Abstract
Characterizing and inferring the buffalograss [Buchloe dactyloides (Nutt.) Engelm.] genome organization and its relationship to geographic distribution are among the purposes of the buffalograss breeding and genetics program. This buffalograss study was initiated to: (1) better understand the buffalograss ploidy complex using various marker systems representing nuclear and organelle genomes; (2) determine whether the geographic distribution was related to nuclear and organelle genome variation; and (3) compare the genetic structure of accessions with different ploidy levels. The 20 buffalograss genotypes (15 individuals from each genotype) that were studied included diploid, tetraploid, pentaploid, and hexaploid using nuclear (intersimple sequence repeat (ISSRs), simple sequence repeat (SSRs), sequence related amplified polymorphism (SRAPs), and random amplified polymorphic DNA (RAPDs)) and cytoplasmic markers (mtDNA and cpDNA). There was a significant correlation between the ploidy levels and number of alleles detected using nuclear DNA (ISSR, SSR, and SRAP, r=0.39, 0.39, and 0.41, P<0.05, respectively), but no significant correlation was detected when mitochondrial (r=0.17, P<0.05) and chloroplast (r=0.11, P<0.05) DNA data sets were used. The geographic distribution of buffalograss was not correlated with nuclear and organelle genome variation for the genotypes studied. Among the total populations sampled, regression analysis indicated that geographic distance could not explain genetic differences between accessions. However, genetic distances of those populations from the southern portion of buffalograss adaptation were significantly correlated with geographic distance (r= 0.48, P<0.05). This result supports the hypothesis that genetic relationship among buffalograss populations cannot be estimated based only on geographic proximity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Buffalograss [Buchloe dactyloides (Nutt.) Englem.] is a native North American turfgrass species. It is a perennial, warm-season (C4), sod-forming turfgrass species that is used for home lawns, parks, cemeteries, airfields, sports turfs, roadsides, and golf courses (Beard 1973). Buffalograss is a member of the Chlorideae tribe and is the only member of the genus Buchloe (Hitchcock 1951). Other genera in the tribe include Bouteloua, Chloris, and Trichloris. Although buffalograss and blue grama [Bouteloua gracilis (H.B.K.) Lag.ex Steud] are associated with one another, they have substantially different morphological and adaptation characteristics. Buffalograss is distributed from Canada to Mexico and from the eastern slope of the Rocky Mountains to the Mississippi River Valley. It is a cross pollinated species and highly heterogeneous, with no evidence of self-pollination (Wu and Lin 1984).
Buffalograss is comprised of a polyploid series of diploid, tetraploid, pentaploid, and hexaploid with a basic chromosome number of 10 (Reeder 1971; Huff et al. 1993; Johnson et al. 1998). The different ploidy levels are morphologically indistinguishable and their genome relationships are unknown. Diploids have been reported to occur only in the central Mexico and southeastern Texas, tetraploids in the southern proportions of the North American Great Plains, and hexaploids are found throughout the region (Huff et al. 1993; Johnson et al. 2001).
Polyploid evolution has received attention due to its ubiquity in plants (Grant 1981; Masterson 1994). Polyploidization may be a significant means of speciation (Leitch et al. 1998). Duplicated genes caused by polyploidy retain their original or similar function or one copy may become silenced (i.e., mutational and epigenetic interactions) and polyploidization will affect DNA structure, allowing greater diversity at higher ploidy levels (Wendel 2000). Gene diversification in polyploids can, therefore, lead to increased polymorphism in nuclear and cytoplasmic markers.
Comparing geographically classified germplasm is of interest in evolutionary biology and plant breeding programs. Hence, the study of geographical distribution of buffalograss is important to study, monitor, and manage germplasm. The proportioning of a large geographic region into more homogeneous areas allows for the evaluation of potential buffalograss germplasm geographic variation. Budak et al. (2004a) suggested two strategies for a better understanding of buffalograss ploidy level distributions. The first strategy was an improved understanding of evolutionary and historical development of the genotypes. The second strategy was elucidating environmental covariates with the emphasis on physiological characteristics.
Molecular marker analysis has contributed to the understanding of buffalograss genetic structure, diversity and phylogenetic relationships (Huff et al. 1993; Peakall et al. 1995; Budak et al. 2004a; b). Cloning and sequencing of resistance gene candidates (Budak et al. 2004c) and chloroplast and mitochondrial genes in buffalograss (Budak et al. 2005) were also reported. However, these nuclear and organelle DNA markers and cytogenetics techniques have not been used extensively to contribute to a better understanding of buffalograss variation in ploidy and geographic distributions.
This buffalograss study was initiated to: (1) better understand the ploidy level with various marker systems representing nuclear and organelle genomes; (2) determine whether the geographic distribution was related to nuclear and organelle genome variation; and (3) compare the genetic structure of accessions with different ploidy levels.
Materials and methods
Plant materials
Fifteen individual plants from each seeded and vegetative genotype representing hexaploid, pentaploid, tetraploid, and diploid genotypes were examined (Table 1). Fifteen individuals were also selected from seeded (‘Cody’, ‘Bowie’, and ‘SWI 2000’) and vegetative (‘Legacy’, ‘Prestige’, and ‘378’) biotype cultivars. ‘Cody’, ‘Bowie’, and ‘SWI 2000’ were planted from approximately 1,500 pure live seeds to insure that the resulting plant populations represented the diversity of each seeded cultivar. Vegetative plugs of ‘Legacy’, ‘Prestige’, and ‘378’ were obtained from the John Seaton Anderson Turfgrass Research Facility located near Mead, NE, USA. Fifteen individual plants from Blue grama [Bouteloua gracilis (H.B.K.) Lag. Ex Steud.], zoysiagrass (Zoysia japonica Steud.), and bermudagrass [Cynodon dactylon (L.) Pers.] were selected as out-group species for the genetic and statistical comparisons. All these above-mentioned grasses and buffalograss belong to the Eragrostoideae subfamily and were reported high level of similarity by Yaneshita et al. (1993). The genotypes were planted in 15 cm diameter pots containing a soil mixture of 35% peat, 32% vermiculite, 9% soil, and 24% sand by volume. Soil was saturated bi-weekly with a nutrient solution (21N–1.5P–12.5K) containing 200 mg L−1 nitrogen. The greenhouse was maintained at 25±1°C with supplemental light supplied by metal halide lamps on a 15/9 h photoperiod (Sylvania Co., Danver, MA, USA).
DNA extraction
Genomic DNA of each line was isolated by a sap-extraction method from 100 mg of fresh tissues. Leaves of 2-week-old seedlings were placed between the two rollers of a sap-extraction apparatus (Ravenel Specialties, Seneca, SC, USA), and 1 ml of extraction buffer (50 mM Tris–HCl, 25 mM EDTA, 1 M NaCl, 1% CTAB, 1 mM 1, 10-phenathroline, and 0.15% 2-mercaptoethanol) was slowly added to the rollers, immediately mixing with the sap for collection in 1.5-ml microcentrifuge tubes. The extract was incubated at 60°C for 1 h, and then mixed with an equal volume of chloroform–isoamyl alcohol (24:1). After centrifuging at 12,000 rpm, the supernatant was transferred to a new tube and isopropanol was added for 30-min incubation at room temperature to precipitate the DNA. The pellet was dried, resuspended in 200 μl of TE buffer (10 mM Tris–HCl, 0.1 mM EDTA, pH 8.0) plus 20 μg of RNase, and then incubated at room temperature overnight. The DNA solution was mixed with 20 μl of 8 M ammonium acetate and 400 μl of cold absolute ethanol for 30 min, centrifuged for 10 min, and then air-dried at room temperature. The DNA was then resuspended in 200 μl of TE buffer, and DNA concentration was quantified by spectrophotometry (TKO100 Fluorometer, Hoefer Scientific Instruments, San Francisco).
PCR amplifications of cytoplasmic genome
Mitochondrial and chloroplast genome regions were amplified using standard primers (Table 2). Restriction enzymes used for both genomes amplification in this study were HaeIII, EcoRI, EcoRV, MboI, DraI, TagI, RsaI, MseI, and MspI. The PCR reaction mixtures were carried out as described by Budak et al. (2004a; b). Cytoplasmic genome amplifications were done in a MJ Research PTC-100 thermocycler programmed for: one cycles of 2 min at 94°C, 35 cycles of 1 min at 94°C, 1 min at 54°C, 2 min at 72°C. Ten microliters of PCR products were used based on the expected numbers and sizes of restriction fragments that were separated on 2.5% agarose gels with the ethidium bromide. Amplified fragments were photographed using a Gel Doc 2000 (Bio-Rad) (Hercules, CA, USA).
PCR amplification of nuclear genome
A combination of co-dominant and dominant markers, intersimple sequence repeat (ISSRs), simple sequence repeat (SSRs), sequence related amplified polymorphism (SRAPs), and random amplified polymorphic DNA (RAPDs) used in this study is presented in Table 3. Evaluation and amplifications of nuclear genome variation of the germplasm tested were carried out as reported by Budak et al. (2004a, b).
Scoring gels and data analysis
Presence or absence of each nuclear and cytoplasmic markers fragment was coded as “1” and “0”, where “1” indicated the presence of a specific allele, and “0” indicated its absence. Average genetic diversity (D) as a measure of genetic variation was estimated using D = 1- (1/L)Σ l Σ i P 2 li , where P i is the frequency of the ith allele at the l locus where L is the number of loci (Weir 1996). The genetic similarity coefficients (GS) or the Dice coefficients (Sneath and Sokal 1973) were measured between genotypes to obtain a GS matrix based on nuclear and cytoplasmic banding patterns. Genetic similarity between two genotypes within one locus was calculated using the formula GS ij =2N ij /(N i +N j ), where N i and N j represents the total number of bands present in cultivar i and j, respectively, and N ij refers to the total number of common bands by the same cultivars (Nei and Li 1979). The distance matrix and dendrogram were constructed using the Numerical Taxonomy Multivariate Analysis System (NTSYS-pc) version 2.1 (Exeter Software, Setauket, NY, USA) software package (Rohlf 2000).
Cluster analysis was performed using PROC CLUSTER (SAS, Cary, NC, USA) with distance matrices to generate composite groups based on a combination of intersite geographic distance and assemblage dissimilarity. Correlations between ploidy level and number of markers scored in each sample were calculated using PROC CORR (SAS, Cary, NC, USA). The number of bands was detected based on the observed total number of bands in all genotypes. Regression analysis using PROC REG (SAS, Cary, NC, USA) was performed to determine associations between pairwise genetic distance from nuclear to organelle DNA data sets and pairwise geographic distances between populations.
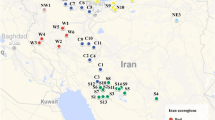
A hierarchical analysis of molecular variance (AMOVA) (Excoffier et al. 1992) was performed to estimate the amount of variation due to differences within and among ploidy levels. The AMOVA was also preformed to estimate the amount of variation within and among geographic regions (Table 3). In this analysis, genotypes were grouped to be southern (G1), northern (G2), and central (G3) types based on their geographic locations (Table 1).
Results and discussion
Elucidating ploidy complex using nuclear and organelle markers
There was a significant linear response between ploidy level and number of alleles detected using the nuclear genome markers, ISSR, SSR, and SRAP, r=0.39, 0.39, and 0.41 (P<0.05), respectively. There was no significant linear response when RAPD markers were used (r=0.21, P<0.05). This response would be expected since RAPDs are not affected by variation in ploidy levels (Weising et al. 1995). Therefore, RAPD markers might be useful when studying high polyploid genotypes because they do not complicate interpretation of RAPD data. This response likely indicates extra copies of homologous chromosomes with the higher ploidy levels. Since different buffalograss ploidy levels are not distinguishable morphologically (Budak et al. 2004a, b), it appears that extra copies of homologous chromosomes at higher ploidy levels do not modify the morphological structure for adaptation to diverse environments.
The increased number of alleles obtained from hexaploids may provide for their broad-based adaptation throughout the Great Plains of North America, when they were compared to diploids that have a very narrow adaptation base. The number of allele from NE 03-65 (diploid genotype) to NE 03-10 (hexaploid genotype) ranged from 30 to 38 using ISSRs. The number of alleles detected from the same genotypes ranged from 28 to 35 with SSRs markers. The SRAPs responded similarly, when 28 markers were used [i.e., the number of allele detected ranged from 31 (diploid) to 37 (hexaploid)], but this pattern was not clear when marker numbers increased from 28 to 52. For instance, NE 03-66 a diploid genotype had 31 alleles while NE 03-10, a hexaploid genotype, also had 30 alleles when the number of markers was increased. Although not conclusive, this response indicates that SRAPs appear not to be influenced by ploidy variation in a similar manner as RAPD markers. The absence of a significant linear response between ploidy levels and alleles might be the result of differences between the molecular markers used. Research with buffalograss by Budak et al. (2004b) found only a low level of similarity among the different marker techniques. It would be suspected that ISSRs and SSRs might detect a greater diversity at higher ploidy levels. This study demonstrated that genetic factors such as chromosome and ploidy levels are strongly correlated with nuclear diversity.
The combined diversity estimates based on several molecular markers cover more genomic regions than a single marker alone, genetic distance estimates based on all molecular markers most likely give the most unbiased distance estimates. In this study, the combined analysis of genomic regions amplified by ISSR, SSR, RAPD, and SRAP, gave genetic distance estimates that averaged 0.67 and ranged from 0.40 to 0.96 and this response indicated tetraploids, pentaploids, and hexaploids grouped together (Table 4). This might indicate that differences between genomes are not high. These results are a further indication that buffalograss might be an autoploid (Johnson et al.1998; Budak et al. 2004a).
Chloroplast (cp) DNA and mitochondrial (mt) DNA analyses had no significant correlation among ploidy levels and the number of alleles detected (r=0.11, and 0.17, P<0.05) based on the primer pairs used, and were not as informative as nuclear genome markers. cpDNA similarities among buffalograss genotype (Table 5) were considerably higher than buffalograss genotype similarities with zoysiagrass, bermudagrass, and blue grama (data not shown). Increasing the number of organelle markers to detect correlation between ploidy levels and the number of alleles might enhance future studies.
There was a discrepancy between the genetic distance estimates based on nuclear, organelle DNA data sets (Tables 4 and 5), which was also found by researchers studying different plant species (Kellogg et al. 1996; Mason-Gramer and Kellogg 1996a, b; Petersen and Seberg 1997; Soltis and Soltis 2003; Redingbaugh et al. 2000). In organelle DNA study, the level of similarity of some genotypes was higher than the combined nuclear DNA data sets (Tables 4 and 5). For instance, although the relationship between Density, a southern type diploid cultivar and SWI 200, a hexaploid northern type of genotype was not high using combined nuclear DNA markers; it was clearly high when cpDNA markers were used. This response is due to the likelihood of the nuclear and chloroplast genomes having different evolutionary histories. This problem is most evident at the polyploid level, since it has been shown that the same morphologically defined polyploid taxon may arise several times. The chloroplast genome is generally uniparentally inherited (Petit et al. 2003), and its evolutionary history may not reflect that of the organism, especially in a species with a high degree of out-crossing (Budak et al. 2005).
Geographical distribution
Geographic distribution of organelle markers should give a more concise picture of migration history than nuclear markers. Organelle markers are uniparentally inherited, and as such, the effective population size needed for study is reduced. Organelle DNA markers showed 197 out of 300 plants representing diploids, tetraploids, pentaploids, and hexaploids from diverse geographical regions did not differ. Buffalograss genotypes from different geographic regions tended to cluster together in this study using both combined nuclear and cytoplasmic DNA marker data sets. For instance, NE 03-7 (40°86′) was grouped with NE 03-46, a tetraploid genotype, (35°32′) clustered at 90% level of similarity. Additionally, NE 03-20, a pentaploid genotype with the geographic location of 41°40′ clustered with a hexaploid genotype NE 03-10 (37°03′) at 90% similarity. Hence, distribution of buffalograss genotypes might be due to geographic origins or breeding origins (Budak et al. 2004b). These results suggest that buffalograss ploidy level influences climatic zone adaptation. The widespread extension of hexaploids and various aneuploids beyond the southern range adaptation of diploids may depend on other genetic factors that interact with ploidy level. Organelle and nuclear genome variation is not geographically structured in buffalograss. This response may be due to environmental factors, sample size, sampling strategies, or a combination of these factors. Regression analysis was used to investigate whether genetic distances could be explained by geographic distance. Among the population sampled in this study, genetic distance was not explained by geographic distance. Genetic distances for those populations from the southern Great Plains were correlated (r=0. 48, P<0.05) with geographic distance. This result indicated that genetic relationship among buffalograss populations cannot be estimated based on geographical proximity alone. Our results agree with Huff et al. (1998), who found no association between geographic distance and GS in little bluestem (Schizachyrium scoparium) using RAPD markers. If genetic relationships could be estimated by geographic proximity, then cluster analysis should group genotypes by their geographic origin. AMOVA results indicated that there was high variation among the ploidy levels and low variation within ploidy levels with the nuclear markers used in this study (Table 6). Organelle marker variation was considerably low and nonsignificant within (8.8%) and among ploidy levels (14.5%). The reduced organelle diversity could be due to uniparental inheritance nature of these markers.
Buffalograss dispersal across its zone of adaptation was based mainly on animal seed dispersal (Quinn et al. 1994; Ortmann et al. 1998). This process would potentially result in a strong genetic differentiation between populations. In this study, distribution of genetic variation indicated higher among and within geographic regions [i.e., southern (G1) (49%), northern (G2) (32%), and central (G3) (25%) than within populations (9%)] (Tables 6 and 7). Some evidence of a decrease in diversity from the south to the north was observed in this study.
Huff et al. (1993) reported the existence of a Texas diploid race in addition to diploid buffalograss accessions from Central Mexico using RAPDs. The existence of different diploid races might indicate that some polyploids could have alloploid origins, since diploid races from different ecological zones could be different diploid progenitors (Peakall et al. 1995). However, co-dominant markers rather than RAPDs (dominant markers) might help dissect the ancestry of polyploids. In addition, co-dominant markers enable an examination of polyploidy dissection, which will help to develop a better understanding of polyploid evolution in buffalograsses.
The selection of promising genotypes appears possible based on genotypes evaluated in this study. Future research with buffalograss and several closely related species [i.e., Buchlomimus nervatus (Swallen) Reeder, Reeder& Rzedowski; Cyclostacya stolonifera (Scribn.) Reeder& Reeder; Opizia stolonifera Presl.; Pringleochloa stolonifera (Fourn.) Scribn., and Soderstromia mexicana (Scribn.) (Reeder and Reeder 1963; Reeder and Rzedowski 1965)] to elucidate buffalograss genome organization, characterization, ecological distribution, and ploidy levels would be desirable.
References
Allouis S, Qi X, Lindup S, Gale MD, Devos K M (2001) Construction of BAC library of pearl millet, Pennisetum glaucum. Theor Appl Genet 102:1200–1205
Arrieta-Montiel M, Lyznik A, Woloszynska M, Janska H, Tohme J, Mackenzie S (2001) Tracing evolutionary and developmental implications of mitochondrial stoichiometric shifting in the common bean. Genetics 158:851–864
Beard JB (1973) Turfgrass: science and culture. Prentice-Hall, Englewood Cliffs, NJ
Budak H, Pedraza F, Baenziger PS, Cregan PB, Dweikat I (2003) Development and utilization of SSR to estimate genetic diversity in a collection of pearl millet germplasm. Crop Sci 43:2284–2290
Budak H, Shearman RC, Parmaksiz I, Gaussoin RE, Riordan TP, Dweikat I (2004a) Molecular characterization of buffalograss germplasm using sequence related amplified polymorphism markers. Theor Appl Genet 108: 328–334
Budak H, Shearman RC, Parmaksiz I, Dweikat I (2004b) Comparative analysis of seeded and vegetative buffalograsses based on phylogenetic relationship using ISSR, SSR, RAPD and SRAP. Theor Appl Genet 109: 280–288
Budak H, Shearman RC, Dweikat I (2004c) Cloning and characterization of resistance gene like sequences in warm season turfgrass species. In: Proceedings of the international conference on mathematics and engineering techniques in medicine and biological science. Las Vegas, pp 225–230
Budak H, Shearman RC, Dweikat I (2005) Evolution of Buchloë dactyloides based on cloning and sequencing of matK, rbcL, and cob genes from plastid and mitochondrial genomes. Genome 48:411–416
Corderio GM, Taylor GO, Henry RJ (2000) Characterization of microsatellite markers from sugarcane (Saccharum sp.), a highly polyploidy species. Plant Sci 155:161–168
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred for metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Grant V (1981) Plant speciation, 2nd edn. Columbia University Press, New York
Grivet G, Heinze B, Vendramin GG, Petit RJ (2001) Genome walking with consensus primers: application to the large single copy region of chloroplast DNA. Mol Ecol Notes 1:345–349
Hitchcock AS (1951) Manual of the grasses of the United States, 2nd edn. US Department of Agriculture Misc Pub 200., Washington
Huff DR, Peakall R, Smouse PE (1993) RAPD variation within and among natural populations of outcrossing buffalograss [Buchloë dactyloides (Nutt) Engelm]. Theor Appl Genet 86:927–934
Huff DR, Quinn JA, Higgins B, Palazzo AJ (1998) Random amplified polymorphic DNA (RAPD) variation among native little bluestem [Schizachyrium scoparium (Michx.) Nash] populations from sites of high and low fertility in forest and grassland biomes. Mol Ecol 7:1591–1597
Johnson PG, Riordan TP, Arumuganathan K (1998) Ploidy level determinations in buffalograss clones and populations. Crop Sci 38:478–482
Johnson PG, Kenworthy KE, Auld DL, Riordan TP (2001) Distribution of buffalograss polyploid variation in the southern Great Plains. Crop Sci 41:909–913
Kellogg EA, Appels R, Mason-Gramer RJ (1996) When gene trees tell different stories: the diploid genera of Triticeae (Gramineae). Syst Bot 21:321–347
Leitch I, Chase MW, Bennett MD (1998) Phylogenetic analysis of DNA C-values provides evidence for a small ancestral genome size in flowering plants. Ann Bot 82:85–94
Li G, Quiros CF (2001) Sequence-related amplified polymorphism (SRAP), a new marker system based on a sample PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet 103: 455–461
Mason-Gamer RJ, Kellogg EA (1996a) Chloroplast DNA analysis of the monogenomic Triticeae: phylogenetic implications and genome specific markers. In: Jauhar PP (ed) Methods of genome analysis in plants. CRC Press, Boca Raton, pp 301–325
Mason-Gamer RJ, Kellogg EA (1996b) Testing for phylogenetic conflict among molecular data sets in tribe Triticeae (Gramineae). Syst Biol 45:524–545
Masterson J (1994) Stomatal size in fossil plants:evidence for polploidy in majority of angiosperms. Science 264:421–424
Nei M, and Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Ortmann J, Schacht WH, Stubbendieck J, Brink D (1998) The "foliage is the fruit" hypothesis: complex adaptations in buffalograss (Buchloe dactyloides). Am Midl Nat 140:252–263
Peakall R, Smouse PE, Huff DR (1995) Evolutionary implications of allozyme and RAPD variation in diploid populations of dioecious buffalograss Buchloe dactyloides. Mol Ecol 4:135–147
Peterson G, Seberg O (1997) Phylogenetic analysis of the Triticeae (Poaceae) based on rpoA sequence data. Mol Phylogenet Evol 7:217–230
Petit RJ, Aguinagalde I, de Beaulieu J-L, Bittkau C, Brewer S, Cheddadi R, Ennos R, Fineschi S, Grivet D, Lascoux M, Mohanty A, Müller-Stark G, Demesure-Musch B, Palmé A, Martín JP, Rendell S, Vendramin GG (2003) Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300:1563–1565
Quinn JA, Mowrey DP, Emanuella SM, Whalley RDB (1994) The “foliage is the fruit” hypothesis: Buchloe dactiloides (Poaceae) and shortgrass prairie of North America. Am J Bot 81:1545:1554
Redingbaugh MG, Jones TA, Zhang Y (2000) Ubiquity of the St chloroplast genome in St-containing Triticeae polyploids. Genome 43:846–52
Reeder JR (1971) Notes on Mexican grasses IX Miscellaneous chromosome numbers. Britannia 23:105–117
Reeder JR, Reeder CG (1963) Notes on Mexican grasses II Cyclostachya, a new dioecious genus. Bull Torrey Bot Club 90:193–201
Reeder JR, Rzedowski J (1965) Notes on Mexican grasses III Buchlomimus, another dioecious genus. Britannia 17:26–33
Roder MS, Korzun V, Wendehake K, Plaschke J, Tixier M, Lorey P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Rohlf JF (2000) NTSYS-pc: numerical taxonomy and multivariate analysis system. Exeter Software, Setauket, New York
Sneath PHA, Sokal RR (1973) Numerical taxonomy. Freeman, San Francisco
Soltis DE, Soltis PS (2003) The role of phylogenetics in comparative genetics. Plant Physiol 132:1790–800
Weir BS (1996) Genetic analysis II. Sinauer Publishers, Sunderland, MA
Weising K, Nybon H, Wolff K, Meyer W (1995) DNA fingerprinting in plants and fungi. CRC Press, Boca Raton
Wendel JW (2000) Genome evolution in polyploids. Plant Mol Biol 42:225–249
Wu L, Lin H (1984) Salt tolerance and salt uptake in diploid and polyploid buffalograsses (Buchloë dactyloides). J Plant Nutr 17:1905–1928
Wu J, Konstantin VK, Strauss SH (1998) Abundant mitochondrial genome diversity, population differentiation and convergent evolution in pines. Genetics 150:1605–1614
Yaneshita M, Ohmura T, Sakamura T, Ogihara Y (1993) Phylogenetic relationships of turfgrasses as revealed by restriction fragment analysis of chloroplast DNA. Theor Appl Genet 87:129–135
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by B. Friebe
Rights and permissions
About this article
Cite this article
Budak, H., Shearman, R.C., Gulsen, O. et al. Understanding ploidy complex and geographic origin of the Buchloe dactyloides genome using cytoplasmic and nuclear marker systems. Theor Appl Genet 111, 1545–1552 (2005). https://doi.org/10.1007/s00122-005-0083-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-005-0083-3