Abstract
In many plant/pathogen interactions, host susceptibility factors are key determinants of disease development promoting pathogen growth and spreading in plant tissues. In the Fusarium head blight (FHB) disease, the molecular basis of wheat susceptibility is still poorly understood while it could provide new insights into the understanding of the wheat/Fusarium graminearum (Fg) interaction and guide future breeding programs to produce cultivars with sustainable resistance. To identify the wheat grain candidate genes, a genome-wide gene expression profiling was performed in the French susceptible wheat cultivar, Recital. Gene-specific two-way ANOVA of about 40 K transcripts at five grain developmental stages identified 1309 differentially expressed genes. Out of these, 536 were impacted by the Fg effect alone. Most of these Fg-responsive genes belonged to biological and molecular functions related to biotic and abiotic stresses indicating the activation of common stress pathways during susceptibility response of wheat grain to FHB. This analysis revealed also 773 other genes displaying either specific Fg-responsive profiles along with grain development stages or synergistic adjustments with the grain development effect. These genes were involved in various molecular pathways including primary metabolism, cell death, and gene expression reprogramming. An increasingly complex host response was revealed, as was the impact of both Fg infection and grain ontogeny on the transcription of wheat genes. This analysis provides a wealth of candidate genes and pathways involved in susceptibility responses to FHB and depicts new clues to the understanding of the susceptibility determinism in plant/pathogen interactions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat (Triticum aestivum L.) has become the second largest cereal crop worldwide just after rice. Used as a main source of protein and starch for human consumption, animal feed, industrial raw materials, and biofuels, its yield is threatened by several diseases such as Fusarium head blight (FHB) primarily caused by the Fusarium graminearum (Fg) fungus (teleomorph Gibberella zeae, Schwabe). FHB is a devastating disease affecting many cereals with small grains (McMullen et al. 1997) and one of the main causes of starch and storage protein losses in grains, which leads to severe decrease in grain quality (Sutton 1982). Furthermore, FHB constitutes a serious threat to human and animal health because of the production and accumulation of mycotoxins such as deoxynivalenol (DON) (Tuite et al. 1990). The ineffectiveness of the current tools in the prevention and the control of the FHB disease, as well as the absence of major resistance genes despite the hundreds of identified quantitative trait loci (QTLs) (Buerstmayr et al. 2003: Li et al. 2011; Zhu et al. 2012), requires to deepen our knowledge regarding the biology of this specific interaction and on the molecular cross-talk promoting Fg invasion. Unlike resistant cultivars, in the susceptible ones, Fg fungus could take advantage of host susceptibility factors to successfully complete its infectious cycle (Chetouhi et al. 2015). Up to now, the factors contributing to the Fg growth in wheat are still poorly understood while several host genes necessary for pathogen growth were already reported in other pathosystems (Jia et al. 2011; González-Lamothe et al. 2012). A loss of functional mutations of such genes has already been successful in providing durable and broad-spectrum plant resistance, making the susceptibility genes (S-genes) a promising source of resistance in breeding strategies. More than 30 potential host susceptibility factors, validated through either transient knockdown or over-expression of the corresponding genes, were reported in the literature (Pavan et al. 2010). Among S-genes, PMR6 and MLO genes were coding for susceptibility factors promoting growth of powdery mildews; ADH1 gene modulates the susceptibility of Hordei spp. plant to Bulmeria graminis fungus (Pathuri et al. 2011); and, recently, IAA-Asp gene was identified as susceptibility gene which promotes the development of Botrytis cinerea in Arabidopsis thaliana (González-Lamothe et al. 2012). Furthermore, studies have shown that some S-genes may be essential for the development of the plant as well as for the pathogen development and its installation. One example is the rice Xa13 gene which is essential for the growth of the Xanthomonas oryzae bacteria and for plant pollen development (Chu et al. 2006). Recently, Brewer et al. (2014) showed that the homoserine kinase DMR1 mediated susceptibility mechanisms that occur during infection of A. thaliana by both F. culmorum and F. graminearum.
Although increasing knowledge is available, identifying S-genes still requires a better understanding of the molecular determinism of the plant-pathogen interacting system, including genome-wide approaches. Among transcriptomic studies performed on the wheat-Fg pathosystem, most provide a limited picture of the whole infection dynamics, prioritizing either on different stages of disease establishment or on organ-specific responses (Golkari et al. 2007; Bernardo et al. 2007; Erayman et al. 2015). Although grains constitute the main target of FHB disease, no study has been interested in identifying and understanding Fg-induced transcriptome adjustments along with the fluctuating molecular context of grain development. In healthy plants, grain ripening involves a fine-tuned control of cellular processes (Rogers and Bendich 1994; Evers and Millar 2002; Nadaud et al. 2010) that impacts grain physiology. The formation of the grain is defined by three specific phases: (i) the cell division and differentiation stage (0–165 °Cd after anthesis) includes the formation of the main cell types (transfer cells, aleurone, starchy endosperm, and cells surrounding the embryo); (ii) the grain filling stage (195–450 °Cd after anthesis ) starts with the deposition of starch and gluten proteins in endosperm cells; and (iii) the desiccation/maturation phase (Wan et al. 2008). The transition between these contrasted phases is finely regulated by gene expression and comes along with increasing grain size and weight. Abiotic stresses, such as heat or drought, are known to broadly impact wheat grain physiology (Ashraf 2014), but little is known about the molecular events occurring in Fg-infected grains. Recent publications refer to a possible link between the development of the grain and the development of FHB (Dornez et al. 2010; Chetouhi et al. 2015 ; Walter et al. 2015), while the whole molecular mechanisms involved in the colonization of the grain are still unknown.
Here, we report a comprehensive view of the mechanisms and main metabolic pathways impacted by FHB disease. Molecular changes were specifically traced in grain tissues of a susceptible wheat cultivar to gain new insights into plant susceptibility and related susceptibility factors. Using the GeneChip® Wheat Genome NimbleGen micro-array, we performed a genome-wide expression profile analysis at different pivotal stages of the wheat grain formation: the cell division stage (50 °Cd), the cell differentiation stage (150 °Cd), the beginning of the grain filling stage (250 °Cd), and the end of the grain filling (350, 450 °Cd). A special attention has been paid for detecting development-dependent gene expression remodeling as a way to provide clues about the identification of susceptibility genes.
Materials and methods
Plant growth and Fusarium graminearum inoculation
During 2009 and 2010, a winter wheat cultivar, so-called Recital, was grown in a greenhouse at the INRA Research Center of Clermont-Ferrand (France) without treatment and pesticide protection. Inoculation (i.e., infection) of wheat ears by the F. graminearum Fg1 strain was done at the flowering stage (~14 weeks after sowing), especially at mid-anthesis, when about 50 % of the stamens were visible. The inoculum (50 μl at 105 spores per milliliter) was deposited by spraying on both sides of the ears. A mock treatment was realized by spraying water and served as negative control. After spraying, the control and the Fg-inoculated ears were covered with a bag for 2 h to maintain sufficient moisture and to promote attachment of the Fg1 spores. The infection process was surveyed during 450 °Cd (corresponding to about 26 days) after the Fg treatment and especially at five key grain developmental stages including cell division (50 °Cd), cell differentiation (150 °Cd), and grain filling stages (250, 350, and 450 °Cd) (Fig. 1a). As a whole, ten infected and ten control ears were randomly harvested at each stage, pooled, and immediately frozen in liquid nitrogen. The grains of negative control and Fg-inoculated ears were dissected, ground under liquid nitrogen, and stored at −80 °C.
Dynamics of FHB development on grains of a susceptible wheat genotype. a Typical symptoms during susceptibility response to Fg in wheat grains (cv-Recital) at different development stages of the grain (50, 150, 250, 350, and 450 °Cd), the inoculation was performed at mid-anthesis; b FgIGS relative quantification using qPCR method during the time course infection; c Log2 relative expression of the Tri5 (trichodiene synthase) gene during the time course using the qRT-PCR method. Values are expressed as the ratio of treated versus control plants per fungal mass unit
Total RNA extraction, double-stranded cDNA synthesis, and microarray design
Total RNA was extracted from 250 mg of the grain powder sample for each negative control and Fg-inoculated samples, using the method described by Bogorad et al. with few adjustments (Bogorad et al. 1983). The RNA was treated with TURBO DNase (Ambion, AM1907), and its integrity was assessed using the RNA 6000 Nano Assay bioanalyzer (Agilent). cDNA was synthesized from total RNA using the SuperScript Kit (Invitrogen) and labeled with fluorescent Cy3 and Cy5 dyes using a Two-Color DNA Labeling Kit (NimbleGen). Hybridization was performed using the NimbleGen Hybridization Kit, according to the manufacturer’s instructions. Two technical replicates were achieved using either the Cy3 or Cy5 dye for each biological replicate. A wheat NimbleGen microarray (ref. A-MEXP-1928) was used for gene expression profile generation. Each microarray included 39,019 unigenes with three different probes per unigene, 78 with two probes and 82 with one probe.
Microarray data normalization and statistical analysis
Microarray images were obtained with NimbleGen MS200 Scanner. Scanning arrays were read with Deva software (v1.2.1). Microarrays were analyzed using the limma bioconductor package in the R environment (Smyth 2004). To control for any technical variation, the raw data were first normalized within arrays using the loess package/method and then normalized between arrays using the Aquantile method (Smyth and Speed 2003). To detect transcripts displaying significant abundance changes during the infection dynamics and to identify their connections with changes involved in grain development, we designed a two-way analysis of variance (ANOVA) using a linear model including the infection and the grain developmental stages as grouping factors. The model used was Y ijk = μ + T i + D j + T i × D j + ε ijk , where Y ijk refers to individual transcript abundance value, μ is the general mean, T i is the effect of the treatment (i = 2; control or infected), D j is the effect of the development stage (j = 5, 50, 150, 250, 350, or 450 °Cd), T i × D j is the infection by development stage interaction, and ε ijk is the residual. For each individual transcript, a Q value was calculated from the resulting p values of each effect (T i , D j , and T i × D j ). A 0.05 threshold was defined to deem transcript abundance changes significant (corresponding to a positive false discovery rate at a 5 % level) (ESM Table S1a and b).
Gene ontology annotation of significant genes
The function of differentially expressed genes was annotated by performing a Blast search with the Blast2GO software, against the non-redundant (nr) protein sequence database using default parameters (Conesa et al. 2005). The Blast2GO software (v1.3.3) was used to obtain gene ontology (GO) information retrieved from database matches. GOslim “goslim_plant.obo” was used to achieve specific GO terms by means of a plant-specific version of the GO. GO-annotated datasets were represented at level 2. unigenes belonging to each group and to each cluster were used to collect the corresponding probe set in the Wheat Affymetrix GeneChip (http://www.affymetrix.com). The corresponding GOiD for each probe set were then identified from the Blast2Go home page (http://bioinfo.cipf.es/b2gfar/affychips:wheat) and used for GO enrichment analysis. In our analysis, the wheat Affymetrix chip annotated by the Blast2Go team was used as a reference (annotated array) to perform Fisher’s exact tests. GO terms with a p value lower than 0.05 were considered to be significantly different and enriched within the cluster (ESM Table S2a–d).
Mapping of significant genes on wheat chromosomes
The localization of 1309 significant genes on the different wheat chromosomes was obtained by aligning the sequence of each significant unigene against wheat chromosome survey sequences generated by the International Wheat Genome Sequencing Consortium (IWGSC) and available on URGI website (https://urgi.versailles.inra.fr/blast). Blastn results were parsed using a cutoff of 95 % identity over a minimum of 200 bp.
PCR analyses on Fusarium graminearum and wheat targets
Relative F. graminearum biomass quantification and FgTri5 gene expression were performed on genomic DNA and total RNA extracts, respectively, and analyzed using qPCR as described in Chetouhi et al. (2015). Wheat targets were analyzed using qRT-PCR on RNA extracts obtained from three independent biological replicates of wheat infected grains. The RNase L inhibitor (TaRLI) gene that displays no expression variation between Fg-inoculated and control wheat samples was used as a reference gene (Giménez et al. 2011) and was amplified along with the target gene allowing normalization of gene expression. Real-time PCR products were detected using the SYBR Green Supermix (Roche) following the manufacturer’s recommendations. One microgram of total RNA was reverse-transcribed into cDNA with oligo(dT) using iScript reverse transcriptase (Bio-Rad) after treatment with amplification grade DNase I (Invitrogen). Four microliters of a 30-fold dilution of this cDNA and 250 nM of each primer (ESM Table S3) were used as a template for qRT-PCR. The PCR cycling conditions included an initial denaturation step of 95 °C for 10 min followed by 55 cycles of 95 °C for 15 s and 60 °C for 1 min. A melting curve analysis was performed at the end of the PCR run over the range 55–95 °C, increasing the temperature stepwise by 0.5 °C every cycle. Baseline and threshold cycles (Ct) were determined automatically using Optical System Software (LightCycler, Roche). Zero template controls were included for each primer pair, and each PCR was performed in triplicate. The PCR efficiencies of target and reference genes were determined by generating standard curves, and the relative expression values were calculated using the ΔCt method (Pfaffl 2001).
Results
Fusarium graminearum settled in grain of a susceptible wheat genotype along with a burst of mycotoxin synthesis and with two distinct spurts of growth
FHB disease was tested using the susceptible French wheat cultivar so-called Recital and the mycotoxigenic fungus, F. graminearum. In this experiment, the dynamics of Fg infection on wheat spikes was surveyed over five pivotal stages of the grain development from 50 to 450 °Cd. The earliest visible symptoms were noticed at the cell differentiation stage of the grain (150 °Cd) while severe damages, including the bleaching of the spikelets and shrunken grains, were detected at the grain filling stage (250 °Cd; Fig. 1a; Fig. S1). The quantification of Fg fungal mass was traced by qPCR method using FgIGS and revealed that Fg did not grow continuously over the infection process. Fg experienced two spurts of growth, the first one occurring at the onset of the symptoms, concomitantly with the beginning of the grain filling stage (>4-log increase between 150 and 250 °Cd, Fig. 1b), and the second one at the end of the latter stage (five times increase between 350 and 450 °Cd, Fig. 1b). Besides these events, no significant change in Fg growth happened even over periods lasting up to 5 days (duration corresponding to about 100 °Cd). Fgtri5 gene expression per fungal mass unit increased up to 17 times during 50 and 150 °Cd just before the first spurt of growth while no expression signal has been detected in the range of 250 to 450 °Cd (Fig. 1c). Our results evidenced that the infection by F. graminearum involved at least three main phases on wheat grain: (i) a first phase until reaching 150 °Cd, during which Fg is likely to enter the grain along with substantial mycotoxin production; (ii) a second phase at the turning point of 150 to 250 °Cd, characterized by a strong and rapid Fg growth; and (iii) a last phase starting from 250 °Cd during which Fg develops independently to the Tri5 gene expression.
Large-scale grain transcriptomics revealed different Fg-responsive patterns connected with grain development stages
A wheat (Triticum aestivum) NimbleGen 40 K unigene microarray was used to assess changes in gene expression profiles upon Fg infection during grain development. This microarray included 39,179 wheat genes that correspond to nearly 42 % of the whole set of genes assigned to the three component genomes (A, B, and D) of the hexaploid wheat (Brenchley et al. 2012). To depict a comprehensive picture of the Fg-responsive genes and their connections with grain development stages, we designed two-way ANOVAs to compute the significance of the variance across Fg treatment (T), development stages (D), and their interaction effects (T × D). This model was used to efficiently trace different populations of responsive genes: (i) genes displaying Fg-responsive pattern without any impact from the development stage (T genes); (ii) genes displaying Fg-responsive pattern that occurs along with the basal adjustments of grain development (T + D genes; unigenes showing both significant T and D effects); and (iii) genes displaying Fg-responsive patterns that depend on the grain development stage (T × D genes, unigenes showing a significant T × D effect at least).
Statistical analyses were computed on the 39,179 unigenes. Using a pFDR significance threshold of 5 %, we identified a total of 13,813 unigenes that display at least one significant effect in the two-way linear model, with 12,504 unigenes deemed significant for the D effect alone (Fig. 2) (ESM Table S1a and b). These D genes reached up to 30 % of the 39,179 unigenes and depict differentially expressed genes involved especially in wheat grain development. This proportion of D genes corroborates previously published results showing that 24 % of the total NimbleGen chip unigenes were involved in wheat grain development (Capron et al. 2012). Focusing on Fg-related responses, we identified 1309 unigenes (~3.5 % of the whole set of wheat unigenes) that displayed a significant change upon Fg inoculation. These included 536 T genes, 601 T + D genes, and 172 T × D genes (Fig. 2). For each gene population, the Blast2GO software was used to collect functional GO annotations. As a whole, 646 GO annotations were identified for the T genes, 695 for the T + D genes, and 188 for the T × D genes (ESM Table S2a–c). A Fisher’s exact test (significant threshold for BH adjusted p values set to 0.05) was applied to test whether these GO showed any enrichment for a given biological process. This evidenced a significant enrichment for 35 GO biological processes; 20 were found in T genes, 21 in T + D genes, and 6 in T × D genes. Interestingly, part of the enriched GO biological processes was specific to each gene population (ESM Table S2d). T genes were mostly enriched for GO terms associated with defense responses to fungus, general stress responses, translational initiation, and brassinosteroid metabolic process. T + D genes grouped GO terms mainly related to GTP catabolic process, histone lysine methylation, response to growth hormone stimulus, and embryo development. T × D genes were enriched toward responses to pathogen and drug transmembrane transport.
To check up on the microarray data, a number of genes exhibiting significant changes after Fg treatment were selected from different functional categories, and their expression was measured in control and in infected tissues using quantitative RT-PCR. The qRT-PCR profiles of these genes exhibited the same trends as those observed in the microarray (Fig. S2).
T genes were mainly defined by increasing transcript abundances upon F. graminearum infection
Considering the 536 T genes, 492 genes were up-regulated in response to Fg infection while no change has been detected between consecutive developmental stages (ESM Table S4a). These set of genes belonged to roughly 20 GO terms including defense response to fungus, glutathione metabolic process, ATP catabolic process, response to jasmonic acid stimulus, killing of cells of other organisms, and brassinosteroid metabolic process (ESM Table S4b). They also gathered 79 genes without functional annotation and 40 hypothetical proteins. Noticeably, 40 other genes were involved in mycotoxin detoxification (e.g., 3 genes encoding UDP-glycosyltransferases, 15 glutathione S-transferases, 7 cytochromes p450, and 6 PDRs), 28 genes encoded different ribosomal proteins, 4 corresponded to pathogenesis related protein, 4 encoded disease resistance proteins, 4 referred to peroxidases, and 5 referred to cytokinin-O-glucosyltransferases. Regarding the 44 remaining T genes displaying a decrease of transcript abundance (ESM Table S4b) in response to Fg infection, they were mainly involved in translational initiation and response to stimulus (ESM Table S4c).
T + D genes revealed four contrasting patterns of gene expression in response to Fg infection
The 601 T + D genes depicted a significant Fg infection effect that occurs along with the basal adjustments of grain development. To deepen into the Fg-responsive patterns, genes were classified in different groups regarding gene expression changes. Among T + D genes, 344 depicted significant T effect along with a fluctuating gene expression during grain development while the 253 others showed a significant linear regression between their transcript abundance change upon Fg inoculation and the development stage, in control samples at least (p < 0.01). Among these 253 unigenes, 232 displayed a negative linear regression (i.e., continuously down-regulated during grain development in healthy grains) whereas 21 displayed a positive linear regression (i.e., up-regulated during grain development in healthy grains) (Table S5a-d). On this basis, we characterized four contrasting groups.
-
Group I: down-regulated genes during development but up-regulated in response to Fg, This group gathered 68 genes that were mainly involved in cell wall macromolecule catabolic process and cellular response to phosphate starvation (ESM Table S5a and e). Among genes belonging to group I, the magnitude of their expression changes tended to increase with the stress duration: slight changes were observed at the early stages of infection while the later time points (i.e., 350 and 450 °Cd) were mainly characterized by ≥2-fold increases of transcript abundance.
-
Group II: down-regulated genes during development and in response to Fg. This group gathered 164 genes and was mainly enriched in GO terms related to regulation of transcription, GTP catabolic process, and protein polymerization (ESM Table S5e). Analogously to the group I, the magnitude of decreasing gene expression tended to strengthen with the stress duration. However, the number of down-regulated genes displaying a ≥2-fold change was lesser than the number of genes showing a <2-fold change.
-
Group III: up-regulated genes during development and in response to Fg. Response to stress, negative regulation of translation, and cell killing were the GO-enriched terms for this set of 18 genes. Interestingly, the magnitude of gene expression changes in response to Fg infection increased between 250 and 350 °Cd and reached all genes at 450 °Cd.
-
Group IV: up-regulated genes during development but down-regulated in response to Fg. This group included only three genes: Ta_S13050011, Ta_S24623180, and Ta_S26027950. These genes were involved in ubiquitin-dependent protein catabolic process or in amine metabolic process (ESM Table S5e). These down-regulated genes displayed a fold change of >2 at 450 °Cd.
T × D genes reveal ontogenic-specific patterns and early responsive genes to F. graminearum infection
T × D genes are expected to depict changes in mRNA levels in response to Fg treatment but with contrasting trends according to the different developmental stages. They included 172 genes showing at least a 1.5-fold change in relative expression between inoculated and control grains in at least one of the five time points of the time course (ESM Table S6). Among these genes, 125 were successfully annotated while the remaining failed to match any known function. Annotated genes included signaling and hormone-related processes (17 %), sugar catabolism, transport and glycolysis (9 %), gene regulation (13 %), drug response (10 %), stress response (6 %), amino and fatty acid metabolism (5 %), secondary metabolism (2 %), and cellular component organization (2 %). In order to obtain an overview of expression profiles and to identify genes with specific patterns of expression along with grain development, we used a hierarchical complete linkage cluster analysis (Fig. 3). Five main clusters were identified (Fig. 3a). Cluster I included genes that were sharply down-regulated in the late responses to Fg. These genes are involved in signaling, gene regulation, and starch synthesis (Fig. 3b). Cluster II contained early-responsive genes displaying high transcript abundance at 50 °Cd only. Eleven genes belonged to this cluster, among which eight were of unknown function and three were involved in signaling and hormone metabolism. Clusters III and V included up-regulated genes belatedly in response to Fg infection. These clusters were enriched in genes involved in drug response, sugar catabolism, transport, glycolysis, and gene regulation. Finally, cluster IV contained genes specifically down-regulated at 250 °Cd but up-regulated at 350 and 450 °Cd (Fig. 3b). No obvious enrichment for any biological process was observed in this cluster.
Genome distribution and chromosome location of Fg-responsive genes
In order to identify chromosome-specific responses to Fg in the wheat grain, the sequence of each significant unigene was blasted against each chromosome arm sequence (Fig. 4) (ESM Table S7). Among the 1309 blasted sequences, 875 matched significantly. This blast showed a 4 % over-representation of Fg-responsive genes in the D component genome, mainly due to chromosomes 7D and 5D. In contrast, the A genome involved a lesser number of genes especially in chromosomes 6 and 7 while the B genome was over-represented only for chromosome 3. This result may be explained by the length of this chromosome. All genes with significant Fg response mapped in chromosomes 5D and 7D are listed in Supplementary Data (ESM Table S8).
Discussion
The identification of regulated genes in the susceptibility responses to FHB in wheat grain represents a major challenge for understanding the molecular determinism of disease occurrence. While few data are available about plant genes involved in the susceptibility to fungal pathogens (Eichmann et al. 2010), many studies suggest their potential as a new breeding strategy to achieve durable and broad spectrum resistance (Pavan et al. 2010). The study of the host molecular mechanisms contributing to the host susceptibility and to the fungal growth on the host tissues is a prerequisite. Here, we report the first use of wheat microarray in the compatible interaction between the wheat grain and Fg pathogen with a particular focus on cellular alterations of a susceptible host as a way to trace putative susceptibility factors. Although several transcriptomic analyses on this hemibiotrophic fungus revealed that the transition from biotrophic phase to necrotrophic phase required the specific expression pattern of several genes (Yang et al. 2013), little is known about the progress of the infection process of Fg, especially on wheat grains. In the current study, we combined the survey of Fg growth, its mycotoxin synthesis, and plant transcriptomic responses to characterize the evolution of FHB in wheat. Our results showed three different phases in the progress of the compatible wheat grain/Fg interaction (Fig. 5). During the first one, Fg involved widely the DON synthesis with slow spreading into plant tissues while wheat grain was slightly impacted, as suggested by the few detected symptoms along with the faint remodeling of grain gene expression. During the second phase, Fg favored spreading through a spurt of growth while reducing DON synthesis. At this stage, wheat experienced increasing symptoms and DON accumulation induced the activation of plant DON detoxification mechanisms through changing expression of 14 cytochrome P450s (Ito et al. 2013), 5 UGTs (Poppenberger et al. 2003), 10 ABC transporters, and 11 PDR genes (Fig. 5) (Table 1). Interestingly, the genes belonging to these families showed a similar pattern in their expression that suggests the co-regulation of all these genes under DON synthesis. The last phase was characterized by a second spurt of Fg growth without any connection to DON synthesis while wheat underwent deep changes in carbohydrate metabolism as suggested by the altered expression of starch synthase, sucrose synthase, alcohol dehydrogenase genes, and other genes involved in the tricarboxylic acid (TCA) cycle (Fig. 5) (Table 1). This result suggests that this fungus growth could be due to plant metabolism inputs and/or signals such as susceptibility factors.
Digging into transcriptome responses yields putative susceptibility factors to Fusarium head blight
The susceptibility of plants to pathogens (bacteria, fungi, virus, and nematode) involves several susceptibility factors (Lapin and Van den Ackerveken 2013). During the last decade, increasing identification of so-called host susceptibility genes suggests their involvement in many plant-pathogen interactions including the wheat/Fg pathosystem (Ma et al. 2006). In the current study, we investigated susceptibility factors using a transcriptomic approach. Our analysis allowed the identification of three groups of genes that were controlled upon Fg infection. The first group called “T genes” includes genes specifically involved in the generic response to Fg infection without any connection with the grain development stages. This mainly gathered genes that respond either to biotic and abiotic stresses such as PR proteins or to toxins such as GST, CytP450, and UGT. These genes may respond to any stress emphasizing that this group includes a number of genes that display low adaptive value to FHB-specific events. Therefore, this group of genes could have limited interest to identify susceptibility factors, especially since the majority of these genes are also activated in resistant genetic backgrounds (Buerstmayr et al. 2003; Kong et al. 2005; Bernardo et al. 2007). The second group of genes showed both development and Fg infection effects so called “T + D genes.” This group includes patterns depicting an impact of the infection along with basal adjustment of grain developmental processes. Interestingly, it gathers genes that exhibit synergistic changes which could sustain grain formation even upon Fg infection. Finally, the third group represents the genes which have development-dependent responses, i.e., Fg-responsive genes whose reaction norm is partly influenced by the molecular context of the grain development stage. These genes called “T × D” may represent the best source for identifying susceptibility genes, because it provides an access to genes actively involved in Fg infection and closely connected to the changing grain metabolic environment. Among the 172 genes displaying T × D interaction, at least 10 unigenes may encode putative susceptibility family genes previously described in the literature. For instance, two of these genes belong to the WRKY transcription factor family such as the WRKY gene identified in barley which promotes plant susceptibility to the fungus Blumeria graminis by the repression of the PAMP-triggered basal defense (Shen et al. 2007). One unigene encoding the MLO protein, a susceptibility gene of wheat to B. graminis fungus (Várallyay et al. 2012), was part of the T × D genes (Table 2). Moreover, two unigenes displaying the same function as the Medicago zinc finger transcription factor were identified in our study. This transcription factor was previously described as a susceptibility gene to Phakopsora pachyrhizi (Ishiga et al. 2013), to Puccinia emaculata, and to Colletotrichum trifolii. This gene plays a role in P. pachyrhizi pre-penetration structure. Two other unigenes orthologous of the A. thaliana Patatin (lipid acyl hydrolase) gene were also identified in our analysis. Patatin gene is required to fungus B. cinerea and bacteria Pseudomonas syringae to infect A. thaliana (La Camera et al. 2009). This gene could provide fatty acid precursors for the biosynthesis of specific oxylipins and differentially affecting resistance to pathogens with distinct lifestyle. A transcription factor from the MYB family was found in our study. This gene may have the same function as MYB3R4 from A. thaliana, identified to be a susceptibility gene to Golovinomyces orontii (Chandran et al. 2013). This gene could promote the enhancement of metabolic demands imposed by the fungus, which acquires all its nutrients from the plant host. Finally, the polygalacturonase gene which confers the susceptibility to B. cinerea fungus in tomato was found in our data (Cantu et al. 2008). Polygalacturonase is one of the cell wall proteins that cooperatively participate in ripening-associated cell wall disassembly. Its suppression reduced the susceptibility of ripening fruit to B. cinerea. Although this list of known susceptibility genes sustains the value of the T × D gene population, complementary approaches will be required to target specific susceptible genes, such as the characterization of different wheat genotypes or tilling mutants (Ma et al. 2006) as well as biochemical investigations to determine the function of each susceptible gene and its contribution to FHB installation and promotion.
FHB fits over grain development processes
Our analysis revealed 172 genes displaying changes in mRNA level in response to Fg treatment but with contrasting response patterns according to the different development stages. This result indicates the high connection of Fg-responsive gene expression with processes related to grain development, suggesting that Fg-specific molecular adjustments depended on grain development or that Fg set up different strategies to fit with different molecular environments of grain formation. One study has previously demonstrated a high relationship between grain development stage and Fg response for the wheat xylanase inhibitors at proteome level (Dornez et al. 2010). They clearly showed that the xylanase inhibitors were more expressed during the soft dough stage (15–25 days post-anthesis (DPA)) than at 5 DPA. TaABCC3, an ABC transporter engaged in the DON response, was also shown to be involved in grain development (Walter et al. 2015), further reinforcing the link between Fg response and grain development. Among the 172 genes displaying T × D interaction, we identified two unigenes that encoded two ABC transporters (Ta_S32696743 (Ta.9295) and Ta_S16058073 (Ta.27443)) corresponding to the TaABCC3 protein (ESM Table S6). In our current analysis, deeper information is emphasized at the transcriptome level and focused on genes displaying a significant Fg by development stage interaction effect. The hierarchical clustering of these genes showed that they displayed different expression profiles between the different time points of the time course (Fig. 3) (ESM Table S6). At the early stages of FHB (50 °Cd), few changes were observed, but the sharpest ones were detected only for hormone-related genes (cluster II, Fig. 3) suggesting that adjustments in cell signaling occur at this time while other early-expected changes such as stress-related processes are altered secondarily from 250 to 450 °Cd (clusters I, III, IV, and V, Fig. 3). This corroborates the already suggested hypothesis that wheat susceptibility to Fg is related to the delay of defense mechanism activation (Ding et al. 2011; Chetouhi et al. 2015). In addition, the phenotypic observations made during the time course experimentation revealed that the grain development was not interrupted (Fig. 1a), which may suggest the role of the grain integrity preservation for appropriate fungal development. Our results suggest that the successful establishment of the FHB on the grain is closely associated with different phases characterized by specific host plant signals and further by the nutrient availability, as already reported for obligate biotrophic and hemibiotrophic fungi (Bouarab et al. 2002).
Genome distribution of significant genes
The chromosome distribution of significant genes revealed that the D genome and especially chromosomes 5 and 7 contributed significantly to the Fg responses (Fig. 4). The highest plasticity detected in the D genome may suggest (i) gene losses in the genomes B and A, (ii) epigenetic modifications which could have led to genome structure reorganization promoting the D genome expression, or/and (iii) the recent apparition of the disease involving a recent adaptation of wheat to this disease. This hypothesis is supported by previous studies evidencing that tetraploid wheat lacking the D genome displayed higher susceptibility to FHB disease and to DON accumulation as compared to the hexaploid wheat cultivars (Limin and Fowler 1985; Ma et al. 2006). Interestingly, differentially expressed genes belonging to the D genome included genes involved in carbohydrate metabolism that were down-regulated while genes involved in DON detoxification and in plant defense were up-regulated (ESM Table S7 and Table S8). At this stage, deepening these results will require the use of ditelosomic lines to identify the genetic effect of different chromosome arms on FHB such as that already done for other characters and stresses (Wanous et al. 2003).
Fusarium graminearum infection induces reprogramming of the host’s primary metabolism, modulation of cell death, and activation of detoxification mechanisms
Transcripts involved in starch and sucrose synthesis were down-regulated specifically from 250 °Cd, suggesting a reduction of de novo carbohydrate synthesis in the wheat grain. This decrease in the synthesis of starch and sucrose in Fusarium-damaged grains could have a drastic impact on the grain filling stage, while healthy grains are gradually filled in starch and storage proteins (Fig. 5) (Table 1) (Chetouhi et al. 2015). This defect in the grain filling stage may explain the decreased size of infected grains. On the other hand, the activation of glycolysis in infected grains suggests that infection by Fusarium promotes high production of glucose which could be used by the fungus for its growth and dissemination in the ear. Transcript abundance changes in starch and sucrose metabolism have already been reported in susceptible wheat genotypes submitted to a 12-h-long Fg infection (Erayman et al. 2015); such early adjustments could in part support the changes we detected at later stages of the infection. Moreover, the induction of glucose production was also previously described in the proteome and transcriptome analyses of several plant/pathogen interactions (Kugler et al. 2013). The repression of genes involved in starch synthesis and the induction of genes involved in glycolysis may suggest the reallocation of carbon assimilates into infected grains from all tissues of the ear (rachis, lemma, and palea). Fg infection seems to induce a metabolic switch at 250 °Cd from aerobic to anaerobic fermentation by a high expression of ADH1 gene (Fig. 5) (Table 1). The expression of this gene co-occurs with the apparition of the aleurone layer in the wheat grain. In mature barley grain, it was shown that the aleurone layer contains only ADH1 homodimers, and its gene expression is regulated by an abscisic acid-gibberellic acid interaction (MacNicol and Jacobsen 2001). The crucial role of fermentative metabolism in plant-pathogen interactions was evidenced several times. In A. thaliana, glycolysis, respiration, and fermentation are up-regulated at the site of powdery mildew attack (Pathuri et al. 2011), in Plasmodiophora brassicae infection (Jubault et al. 2013) and in Agrobacterium tumefaciens infection (Deeken et al. 2006). It is speculated that the fermentation is favored under cellular conditions associated with parasitic nutrient acquisition (Chandran et al. 2010; Bergmann and Fleming 2010). A second hypothesis for a positive regulatory of enzyme involved in ethanol fermentation is the accumulation of sugars in the infected grains, which may induce a hypoxia-like response and consequently may force plant cells to switch to fermentative energy metabolism (Koch et al. 2000). Furthermore, wheat genes involved in the TCA cycle were up-regulated during the infection (Fig. 5) (Table 1), which suggests the use of this pathway by Fg in the effective virulence strategy, as observed in some necrotrophic and/or toxin-producing plant pathogens (Brauc et al. 2012; Tsuge et al. 2013). The genes of polyamines synthesis (agmatine and ornithine genes) were also up-regulated from the second phase of Fg growth. Recently, it was observed that the primary GOGAT cycle was redirected toward the production of ornithine and arginine, resulting in the formation of polyamines (Gardiner et al. 2010). A metabolo-proteomics approach revealed the induction of agmatine-to-polyamine conversion (Gunnaiah et al. 2012) which could hypothetically contribute to Fg dissemination and pathogenecity (Gardiner et al. 2009).
Several genes involved in cell death and in apoptosis regulation were also evidenced in our study. Most genes involved in the inhibition of cell death were up-regulated (e.g., Bax inhibitor-1 protein and mlo gene; Ihara-Ohori et al. 2007) whereas the gene involved in its induction (apoptosis-inducing factor) were down-regulated, which would consequently lead to sustain cell functioning. It suggests that apoptosis-like in wheat (PCD) may play a crucial role in the plant colonization process promoted by Fg fungus. Similar observations were reported in A. thaliana cells treated with DON mycotoxins produced by Fg during its infectious cycle (Diamond et al. 2013). The results suggested that mycotoxins synthesized by Fg fungus during the infectious cycle contribute to the inhibition of plant apoptosis-like programmed cell death, thus allowing the accomplishment of the fungal biological cycle. In addition, the high expression of the mitochondrial alternative oxidase (AOX) could maintain the cell viability and provides protection against DON-induced PCD (Desmond et al. 2008). In soybean, the AOX protects cells against hydrogen peroxide-induced cell death (Amora et al. 2000). The over-expression of wheat AOX (Waox 1a) in Arabidopsis results in a decrease of reactive oxygen species production following an abiotic stress (Sugie et al. 2006). The ubiquitin-proteasome system (E3-ligase, F-box proteins) genes involved in immune response, programmed cell death (Hershko and Ciechanover 1998), and in grain development (Capron et al. 2012) were substantially down-regulated after Fg infection (Fig. 5) (Table 1). This supports the hypothesis that Fg response, grain development, and cell death are possibly connected.
Detoxification of xenobiotic in plants involves chemical modifications of the mycotoxins (DON) by enzymes such as UGTs, GSTs, or CYPs and further, additional steps for removing compounds (Coleman et al. 1997). DON detoxification has frequently been proposed as one of the resistance mechanisms of small grain cereals to FHB (Boutigny et al. 2008). In our study, we found that the detoxification mechanisms in wheat grain were activated. Among the detoxification processes, glucosylation of DON into DON-3Glc is the most unknown mechanism (Lemmens et al. 2005; Schweiger et al. 2010). As revealed in our data, a number of genes potentially involved in detoxification processes were identified among the genes differentially regulated during the time course. They include genes encoding putative UGTs, GSTs, cytochrome P450, and ABC transporters suggesting that DON detoxification mechanisms in wheat grains could be very similar to those observed in Brachypodium spp., barley, and in wheat rachis. These data suggest that despite the susceptibility of the cultivar used in this study (Recital) to FHB, genes involved in detoxification mechanisms are still induced agreeing with the detection of the QTLs involved in detoxification of mycotoxins in this genotype (Gervais et al. 2003).
The analysis of the dynamics of wheat gene expression in grain proves to be helpful in identifying genes that are associated with the FHB disease. Our analysis provides a wealth of candidate genes and pathways involved in the FHB response, but their exact role in the disease is still to be defined. These probably include several potential plant targets of F. graminearum effectors which are believed to enable colonization of the grain tissue. Genes and pathways have now to be characterized further regarding their genetic mechanisms and toward their potential interest in breeding.
References
Amor Y, Chevion M, Levine A (2000) Anoxia pretreatment protects soybean cells against H(2)O(2)-induced cell death: possible involvement of peroxidases and of alternative oxidase. FEBS Lett 477:175–180
Ashraf M (2014) Stress-induced changes in wheat grain composition and quality. Crit Rev Food Sci Nutr 54:1576–1583. doi:10.1080/10408398.2011.644354
Bergmann DC, Fleming AJ (2010) From molecule to model, from environment to evolution: an integrated view of growth and development. Curr Opin Plant Biol 13:1–4. doi:10.1016/j.pbi.2009.12.001
Bernardo A, Bai G, Guo P et al (2007) Fusarium graminearum-induced changes in gene expression between Fusarium head blight-resistant and susceptible wheat cultivars. Funct Integr Genomics 7:69–77. doi:10.1007/s10142-006-0028-1
Bogorad L, Gubbins EJ, Krebbers E et al (1983) Cloning and physical mapping of maize plastid genes. Methods Enzymol 97C:524–554. doi:10.1016/0076-6879(83)97160-4
Bouarab K, Melton R, Peart J et al (2002) A saponin-detoxifying enzyme mediates suppression of plant defences. Nature 418:889–892. doi:10.1038/nature00950
Boutigny A-L, Richard-Forget F, Barreau C (2008) Natural mechanisms for cereal resistance to the accumulation of Fusarium trichothecenes. Eur J Plant Pathol 121:411–423. doi:10.1007/s10658-007-9266-x
Brauc S, De Vooght E, Claeys M et al (2012) Overexpression of arginase in Arabidopsis thaliana influences defence responses against Botrytis cinerea. Plant Biol 1:39–45. doi:10.1111/j.1438-8677.2011.00520.x
Brenchley R, Spannagl M, Pfeifer M et al (2012) Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491:705–710. doi:10.1038/nature11650
Brewer HC, Hawkins ND, Hammond-Kosack KE (2014) Mutations in the Arabidopsis homoserine kinase gene DMR1 confer enhanced resistance to F. culmorum and F. graminearum. BMC Plant Biol 14:317. doi:10.1186/s12870-014-0317-0
Buerstmayr H, Steiner B, Hartl L et al (2003) Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. II. Resistance to fungal penetration and spread. Theor Appl Genet 107:503–508. doi:10.1007/s00122-003-1272-6
Cantu D, Vicente AR, Greve LC et al (2008) The intersection between cell wall disassembly, ripening, and fruit susceptibility to Botrytis cinerea. Proc Natl Acad Sci U S A 105:859–864. doi:10.1073/pnas.0709813105
Capron D, Mouzeyar S, Boulaflous A et al (2012) Transcriptional profile analysis of E3 ligase and hormone-related genes expressed during wheat grain development. BMC Plant Biol 12:35. doi:10.1186/1471-2229-12-35
Chandran D, Inada N, Hather G et al (2010) Laser microdissection of Arabidopsis cells at the powdery mildew infection site reveals site-specific processes and regulators. Proc Natl Acad Sci 107:460–465. doi:10.1073/pnas.0912492107
Chandran D, Rickert J, Cherk C et al (2013) Host cell ploidy underlying the fungal feeding site is a determinant of powdery mildew growth and reproduction. Mol Plant-Microbe Interact 26:537–545. doi:10.1094/MPMI-10-12-0254-R
Chetouhi C, Bonhomme L, Lecomte P et al (2015) A proteomics survey on wheat susceptibility to Fusarium head blight during grain development. Eur J Plant Pathol 141:407–418. doi:10.1007/s10658-014-0552-0
Chu Z, Yuan M, Yao J et al (2006) Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev 20:1250–1255. doi:10.1101/gad.1416306
Coleman J, Blake-Kalff M, Davies E (1997) Detoxification of xenobiotics by plants: chemical modification and vacuolar compartmentation. Trends Plant Sci 2:144–151. doi:10.1016/S1360-1385(97)01019-4
Conesa A, Götz S, García-Gómez JM et al (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinforma Oxf Engl 21:3674–3676. doi:10.1093/bioinformatics/bti610
Deeken R, Engelmann JC, Efetova M et al (2006) An integrated view of gene expression and solute profiles of Arabidopsis tumors: a genome-wide approach. Plant Cell 18:3617–3634. doi:10.1105/tpc.106.044743
Desmond OJ, Manners JM, Stephens AE et al (2008) The Fusarium mycotoxin deoxynivalenol elicits hydrogen peroxide production, programmed cell death and defence responses in wheat. Mol Plant Pathol 9:435–445. doi:10.1111/j.1364-3703.2008.00475.x
Diamond M, Reape TJ, Rocha O et al (2013) The Fusarium mycotoxin deoxynivalenol can inhibit plant apoptosis-like programmed cell death. PLoS ONE 8, e69542. doi:10.1371/journal.pone.0069542
Ding L, Xu H, Yi H et al (2011) Resistance to hemi-biotrophic F. graminearum infection is associated with coordinated and ordered expression of diverse defense signaling pathways. PLoS ONE 6, e19008. doi:10.1371/journal.pone.0019008
Dornez E, Croes E, Gebruers K et al (2010) 2-D DIGE reveals changes in wheat xylanase inhibitor protein families due to Fusarium graminearum DeltaTri5 infection and grain development. Proteomics 10:2303–2319. doi:10.1002/pmic.200900493
Eichmann R, Bischof M, Weis C et al (2010) BAX INHIBITOR-1 is required for full susceptibility of barley to powdery mildew. Mol Plant Microbe Interact 23:1217–1227. doi:10.1094/MPMI-23-9-1217
Erayman M, Turktas M, Akdogan G et al (2015) Transcriptome analysis of wheat inoculated with Fusarium graminearum. Front Plant Sci 20:867. doi:10.3389/fpls.2015.00867
Evers T, Millar S (2002) Cereal grain structure and development: some implications for quality. J Cereal Sci 36:261–284. doi:10.1006/jcrs.2002.0435
Gardiner DM, Kazan K, Manners JM (2009) Nutrient profiling reveals potent inducers of trichothecene biosynthesis in Fusarium graminearum. Fungal Genet Biol 46:604–613. doi:10.1016/j.fgb.2009.04.004
Gardiner DM, Kazan K, Praud S et al (2010) Early activation of wheat polyamine biosynthesis during Fusarium head blight implicates putrescine as an inducer of trichothecene mycotoxin production. BMC Plant Biol 10:289. doi:10.1186/1471-2229-10-289
Gervais L, Dedryver F, Morlais J-Y et al (2003) Mapping of quantitative trait loci for field resistance to Fusarium head blight in an European winter wheat. Theor Appl Genet 106:961–970. doi:10.1007/s00122-002-1160-5
Giménez MJ, Pistón F, Atienza SG (2011) Identification of suitable reference genes for normalization of qPCR data in comparative transcriptomics analyses in the Triticeae. Planta 233:163–173. doi:10.1007/s00425-010-1290-y
Golkari S, Gilbert J, Prashar S, Procunier JD (2007) Microarray analysis of Fusarium graminearum-induced wheat genes: identification of organ-specific and differentially expressed genes. Plant Biotechnol J 5:38–49. doi:10.1111/j.1467-7652.2006.00213.x
González-Lamothe R, El Oirdi M, Brisson N, Bouarab K (2012) The conjugated auxin indole-3-acetic acid-aspartic acid promotes plant disease development. Plant Cell 24:762–777. doi:10.1105/tpc.111.095190
Gunnaiah R, Kushalappa AC, Duggavathi R et al (2012) Integrated metabolo-proteomic approach to decipher the mechanisms by which wheat QTL (Fhb1) contributes to resistance against Fusarium graminearum. PLoS ONE 7, e40695. doi:10.1371/journal.pone.0040695
Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67:425–479. doi:10.1146/annurev.biochem.67.1.425
Ihara-Ohori Y, Nagano M, Muto S et al (2007) Cell death suppressor Arabidopsis bax inhibitor-1 is associated with calmodulin binding and ion homeostasis. Plant Physiol 143:650–660. doi:10.1104/pp.106.090878
Ishiga Y, Upplapapti S, Mysore KS (2013) Expression analysis reveals a role for hydrophobic or epicuticular wax signals in pre-penetration structure formation of Phakopsora pachyrhizi. Plant Signal Behav 8, e26959. doi:10.4161/psb.26959
Ito M, Sato I, Ishizaka M et al (2013) Bacterial cytochrome P450 system catabolizing the Fusarium toxin deoxynivalenol. Appl Environ Microbiol 79:1619–1628. doi:10.1128/AEM.03227-12
Jia H, Millett BP, Cho S et al (2011) Quantitative trait loci conferring resistance to Fusarium head blight in barley respond differentially to Fusarium graminearum infection. Funct Integr Genomics 11:95–102. doi:10.1007/s10142-010-0192-1
Jubault M, Lariagon C, Taconnat L et al (2013) Partial resistance to clubroot in Arabidopsis is based on changes in the host primary metabolism and targeted cell division and expansion capacity. Funct Integr Genomics 13:191–205. doi:10.1007/s10142-013-0312-9
Koch KE, Ying Z, Wu Y, Avigne WT (2000) Multiple paths of sugar-sensing and a sugar/oxygen overlap for genes of sucrose and ethanol metabolism. J Exp Bot 51 Spec No:417–427
Kong L, Anderson JM, Ohm HW (2005) Induction of wheat defense and stress-related genes in response to Fusarium graminearum. Genome 48:29–40. doi:10.1139/g04-097
Kugler KG, Siegwart G, Nussbaumer T et al (2013) Quantitative trait loci-dependent analysis of a gene co-expression network associated with Fusarium head blight resistance in bread wheat (Triticum aestivum L.). BMC Genomics 14:728. doi:10.1186/1471-2164-14-728
La Camera S, Balagué C, Göbel C et al (2009) The Arabidopsis patatin-like protein 2 (PLP2) plays an essential role in cell death execution and differentially affects biosynthesis of oxylipins and resistance to pathogens. Mol Plant-Microbe Interact 22:469–481. doi:10.1094/MPMI-22-4-0469
Lapin D, Van den Ackerveken G (2013) Susceptibility to plant disease: more than a failure of host immunity. Trends Plant Sci 18:546–554. doi:10.1016/j.tplants.2013.05.005
Lemmens M, Scholz U, Berthiller F et al (2005) The ability to detoxify the mycotoxin deoxynivalenol colocalizes with a major quantitative trait locus for Fusarium head blight resistance in wheat. Mol Plant-Microbe Interact 18:1318–1324. doi:10.1094/MPMI-18-1318
Li Z, Zhou M, Zhang Z, Ren L et al (2011) Expression of a radish defensin in transgenic wheat confers increased resistance to Fusarium graminearum and Rhizoctonia cerealis. Funct Integr Genomics 111:63–70. doi:10.1007/s10142-011-0211-x
Limin AE, Fowler DB (1985) Cold hardness in Triticum and Aegilops species. Can J Plant Sci 65:71–77. doi:10.4141/cjps85-010
Ma H-X, Bai G-H, Gill BS, Hart LP (2006) Deletion of a chromosome arm altered wheat resistance to Fusarium head blight and deoxynivalenol accumulation in Chinese spring. Plant Dis 90:1545–1549. doi:10.1094/PD-90-1545
MacNicol PK, Jacobsen JV (2001) Regulation of alcohol dehydrogenase gene expression in barley aleurone by gibberellin and abscisic acid. Physiol Plant 111:533–539
McMullen M, Jones R, Gallenberg D (1997) Scab of wheat and barley: a re-emerging disease of devastating impact. Plant Dis 81:1340–1348
Nadaud I, Girousse C, Debiton C et al (2010) Proteomic and morphological analysis of early stages of wheat grain development. Proteomics 10:2901–2910. doi:10.1002/pmic.200900792
Pathuri IP, Reitberger IE, Hückelhoven R, Proels RK (2011) Alcohol dehydrogenase 1 of barley modulates susceptibility to the parasitic fungus Blumeria graminis f.sp. hordei. J Exp Bot 62:3449–3457. doi:10.1093/jxb/err017
Pavan S, Jacobsen E, Visser RGF, Bai Y (2010) Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol Breed 25:1–12. doi:10.1007/s11032-009-9323-6
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29, e45
Poppenberger B, Berthiller F, Lucyshyn D et al (2003) Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J Biol Chem 278:47905–47914. doi:10.1074/jbc.M307552200
Rogers SO, Bendich AJ (1994) Extraction of total cellular DNA from plants, algae and fungi. In: Gelvin SB, Schilperoort RA (eds) Plant molecular biology manual. Springer, Netherlands, pp 183–190
Schweiger W, Boddu J, Shin S et al (2010) Validation of a candidate deoxynivalenol-inactivating UDP-glucosyltransferase from barley by heterologous expression in yeast. Mol Plant-Microbe Interact 23:977–986. doi:10.1094/MPMI-23-7-0977
Shen Q-H, Saijo Y, Mauch S et al (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315:1098–1103. doi:10.1126/science.1136372
Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:1–25. doi:10.2202/1544-6115.1027
Smyth GK, Speed T (2003) Normalization of cDNA microarray data. Methods San Diego Calif 31:265–273
Sugie A, Naydenov N, Mizuno N et al (2006) Overexpression of wheat alternative oxidase gene Waox1a alters respiration capacity and response to reactive oxygen species under low temperature in transgenic Arabidopsis. Genes Genet Syst 81:349–354
Sutton JC (1982) Epidemiology of wheat head blight and maize ear rot caused by Fusarium graminearum. Can J Plant Pathol 4:195–209. doi:10.1080/07060668209501326
Tsuge T, Harimoto Y, Akimitsu K et al (2013) Host-selective toxins produced by the plant pathogenic fungus Alternaria alternata. FEMS Microbiol Rev 37:44–66. doi:10.1111/j.1574-6976.2012.00350.x
Tuite J, Shaner G, Everson RJ (1990) Wheat scab in soft red winter wheat in Indiana in 1986 and its relation to some quality measurements. Plant Dis 74:959–962. doi:10.1094/PD-74-0959
Várallyay É, Giczey G, Burgyán J (2012) Virus-induced gene silencing of Mlo genes induces powdery mildew resistance in Triticum aestivum. Arch Virol 157:1345–1350. doi:10.1007/s00705-012-1286-y
Walter S, Kahla A, Arunachalam C et al (2015) A wheat ABC transporter contributes to both grain formation and mycotoxin tolerance. J Exp Bot 66:2583–2593. doi:10.1093/jxb/erv048
Wan Y, Poole RL, Huttly AK et al (2008) Transcriptome analysis of grain development in hexaploid wheat. BMC Genomics 9:121. doi:10.1186/1471-2164-9-121
Wanous MK, Munkvold JD, Kruse JD et al (2003) Identification of chromosome arms influencing expression of the HMW glutenins in wheat. Theor Appl Genet 106:213–220. doi:10.1007/s00122-002-1098-7
Xiong L, Yang Y (2003) Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid–inducible mitogen-activated protein kinase. Plant Cell 15:745–759. doi:10.1105/tpc.008714
Yang F, Li W, Jørgensen HJL (2013) Transcriptional reprogramming of wheat and the hemibiotrophic pathogen Septoria tritici during two phases of the compatible interaction. PLoS ONE 8, e81606. doi:10.1371/journal.pone.0081606
Zhu X, Li Z, Xu H et al (2012) Overexpression of wheat lipid transfer protein gene TaLTP5 increases resistances to Cochliobolus sativus and Fusarium graminearum in transgenic wheat. Funct Integr Genomics 12:481–488. doi:10.1007/s10142-012-0286-z
Acknowledgments
This work is part of CC PhD work, funded by the French National Institute for Agronomic Research (INRA). We thank members of the PHACC unit for plant preparation, and Philippe Lecomte and Cyrille Saintenac for fruitful discussions.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Symptoms observed during susceptibility responses to Fg at the 250°Cd developmental stage. (a) and (b) represent control and Fg inoculated ears, (c) and (d) represent control and Fg infected grains, respectively. (PDF 60 kb)
Fig. S2
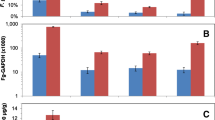
Check up on the microarray data by qRT-PCR. Micro-array (blue) and quantitative RT-PCR (red) data are expressed in a log2 ratio obtained for comparison between Fg-inoculated and control plants at three time points (250°Cd, 350°Cd and 450°Cd). (PDF 57 kb)
ESM 3
(XLSX 17429 kb)
ESM 4
(XLSX 75 kb)
ESM 5
(XLSX 8 kb)
ESM 6
(XLSX 60 kb)
ESM 7
(XLSX 39 kb)
ESM 8
(XLSX 19 kb)
ESM 9
(XLSX 61 kb)
Rights and permissions
About this article
Cite this article
Chetouhi, C., Bonhomme, L., Lasserre-Zuber, P. et al. Transcriptome dynamics of a susceptible wheat upon Fusarium head blight reveals that molecular responses to Fusarium graminearum infection fit over the grain development processes. Funct Integr Genomics 16, 183–201 (2016). https://doi.org/10.1007/s10142-016-0476-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-016-0476-1