2985, as compared to HD2329.
Expression profiling of identified differentially expressed transcripts (DETs) in contrasting wheat cultivars under heat stress. HD2985 (thermotolerant) and HD2329 (thermosusceptible) cultivars of wheat were selected for the study under control and heat stress condition using quantitative real-time PCR (qRT-PCR); 12 unigenes were randomly selected for the validation—ferredoxin NADP reductase (transcript_contig_31), HSP26 (CL4695Contig1), HSP17.8 (CL2208Contig1) serine threonine-protein kinase (CL17268Contig1), HSC70 kDa (CL358Contig1), malate dehydrogenase (CL4446Contig1), novel transcript (CL13346Contig1), novel transcript (CL1679Contig1), hypothetical protein (transcript_contig_11580), novel transcript (transcript_contig_26692), novel transcript (transcript_contig_12003) and RCA-A (CL2092Contig1); control—22 ± 3 °C, HS-treated—42 °C, 2 h; β-actin gene was used for normalizing the data; relative fold expression was calculated by the Pfaffl method (Pfaffl 2001); vertical bars indicate s.e (n = 3)
Temporal expression of DETs under HS
The temporal expression pattern of randomly selected 12 DETs (as explained in an earlier section) was studied in wheat cvs. HD2985 and HD2329 exposed to 42 °C for different durations (0.5, 1, 2 and 4 h). In case of HD2985, we observed an increase in the expression of HSP17.8 in response to HS of 42 °C for 2 h, and further non-significant variations were observed. Similarly, HSP17.8 showed a continuous increase in the relative expression with increase in the duration of HS (Fig. 6). A similar pattern of expression of HSP26 was observed in HD2985, whereas in HD2329, downregulation was observed after 1 h of HS (Fig. 6). Expression profiling of novel transcript (contig_26692) showed a significant increase in the fold expression in response to HS (42 °C) for 30 min, 1, 2 and 4 h in HD2985, as compared to control (22 ± 3 °C). HD2329 showed upregulation of novel transcript (contig_26692) in response to HS for 1 h, and further decrease in the expression was observed. A similar pattern of expression of novel transcripts (CL13346Contig1 and contig_12003) was observed in both the cultivars under HS.
Expression profiling of differentially expressed transcripts (DETs) in wheat in response to different duration of HS (42 °C, 2 h). HD2985 (thermotolerant) and HD2329 (thermosusceptible) cultivars were selected for the study using quantitative real-time PCR (qRT-PCR); 12 DETs were randomly selected for the expression analysis—a HSP17.8, b HSP26, c novel transcript (contig_26692), d novel transcript (CL13346 contig_1), e RuBisCo activase, f novel transcript (contig_12003), g hypothetical protein (F775_24989), h novel transcript (CL1679 contig_1), i malate dehydrogenase, j heat shock cognate 70 kDa protein, k FENR1 (ferredoxin NADP reductase) and l serine threonine-protein kinase; the expression of the β-actin gene was used for normalizing the data; relative fold expression was calculated by the Pfaffl method (Pfaffl 2001); heat stress of 42 °C was given for 30 min, 1, 2, and 4 h inside a microprocessor-regulated chamber; vertical bars indicate s.e (n = 3)
Transcript profiling of RCA-A and transcript_contig_12003 showed significant increase in the relative fold expression in response to HS (42 °C) for 30 min, 1, 2 and 4 h in both the cultivars (Fig. 6). Expression profiling of novel gene (contig_12003), hypothetical protein (contig_11580) and novel gene (CL1679 contig1) showed similar pattern of expression under HS for different durations in both the cultivars (HD2985 and HD2329). Expression profiling of malate dehydrogenase showed downregulation in response to HS for 30 min in both the cultivars, and further increase in the expression was observed. Similarly, HSC70 which has been reported to play a dual role in thermotolerance of plant under HS showed significant increase in the fold expression in response to HS (42 °C) for 30 min, 1, 2 and 4 h in both the cultivars. Ferredoxin NADP reductase which is the last enzyme of the photosynthetic light cycle showed downregulation in response to HS for 4 h (HD2985), whereas HD2329 showed early downregulation under HS (2 h onward). Serine threonine-protein kinase (STK) showed upregulation in HD2985 and downregulation in HD2329 in response to HS for different durations (Fig. 6).
Validation of unigenes expression (RNA-seq) through quantitative real-time PCR
In order to confirm the accuracy and reproducibility of the transcriptome results, 12 unigenes (as listed earlier) were selected for the validation using RT-qPCR. Total RNA samples from the leaves of control and HS-treated wheat cv. HD2985 were used as templates. The log2 fold change (FC) values of unigenes observed by RNA-seq and RT-qPCR was compared by histogram and scatter plot diagram (Fig. 7a, b). The expression profile of the candidate unigenes revealed by RT-qPCR data were consistent with those derived from the sequencing. Linear regression analysis of the log2 fold change (FC) of the gene expression ratios between RNA-seq and RT-qPCR showed significantly positive correlation, confirming our RNA-seq analysis.
Validation of unigene expression through quantitative RT-PCR. a Expression (log2 FC) of 12 randomly selected unigenes as observed through RNA-seq and quantitative real-time PCR. b Cluster map analysis; positive correlation was established between the expression of unigenes observed through RNA-seq and qRT-PCR
Validation of expression at protein level under HS
Biological activity and accumulation pattern of protein in wheat leaves under HS
In order to validate the activity and accumulation pattern of protein specific to the DET identified, we selected RuBisCo—the most abundant enzyme in leaves for this characterization. Samples from post-anthesis (10 DAA), milky-ripe and mealy-ripe stages were used for the analysis. Activity assay in leaves of HD2985 under control condition (22 ± 3 °C) showed maximum RuBisCo activity during milky-ripe, as compared with post-anthesis and mealy-ripe stages (Fig. 8a). Significant decrease in the RuBisCo activity was observed in plants exposed to HS (42 °C, 2 h); percent decrease was observed maximum during the milky-ripe stage. The post-anthesis stage was observed more thermostable, as compared to the grain-filling stage. Accumulation pattern of RuBisCo was studied in HD2985 by immunoblot assay (Fig. 8b). The accumulation of RuBisCo was observed maximum during the milky-ripe stage compared with other stages as evident from the distinct band. We observed a significant decrease in the intensity of band under HS. This makes us to conclude that HS has not much effect on the accumulation pattern of the RuBisCo protein (as evident from immunoblot assay), though activity of RuBisCo decreases with HS.
Effect of heat stress on ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCo) activity and accumulation pattern in wheat during post-anthesis (10 DAA) and grain-filling stages. a Alterations in the RuBisCo activity during different stages of grain-filling, i.e. post-anthesis, milky-ripe and mealy-ripe stages. b Immunoblot assay of RuBisCo in wheat cv. HD2985 during post-anthesis, milky-ripe and mealy-ripe stages; C—22 ± 3 °C, HS—42 °C, 2 h
Development of heat-responsive simple sequence repeat (SSR) markers
Data mining for the identification of SSR markers
De novo assembly and data analysis of wheat cv. HD2985 using SSR identification tools showed the presence of 2008 and 2483 SSRs in control (22 ± 3 °C) and HS-treated (42 °C, 2 h) samples as shown in Online Resource 13 (ESM_13.xls). The sequence of the identified SSRs in control and HS-treated samples of wheat cv. HD2985 has been depicted in the Online Resource 14 (ESM_14.doc). Maximum of trinucleotide repeat patterns were observed in both the samples. The trinucleotide repeat patterns constitute 62.8% of the total SSRs identified (Table 4). Hexanucleotide repeat pattern was observed very less in the identified SSRs, as compared to other patterns (constitute ~0.7% of total identified SSRs); 243 SSRs were observed to be localized on the HS-associated transcripts mined from the NGS data. The identified SSRs were characterized for its polymorphism in 30 different genotypes of wheat collected from our mini-core subset developed for the thermotolerance. Fourteen of the identified SSRs showed polymorphism as evident from the PIC value (Table 5); significant PIC value ranging from 0.421 (SSR-A211) to 0.743 (SSR-A240) was observed. It shows the new allelic variations in the genotypes.
In order to established the genetic relationship between the 30 diverse wheat lines selected for the screening using the identified HS-associated SSR markers, a dendrogram was drawn. The lines used for the screening were classified into two broad families, i.e. A and B; family A was classified into four different subfamilies—1A, 1B, 1C and 1D (ESM_15.TIFF). The identified SSRs were analysed for the motif length. We observed 4000 SSRs to have motif length more than 15 followed by 3000 SSRs to have motif length ~10; significant variations were observed in the motif length of the identified SSRs from the control and HS-treated wheat (ESM_16.TIFF).
Validation of identified SSR markers for their heat-responsive nature
The identified DETs with localized SSRs were used for the primer designing keeping the 150 bp flanking regions from the localized SSRs. The primers were first optimized for the amplification in two contrasting wheat cvs. HD2985 (thermotolerant) and HD2329 (thermosensitive). We observed a uniform single band of desired size on 3% metaphor gel with 200 primers out of set of 243 primers. Thirty lines of wheat (selected from our mini-core) were used for the screening using the selected SSR markers. We observed polymorphic bands in all the selected 30 genotypes with the primers SSR-A211, SSR-A221, SSR-A223 and SSR-A240 which validates the heat-responsive nature of these selected markers (Fig. 9).
Validation of identified genic SSRs localized on the differentially expressed transcripts for polymorphism in different lines of wheat. Thirty lines of wheat were selected from the mini-core set developed for the thermotolerance; primers were designed keeping 150 bp flanking regions from the localized SSRs
Discussion
Heat stress has a drastic effect on the carbon assimilatory processes, as it disrupts the photosynthetic apparatus of the plant and disturbs the source to sink ratio (Kumar et al. 2013b; Farooq et al. 2011). Visual observation of HS-treated leaves in thermotolerant and thermosusceptible wheat cvs. showed disintegration of the chlorophyll; effect was observed more pronounced during the mealy-ripe stage, as compared with other stages of growth. Our findings are in conformity with the observation of Wang et al. (2014a, b). Physiochemical validation showed significant changes in the photosynthetic rate of wheat cv. HD2985 under HS condition, as compared to control at different stages of growth. We observed non-significant differences in the WUE of wheat cv. HD2985 under control and HS-treated conditions during pollination, and mealy-ripe stages. Maximum WUE was observed in HS-treated HD2985 during pollination. Blum (2009) reported that effective use of water (EUW) is a rather more appropriate term to correlate with the yield of the plant, as compared to WUE. In most of the cases, WUE has been reported to be positively correlated with the growth, tolerance level and yield of the plant (Chamekh et al. 2016; Kantar et al. 2011).
Different omics platform have been used in the past to decipher the mechanism of thermotolerance in wheat; though information at transcriptome level is limited. Assembly of genome or transcriptome of wheat for the identification of genes associated with the abiotic stress response is quite tedious in view of the complexity of the genome and availability of very limited information on the public domain. In the past, the library construction approach was the method of choice to identify DEGs in the crops (Khurana et al. 2011). Ergen and Budak (2009) constructed six subtractive cDNA libraries using leaf and root tissues of wild emmer wheat genotypes TR39477 (tolerant) and TTD-22 (sensitive), and modern wheat variety Kiziltan exposed to drought stress and sequenced over 13,000 expressed sequence tags (ESTs). Array chips were also used for many years to explore differentially expressed genes associated with stress tolerance. Ergen et al. (2009) reported the expression pattern of several unique genes in roots and leaves associated with IP(3)-dependent signal transduction pathways, ethylene- and abscisic acid (ABA)-dependent signalling in a drought-tolerant wheat genotype, as compared to sensitive using the Affymetrix GeneChip(R) Wheat Genome Array hybridization. Currently, high-throughput NGS is, however, contributing significantly in the identification of genes associated with the abiotic stresses (Budak et al. 2015). Various crops like chickpea (Molina et al. 2011), rice (Zhang et al. 2012), sorghum (Dugas et al. 2011), soybean (Hao et al. 2011), etc., have been subjected to high-throughput NGS for identification of abiotic stress-related genes. NGS of expressed mRNAs (RNA-seq) is gradually transforming the field of transcriptomics (Wang et al. 2009; Akpinar et al. 2013). We, however, used high-throughput deep sequencing technology for whole transcriptome analysis of control and HS-treated wheat cv. HD2985 using the technology of Illumina HiSeq™ 2000 and Roche GS-FLX 454 platforms. Earlier, Roche GS-FLX 454 was the most widely used platform for de novo transcriptome sequencing due to its long read length; in due course of time, it was confirmed that the relatively short reads can be effectively assembled by the Illumina transcriptome or whole genome de novo sequencing and assembly with the advantage of paired-end sequencing (Maher et al. 2009); now, it has been exhaustively used for the sequencing in case of model and non-model plants with complex genome.
We constructed 52,567 (control) and 59,658 (HS-treated) unigenes for the thermotolerant wheat cv. with 1216 unigenes showing differential expression under the HS. There were 10,640 entries mapping to GO terms, indicating putative functional categories of the transcripts. Xiao et al. (2013) using a high-throughput RNA sequencing based on NGS technology (Illumina HiSeq 2000) identified 165,499 unigenes in wheat infested with Fusarium blight. Wan et al. (2008) also reported 14,550 transcripts to be differentially expressed in wheat during the period between 6 and 42 days after anthesis. K-means clustering in the present investigation identified 16 expression clusters suggesting higher number of modulated genes under the HS. In a recent study of gene expression of barley and Brachypodium distachyon, many genes responsive to abiotic stress were detected as co-expressed across a large body of expression datasets (Mochida et al. 2006; Priest et al. 2014). Sarkar et al. (2014) studied the co-expression network analysis in rice under HS and reported a higher number of upregulated genes compared to downregulated genes. In our lab, we also identified a total of 21,529 unigenes in control and HS-treated wheat cv. HD2329 (thermosusceptible) through de novo transcriptomic approach (Kumar et al. 2015).
The identified unigenes showing differential expression under HS has been predicted to involve in defence-related, metabolic or secondary metabolite biosynthesis pathways. Qin et al. (2008) reported that differences in the heat tolerance in different wheat genotypes are associated with multiple processes and mechanisms involving HSPs, transcription factors, and other stress-related genes. Signalling pathways has a very important role to play in triggering the tolerance mechanism of any organism under stress (Budak 2012). We observed very high digital fold expression (Log2 FC) of calcium signalling pathway-associated genes in wheat cv. HD2985 under the HS. It has been reported that calcium bursts take place inside the cytosol in response to HS, which is further amplified by Ca2+ binding proteins (CBP), and, in turn, upregulate the expression of HSPs (Goswami et al. 2015; Liu et al. 2008).
We identified some of the heat-responsive genes involved in metabolism, stress response and photosynthesis such as FENR1, HSP26, HSP17, HSFA6A, RCA-A, COX3, etc. Transcript validation showed very high fold expression of catalytic chaperones like small HSPs (HSP17.8 and HSP26) followed by novel transcripts and hypothetical protein under the HS. Similar observations were reported by Le et al. (2011) in soybean and Kumar et al. (2013a) in wheat. Similarly, few novel stress-inducible genes like DREB transcription factor and putative membrane protein (TMPIT1) genes were identified and cloned from wild emmer wheat; characterization showed very significant variations in the expression in contrasting wheat cvs. (Lucas et al. 2011a, 2011b). Transcripts involved in pathways and redox regulation showed less increase in the expression, as compared to catalytic chaperone under HS. We observed very high fold increase in the expression of Rca (a catalytic chaperone of RuBisCo) in wheat under HS which is in conformity with the findings of Wan et al. (2008). A similar observation for Rca was made by Xue et al. (2015) in wheat under HS.
The predicted protein of the identified transcript was also validated in wheat under HS. We selected the RuBisCo enzyme—the most abundant protein involved in the carbon metabolism process for their accumulation pattern and activity assay under HS. We observed a decrease in the activity of the RuBisCo enzyme under HS (42 °C, 2 h), though significant difference was not observed in the protein accumulation pattern under HS. It may be due to the abundance of the RuBisCo protein in the leaves. Heat stress deactivates the RuBisCo enzyme by aggregating the Rca—a catalytic chaperone—involved in activating the RuBisCo enzyme. RuBisCo has been observed to deactivate within 7 s of imposed HS treatment (Kumar et al. 2016). It makes us to conclude that the milky-ripe stage of wheat is critical for maintaining source (photosynthesis) to sink (grain-filling) ratio, as slight variations in the temperature causes reduction in the activity of RuBisCo and ultimately leads to decrease in the photosynthetic rate and accumulation of photosynthates in the seeds during grain-filling. Even part of the transportable form of carbohydrate is accumulated in the stem as reserve (contingency planning) to be utilized during the stress condition (Khatun et al. 2015).
Limited information regarding the heat-responsive microsatellite markers in wheat is available on public domain. Akpinar et al. (2017) reported 68,592 SNPs from the Ae. tauschii genotype MvGB589 mapped on to the chromosome 5D through genomic and transcriptomic approaches. Approximately ~4000 SSR/microsatellite were identified by mining the transcriptome data of wheat cv. HD2985 under control (22 ± 3 °C) and HS-treated (42 °C, 2 h) conditions. Based on the polymorphic study, four of the SSRs were validated to be heat-responsive in nature. Fox et al. (2014) identified ~22,000 SSR markers in Triticum monococcum using de novo transcriptomic approach. Similarly, Wang et al. (2014a, 2014b) identified ~4493 SSRs in Medicago sativa using RNA-seq approach. Similarly, Budak et al. 2005 established significant correlation between the ploidy levels and number of alleles detected using nuclear DNA (ISSR, SSR and SRAP) in buffalo grass. Maximum of trinucleotides repeat patterns were observed in the identified SSRs which is in conformity with the observation of Wang et al. (2014a, 2014b). The identified heat-responsive SSRs will play a very important role in screening wheat germplasm for thermotolerance.
Conclusions
The findings in the present investigation demonstrated the use of short-read sequence data to rapidly characterize the transcriptome of HS-treated wheat and have contributed significantly to the corpus of wheat transcript data. Most of the DEGs were associated with heat stress response, protein-folding, the oxidation-reduction process, photosynthesis, etc. We observed many candidate genes associated with heat-responsive TFs, HSPs and the carbon assimilatory pathway. Our results showed the presence of ~2000 SSRs in HD2985; four markers were validated to be heat-responsive in nature through polymorphic study. Expression analysis showed very high expression of catalytic chaperones (HSP17, HSP26) along with metabolic pathway-related genes in wheat cv. HD2985, which may be the reason behind the thermotolerant behaviour of HD2985. The identified novel heat-responsive genes and SSRs will pace-up the screening programs of wheat germplasm for thermotolerance. The contribution is significant for the progress of functional genomic studies and breeding programs.
References
Akpinar BA, Lucas SJ, Budak H (2013) Genomics approaches for crop improvement against abiotic stress. Sci World J. doi:10.1155/2013/361921
Akpinar BA, Lucas S, Budak H (2017) A large-scale chromosome-specific SNP discovery guideline. Funct Integra Genomics 17(1):97–105
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410. doi:10.1016/S0022-2836(05)80360-2
Bita C, Gerats T (2013) Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front Plant Sci 4:273
Blum A (2009) Effective use of water (EUW) and not water-use efficiency (WUE) is the target of crop yield improvement under drought stress. Field Crops Res 112(2):119–123
Bokszczanin KL, Fragkostefanakis S, Bostan H, Bovy A, Chaturvedi P, Chiusano ML et al (2013) Perspectives on deciphering mechanisms underlying plant heat stress response and thermotolerance. Front Plant Sci 4:315
Brautigam A, Gowik U (2010) What can next generation sequencing do for you? Next generation sequencing as a valuable tool in plant research. Plant Biol 12:831–841
Budak H (2012) Plant abiotic stress signalling. Plant Siganl Behav 7(11):1450–1455
Budak H, Shearman RC, Gulsen O, Dweikat I (2005) Understanding ploidy complex and geographic origin of the Buchloe dactyloides genome using cytoplasmic and nuclear marker systems. Theor Appl Genet 111(8):1545–1552
Budak H, Akpinar BA, Unver T, Turktas M (2013a) Proteome changes in wild and modern wheat leaves upon drought stress by two-dimensional electrophoresis and nanoLC-ESI–MS/MS. Plant Mol Biol 83(1–2):89–103
Budak H, Kantar M, Kurtoglu KY (2013b) Drought tolerance in modern and wild wheat. Sci World J. doi:10.1155/2013/548246
Budak H, Hussain B, Khan Z, Ozturk NZ, Ullah N (2015) From genetics to functional genomics: improvement in drought signaling and tolerance in wheat. Front Plant Sci 6:1012
Chamekh Z, Ayadi S, Karmous C, Trifa Y, Amara H, Boudabbous K et al (2016) Comparative effect of salinity on growth, grain yield, water use efficiency, δ 13 C and δ 15 N of landraces and improved durum wheat varieties. Plant Sci 251:44–53
Chapman JA, Mascher M, Buluç A, Barry K, Georganas E, Session A, Strnadova V, Jenkins J, Sehgal S, Oliker L, Schmutz J (2015) A whole-genome shotgun approach for assembling and anchoring the hexaploid bread wheat genome. Genome Biol 16(1):26
Chen J, Burke JJ, Velten J, Xin Z (2006) FtsH11 protease plays a critical role in Arabidopsis thermotolerance. Plant J 48:73–84
Chopra R, Burow G, Farmer A, Mudge J, Simpson CE, Burow MD (2014) Comparisons of de novo transcriptome assemblers in diploid and polyploid species using peanut (Arachis spp.) RNA-seq data. PLoS One 9(12):e115055
Dong L, Liu H, Zhang J, Yang S, Kong G, Chu JS et al. (2015) Single molecule real-time transcript sequencing facilitates common wheat genome annotation and grain transcriptome research. BMC Genomics 16(1):1039
Dugas DV, Monaco MK, Olson A et al (2011) Functional annotation of the transcriptome of Sorghum bicolor in response to osmotic stress and abscisic acid. BMC Genomics 12:514. doi:10.1186/1471-2164-12-514
Ergen NZ, Budak H (2009) Sequencing over 13 000 expressed sequence tags from six subtractive cDNA libraries of wild and modern wheats following slow drought stress. Plant Cell Environ 32(3):220–236
Ergen NZ, Thimmapuram J, Bohnert HJ, Budak H (2009) Transcriptome pathways unique to dehydration tolerant relatives of modern wheat. Funct Integra Genomics 9(3):377–396
Farooq M, Bramley H, Palta JA, Siddique KHM (2011) Heat stress in wheat during reproductive and grain-filling phases. CRC Crit Rev Plant Sci 30:491–507
Fox SE, Geniza M, Hanumappa M et al (2014) De novo transcriptome assembly and analyses of gene expression during photomorphogenesis in diploid wheat Triticum monococcum. PLoS One 9:e96855. doi:10.1371/journal.pone.0096855
Goswami S, Kumar RR, Rai RD (2014) Heat-responsive microRNAs regulate the transcription factors and heat shock proteins in modulating thermo stability of starch biosynthesis enzymes in wheat (Triticum aestivum L.) under the heat stress. Aust J Crop Sci 8:697–705
Goswami S, Kumar RR, Sharma SK et al (2015) Calcium triggers protein kinases-induced signal transduction for augmenting the thermotolerance of developing wheat (Triticum aestivum) grain under the heat stress. J Plant Biochem Biotechnol 24:441–452. doi:10.1007/s13562-014-0295-1
Gotz S, García-Gómez JM, Terol J et al (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36:3420–3435. doi:10.1093/nar/gkn176
Hao QN, Zhou XA, Sha AH et al (2011) Identification of genes associated with nitrogen-use efficiency by genome-wide transcriptional analysis of two soybean genotypes. BMC Genomics 12:525
Haq S, Jain R, Sharma M, Kachhwaha S, Kothari SL (2014) Identification and characterization of microsatellites in expressed sequence tags and their cross transferability in different plants. Int J Genomics 2014:1–12. doi:10.1155/2014/863948
Hasanuzzaman M, Nahar K, Alam MM, Roychowdhury R, Fujita M (2013) Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int J Mol Sci 14(5):9643–9684
Hatfield JL, Prueger JH (2015) Temperature extremes: effect on plant growth and development. Weather Clim Extremes 10:4–10
Hays DB, Do JH, Mason RE et al (2007) Heat stress induced ethylene production in developing wheat grains induces kernel abortion and increased maturation in a susceptible cultivar. Plant Sci 172:1113–1123
International Wheat Genome Sequencing Consortium (IWGSC) TIWGSC, Lobell DB, Schlenker W et al (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Sci 345:1251788. doi:10.1126/science.1251788
Kantar M, Lucas SJ, Budak H (2011) Drought stress: molecular genetics and genomics approaches. Adv Bot Res 57:445–493
Khatun S, Ahmed JU, Mohi-Ud-Din M (2015) Variation of wheat cultivars in their relationship between seed reserve utilization and leaf temperature under elevated temperature. J Crop Sci Biotechnol 18:97–101. doi:10.1007/s12892-014-0117-y
Khurana P, Chauhan H, Khurana N (2011) Characterization and expression of high temperature stress responsive genes in bread wheat (Triticum aestivum L.) Czech J Genet Plant Breed 47:S94–S97
Kumar RR, Rai RD (2014) Can wheat beat the heat: understanding the mechanism of thermotolerance in wheat (Triticum aestivum L.) Cereal Res Commun 42:1–18. doi:10.1556/CRC.42.2014.1.1
Kumar RR, Sharma SK, Goswami S et al (2013a) Characterization of differentially expressed stress-associated proteins in starch granule development under heat stress in wheat (Triticum aestivum L.) Indian J Biochem Biophys 50:126–138
Kumar RR, Sharma SK, Goswami S et al (2013b) Transcript profiling and biochemical characterization of mitochondrial superoxide dismutase (mtSOD) in wheat (Triticum aestivum) under different exogenous stresses. Aust J Crop Sci 7:414
Kumar RR, Goswami S, Sharma SK et al (2015) Harnessing next generation sequencing in climate change: RNA-Seq analysis of heat stress-responsive genes in wheat (Triticum aestivum L.) Omi A J Integr Biol 19:632–647
Kumar RR, Goswami S, Singh K et al (2016) Identification of putative RuBisCo Activase (TaRca1)—the catalytic chaperone regulating carbon assimilatory pathway in wheat (Triticum aestivum) under the heat stress. Front Plant Sci. doi:10.3389/fpls.2016.00986
Le DT, Nishiyama R, Watanabe Y et al (2011) Genome-wide expression profiling of soybean two-component system genes in soybean root and shoot tissues under dehydration stress. DNA Res 18:17–29
Liu HT, Gao F, Li GL et al (2008) The calmodulin-binding protein kinase 3 is part of heat-shock signal transduction in Arabidopsis thaliana. Plant J 55:760–773
Long SP, Bernacchi CJ (2003) Gas exchange measurements, what can they tell us about the underlying limitations to photosynthesis? Procedures and sources of error. J Exp Bot 54:2393–2401
Lucas S, Dogan E, Budak H (2011a) TMPIT1 from wild emmer wheat: first characterization of a stress-inducible integral membrane protein. Gene 483(1):22–28
Lucas S, Durmaz E, Akpınar BA, Budak H (2011b) The drought response displayed by a DRE-binding protein from Triticum dicoccoides. Plant Physiol Biochem 49(3):346–351
Maher CA, Palanisamy N, Brenner JC et al (2009) Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci U S A 106:12353–12358
Martin JA, Wang Z (2011) Next-generation transcriptome assembly. Nat Rev Genet 12:671–682
Matsui K, Umemura Y, Ohme-Takagi M (2008) AtMYBL 2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 55:954–967. doi:10.1111/j.1365-313X.2008.03565.x
Mochida K, Kawaura K, Shimosaka E et al (2006) Tissue expression map of a large number of expressed sequence tags and its application to in silico screening of stress response genes in common wheat. Mol Gen Genomics 276:304–312
Molina C, Zaman-Allah M, Khan F et al (2011) The salt-responsive transcriptome of chickpea roots and nodules via deepSuperSAGE. BMC Plant Biol 11:31
Mortazavi A, Williams BA, McCue K et al (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Mundry M, Bornberg-Bauer E, Sammeth M, Feulner PGD (2012) Evaluating characteristics of de novo assembly software on 454 transcriptome data: a simulation approach. PLoS One 7:e31410. doi:10.1371/journal.pone.0031410
Mutz K-O, Heilkenbrinker A, Lonne M et al (2013) Transcriptome analysis using next-generation sequencing. Curr Opin Biotechnol 24:22–30. doi:10.1016/j.copbio.2012.09.004
Pertea G, Huang X, Liang F et al (2003) TIGR Gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19:651–652
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi:10.1093/nar/29.9.e45
Priest HD, Fox SE, Rowley ER et al (2014) Analysis of global gene expression in Brachypodium distachyon reveals extensive network plasticity in response to abiotic stress. PLoS One 9:e87499. doi:10.1371/journal.pone.0087499
Qin D, Wu H, Peng H et al (2008) Heat stress-responsive transcriptome analysis in heat susceptible and tolerant wheat (Triticum aestivum L.) by using wheat genome array. BMC Genomics 9:432
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Sarkar NK, Kim YK, Grover A (2014) Coexpression network analysis associated with call of rice seedlings for encountering heat stress. Plant Mol Biol 84:125–143
Wahid A, Gelani S, Ashraf M, Foolad MR (2007) Heat tolerance in plants: an overview. Environ Exp Bot 61:199–223. doi:10.1016/j.envexpbot.2007.05.011
Wan Y, Poole RL, Huttly AK et al (2008) Transcriptome analysis of grain development in hexaploid wheat. BMC Genomics 9:121. doi:10.1186/1471-2164-9-121
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63
Wang Y, Zhang H, Hou P, Su X, Zhao P, Zhao H, Liu S (2014a) Foliar-applied salicylic acid alleviates heat and high light stress induced photoinhibition in wheat (Triticum aestivum) during the grain filling stage by modulating the psbA gene transcription and antioxidant defense. Plant Growth Regul 73(3):289–297
Wang X, Cai J, Liu F et al (2014b) Multiple heat priming enhances thermo-tolerance to a later high temperature stress via improving subcellular antioxidant activities in wheat seedlings. Plant Physiol Biochem 74:185–192
Xiao J, Jin X, Jia X et al (2013) Transcriptome-based discovery of pathways and genes related to resistance against Fusarium head blight in wheat landrace Wangshuibai. BMC Genomics 14:197
Xue GP, Drenth J, McIntyre CL (2015) TaHsfA6f is a transcriptional activator that regulates a suite of heat stress protection genes in wheat (Triticum aestivum L.) including previously unknown Hsf targets. J Exp Bot 66(3):1025–1039
Zerbino DR, Birney E (2008) Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829
Zhang T, Zhao X, Wang W et al (2012) Comparative transcriptome profiling of chilling stress responsiveness in two contrasting rice genotypes. PLoS One 7(8):e43274. doi:10.1371/journal.pone.0043274
Zhao Q-Y, Wang Y, Kong Y-M et al (2011) Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics 12:S2
Acknowledgements
The research work was financially supported by the Indian Council of Agriculture Research (ICAR) under the National Initiative for Climate Resilient Agriculture (NICRA) project (sanction no 12/115 TG3079), and the Department of Science and Technology, Government of India (GOI) under SERB start-up grant (sanction no SERB/SB/SO/PS/07/2014). We are highly thankful to the scientists of IASRI, New Delhi, for analysing the raw data and executing the bioinformatics characterisation under the CABin project (21-56) TG3064.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary materials
ESM 1
Layout of the experiment used for the de novo sequencing and validation of heat-responsive genes in present investigation; Distribution of pots used for the sowing and heat stress treatment of contrasting wheat cvs. HD2985 and HD2329. (GIF 197 kb)
ESM 2
Workflow chart depicting the de novo sequencing of control and HS-treated wheat cv. HD2985. (GIF 1908 kb) (DOCX 134 kb)
ESM 3
List of primers used for the validation of randomly selected differentially expressed transcripts (DETs) in wheat under heat stress using quantitative real-time PCR (qRT-PCR). (DOCX 16 kb)
ESM 4
List of primers designed for the SSR validation along with the respective transcripts identified through de novo transcriptomic approach (PDF 6488 kb)
ESM 5
Visual observation of leaf discs collected from control and HS-treated wheat cv. HD2985 at different stages of growth, (a) Pollination (control), (b) Pollination (HS-treated), (c) Milky-ripe (control), (d) Milky-ripe (HS-treated), (e) Mealy-ripe (control), (f) Mealy-ripe (HS-treated); Discs were extracted from the flag leaves used for the IRGA analysis, C - 22 ± 3 °C, HS - 42 °C, 2 h. (GIF 1908 kb)
ESM 6
De novo assembly statistics for whole transcriptome analysis of wheat cv. HD2985. (DOCX 14 kb)
ESM 7
Annotation of unigenes identified from the de novo sequencing of the control and heat shock treated wheat cv. HD2985 using NCBI-nr reference sequence database, Gene Ontology database and pfam database. (XLSX 2401 kb)
ESM 8
Annotation of unigenes identified from the de novo sequencing of the control and heat shock treated wheat cv. HD2985 using Uniprot database (http://www.uniprot.org/). (XLSX 1244 kb)
ESM 9
TriFLDB annotation for the identified unigenes from wheat cv. HD2985 under control and HS-conditions. (XLSX 698 kb)
ESM 10
PlantCyc annotation for identified unigenes from wheat cv. HD2985 under control and HS-conditions. (XLSX 611 kb)
ESM 11
Differential expressions of identified unigenes in control and HS-treated samples of wheat cv. HD2985. (XLSX 2241 kb)
ESM 12
Expression clusters analysis of identified unigenes in control and HS-treated samples of wheat cv. HD2985. (XLSX 214 kb)
ESM 13
List of microsatellite markers observed on differentially expressed transcripts. (a) SSRs identified in control sample, (b) SSRs identified in HS-treated sample of wheat cv. HD2985 through transcriptomic approach; C - 22 ± 3 °C, HS - 42 °C, 2 h. (XLSX 270 kb)
ESM 14
Sequence of SSR markers identified in control and HS-treated wheat cv HD2985 through de novo transcriptomic approach. (a) Microsatellite marker sequence identified in control sample, SSR sequences identified in HS-treated sample of wheat cv. HD2985; C - 22 ± 3 °C, HS - 42 °C, 2 h. (FA 354 kb)
ESM 15
Dendrogram showing genetic relationship among 30 wheat genotypes generated from SSR markers; Scale at the bottom of the dendrogram indicates the level of similarity between the genotypes. (GIF 170 kb)
ESM 16
Motif length analysis of the identified Simple Sequence Repeats (SSRs). SSRs were identified from wheat cv. HD2985 under control and HS-conditions; C - 22 ± 3 °C, HS - 42 °C, 2 h. (GIF 248 kb)
Rights and permissions
About this article
Cite this article
Kumar, R.R., Goswami, S., Shamim, M. et al. Exploring the heat-responsive chaperones and microsatellite markers associated with terminal heat stress tolerance in developing wheat. Funct Integr Genomics 17, 621–640 (2017). https://doi.org/10.1007/s10142-017-0560-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-017-0560-1