Abstract
X1-homologous genes (XHS) encode plant specific proteins containing three basic domains (XH, XS, zf-XS). In spite of their physiological importance, systematic analyses of ZmXHS genes have not yet been explored. In this study, we isolated and characterized ten ZmXHS genes in a whole-of-genome analysis of the maize genome. A total of ten members of this family were identified in maize genome. The ten ZmXHS genes were distributed on seven maize chromosomes. Multiple alignment and motif display results revealed that most ZmXHS proteins share all the three conserved domains. Putative cis-elements involved in abiotic stress responsive, phytohormone, pollen-specific and quantitative, seed development and germination, light and circadian rhythms regulation, Ca2+-responsive, root hair cell-specific, and CO2-responsive transcriptional activation were observed in the promoters of ZmXHS genes. Yeast hybrid assay revealed that the XH domain of ZmXHS5 was necessary for interaction with itself and ZmXHS2. Microarray data showed that the ZmXHS genes had tissue-specific expression patterns in the maize developmental steps and biotic stresses response. Quantitative real-time PCR analysis results indicated that, except ZmXHS9, the other nine ZmXHS genes were induced in the seedling leaves by at least one of the four abiotic stresses applied.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
X1-homologous genes (XHS) encode plant specific proteins containing three basic domains XH, XS, and zf-XS which were named after Arabidopsis SGS3 (Suppessor of Gene Silencing 3) and rice homolog X1 (Mourrain et al. 2000; Bateman 2002; Qin et al. 2009). Among these protein domains, zf-XS motif is a C2H2 type zinc finger domain, the XS motif is an RNA-binding domain and XH motif refers to X-homolog domain with unknown function (Bateman 2002). Furthermore, these protein domains, some of SGS3-like proteins also include a coil–coil domain localized between the XS and XH domains (Ausin et al. 2009; Zheng et al. 2010).
The main region of rice X1 and Arabidopsis SGS3, XS domain, is around 140 amino acid residues in length. The XS domain contains an absolutely conserved aspartate that could present an enzymatic active site (Bateman 2002). The XS domain containing proteins are predicted by ncoils to contain coiled-coils (Lupas et al. 1991), which indicate that they could oligomerise. Like most coiled-coil proteins (which could form a dimeric or a trimeric structure), different members of the XS domain family may oligomerise via their coiled-coils to form a variety of complexes (Bateman 2002). The XH domain is between 124 and 145 residues in length. The XH domain contains one absolutely conserved glutamate which could potentially be an active site or other functionally vital region. XS and XH domains are found in most of these proteins and two domains may interact (Bateman 2002; Xie et al. 2012a). The zf-XS domain is an N-terminal cysteine/histidine zinc binding domain and usually accompanies a XS domain (Bateman 2002).
Up to now, some experimental data have been reported indicating the functions of this family, such as SGS3 in Arabidopsis, OXHS in rice and SISGS3 in tomato (Mourrain et al. 2000; Glick et al. 2008; Qin et al. 2009). SGS3 is essential for post-transcriptional gene silencing and natural virus resistance (Mourrain et al. 2000; Xie et al. 2012b), which are vital for endogenous gene silencing from DNA viruses (Muangsan et al. 2004; Qin et al. 2009) and the production of trans-acting siRNAs in Arabidopsis (Peragine et al. 2004). SGS3 acts upstream of RDR6-dependent dsRNA production (Yoshikawa et al. 2005) and the XS domain probably acts as an RNA recognition motif (Bateman 2002; Zhang and Trudeau 2008; Elmayan et al. 2009). Along with the function of SGS3 in Arabidopsis antiviral response, SISGS3 is targeted by the tomato yellow leaf curl geminivirus PTGS suppressor protein V2 (Glick et al. 2008). Nine rice OXHS genes are responsive to at least one of the abiotic stresses including salt, drought, cold, and abscisic acid treatment (Qin et al. 2009).
These reports in plants suggested that XHS family genes may be crucial not only for plant development but also for response and adaptation to stresses. So, in order to indicate the functions of ZmXHS gens in this family, transcript expression levels of the family genes were investigated in various maize tissues and in the seedling leaves under various abiotic and biotic stresses. The results presented in this study gave an important reference for function studies of XHS family genes in maize.
Materials and methods
Isolation of XHS gene in maize
The sequences of 14 Arabidopsis and 11 rice XHS proteins were downloaded from the TAIR database (http://www.arabidopsis.org) and TIGR database (http://rice.plantbiology.msu.edu) (Qin et al. 2009). To obtain all the XHS genes in maize, BLASTP searches were performed in the Maize sequence database (http://www.maizesequence.org/index.html) and Gramene database (http://www.gramene.org/Multi/blastview) with the Arabidopsis and rice XHS proteins as queries. First, all the corresponding protein sequences of the putative XHS family members were downloaded if it satisfied with E < 10-10 and including XH domain (accession no. PF03469), XS domain (accession no. PF03468), or zf-XS domain (accession no. PF03470). And then, all the candidate proteins were confirmed with the Pfam database (http://www.sanger.ac.uk/Software/Pfam/search.shtml) (Sonnhammer et al. 1997). The full-length cDNA sequences of ZmXHS genes in maize were downloaded from the Maize sequencing database.
Gene structure analysis of maize XHS genes
The information of all ZmXHS genes, including chromosomal location, full-length DNA sequences were obtained from the B73 maize sequencing database (http://www.maizesequence.org/index.html). Exons and introns structures of ZmXHS genes were confirmed by using GSDS (http://gsds.cbi.pku.edu.cn/) (Guo et al. 2007).
Motif display analysis of ZmXHS proteins
Multiple Expectation Maximization for Motif Elicitation (MEME) utility program (Bailey et al. 2009) was used to display domains of ZmXHS, OsXHS, AtXHS, and SbXHS proteins. The matrix for phylogenetic analysis included the 14, 11, 19, and 10 XHS genes from Arabidopsis, rice, sorghum and maize.
Promoter regions analysis of ZmXHS genes
To investigate cis-elements in promoter sequences of ZmXHS genes, 2 kb of B73 genomic DNA sequences upstream of the initiation codon (ATG) were downloaded from NCBI. Promoter structure was predicted using Promoter 2.0 software (http://www.cbs.dtu.dk/services/Promoter/) and PLACE (Plant cis-acting Regulatory DNA Elements, with more than 6 bp) (http://www.dna.affrc.go.jp/PLACE/) (Higo et al. 1999).
Yeast two-hybrid assays
A Gal4-based two-hybrid system was used as described by the manufacturer (Clontech). The full-length and truncated ZmXHS5 (ZmXHS5-1, lacking the XH domain; ZmXHS5-2, XH domain alone) coding region were amplified by PCR with primers containing restriction sites and cloned into the pGADT7 cloning vector (Clontech) to make the bait plasmid pGADT7-ZmXHS. The coding sequences of the ZmXHS2 and ZmXHS5 were cloned into the DNA binding domain vector pGBKT7. The primers and restriction sites used to create these constructs are listed in Supplementary Table 1. Each pGBKT7-ZmXHS was separately co-transformed with pGADT7-ZmXHS5/ZmXHS5-1/ZmXHS5-2 into Saccharomyces cervisiae AH109 by the LiAc-mediated yeast transformation method according to the Yeast Protocols Handbook of Clontech. Co-transformed yeast cells were selected on synthetic medium lacking Leu and Trp and interaction was selected on synthetic medium lacking Leu, Trp, His, and Ade.
Plant materials stress treatment
Seeds of the maize (Zea mays) B73, were surface-sterilized, germinated and hydroponically grown to the three-leaf stage in pots filled with vermiculite, with four seeds per pot. The pots were placed in a greenhouse at 28 °C under 16 h light and 20 °C under 8 h dark and watered once every 3 days. For ABA treatment, leaves were sprayed with 100 μM (+)-cis, trans-ABA and then rapped up with preservative film, followed by sampling at 0, 1, 3, 6, 12, and 24 h. Abiotic treatments concluded cold, NaCl and PEG. For cold stress, the seedlings were put into a growth chamber at 4 °C and sampled at 0, 1, 3, 6, 12, and 24 h, respectively. For NaCl and PEG treatment, the seedlings were carefully removed from the vermiculite and washed clean with tap water and then dried using filter paper. Roots of the seedlings were then submerged in 200 mM NaCl or 20 % PEG (molecular weight 6,000) solution, air conditioned by a pump and sampled at 0, 1, 3, 6, 12, and 24 h, respectively (Zhang et al. 2012).
Microarray data collection and analyses of expression profiles
The expression behaviors of ZmXHS genes were examined in a set of maize transcriptome data at PLEXdb (http://www.plexdb.org). The microarray data of genome-wide gene expression datas of maize inbred line B73 (GSE27004) and the data of transcriptomic analysis of induced senescence in maize were provided by Kaeppler from University of Wisconsin (Sekhon et al. 2011, 2012). Gene expression data from the Affymetrix GeneChip array data during three kinds of fungal infection were downloaded from GEO with accession numbers, GSE31188, GSE19501, and GSE29747, respectively (Ghareeb et al. 2011; Voll et al. 2011). The data were analyzed by GeneSpring 12.5 software. Heat map was used to present the number of ZmXHS genes, and the map was generated using MultiExperiment Viewer (MeV, version 4.8.1) software. The data were adjusted by median centering of genes. The data were clustered by complete linkage clustering method, and a euclidean distance metric was used.
Transcript level analysis ZmXHS genes
Total RNA was prepared using the TRIZOL reagent (Invitrogen, Karlsruhe, Germany) and then purified using the DNase I (RNase free) (TaKaRa, Dalian) according to the manufacturer's protocol. After an initial photometric determination of total RNA concentration, ethidium bromide fluorescence of ribosomal bands on the electrophoresis-gel was used for the control of total RNA integrity and the adjustment of equal RNA amounts. Real-time PCR was performed in an optical 96-well plate with an ABI StepOnePlus Real-Time PCR System. Gene-specific primers were designed for all ten ZmXHS genes (Table 2) and ZmGAPDH transcript served as the control (Kozak 1999). Each reaction contained 10 μl of SYBR Premix Ex Taq (TaKaRa, Dalian), 1.0 μl of cDNA samples, 0.4 μl ROX Reference Dye II and 10 μM gene-specific primers in a final volume of 20 μl. The thermal cycle used was as follows: 95 °C of 30 S, 40 cycles of 95 °C for 5 s and 60 °C for 60 s. For melt curve, 95 °C 15 S, 60 °C for 60 s and 95 °C 15 S. The analysis of real-time PCR data used 2−ΔΔC T method (Livak and Schmittgen 2001).
Results
Identification of XHS family genes in maize
After carefully surveying the maize genome, ten members were defined as ZmXHS genes (Table 1). There was no ZmXHS gene homologous to OXHS3 and OXHS10 but two OXHS8 homologous genes named ZmXHS8a and ZmXHS8b. The length of ZmXHS proteins ranged from 253 aa (amino acids) to 691 aa. BLAST analysis against the Pfam database showed that all of them including XH domain (accession no. PF03469), XS domain (accession no. PF03468), or zf-XS domain (accession no. PF03470) (Table 1).
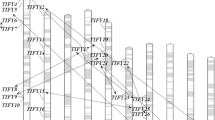
The ten ZmXHS genes were distributed on seven maize chromosomes: chromosome 1 contained two genes; chromosome 3 contained three genes, and chromosome 4, 6, 8, 9, 10 contained one gene, respectively (Fig. 1). Based on phylogenetic results (Supplementary Fig. 1), three paralogs were identified in ZmXHS genes including ZmXHS1/ ZmXHS2, ZmXHS6/ZmXHS7, and ZmXHS8a/ZmXHS8b (Fig. 1). The full-length cDNA sequences were compared with the corresponding genomic DNA sequences to determine the numbers and positions of exons and introns within each ZmXHS gene by using GSDS (http://gsds.cbi.pku.edu.cn/chinese.php) (Guo et al. 2007). The genome structure of the ZmXHS genes showed that most ZmXHSs had six exons, except for ZmXHS8a, 8b, 9 which had three (Fig. 2; Table 2).
Motif analysis and protein architecture
To identify the conserved domains distribution in XHS proteins, the MEME web server was employed to analyze the protein sequences from Arabidopsis, rice, sorghum and maize. Three putative conserved domains were detected in the ZmXHS family, including XH (PF03469), XS (PF03468), and zf-XS (PF03470) which have been identified in the X1 and SGS3 proteins. According to the configuration of the putative domains, the 10 ZmXHS proteins can be divided into four types (Fig. 3). The protein sequences in type I, including ZmXHS1, ZmXHS2, ZmXHS4, ZmXHS5 and ZmXHS6 contained the XH domain, XS domain, and zf-XS domain. Three proteins ZmXHS8a, ZmXHS8b, ZmXHS9 each containing only the zf-XS domain were classified as type II. Type III protein ZmXHS7 contained XH domain and XS domain, while Type IV protein ZmXHS11 contained XS domain and zf-XS domain.
All these three putative conserved domains were present in most of the XHS members of rice, Arabidopsis and sorghum. According to the configuration of the putative domains, the XHS proteins of rice, Arabidopsis and sorghum can be divided into three or four types, respectively (Supplementary Fig. 2) (Qin et al. 2009).
The XH domain of ZmXHS5 is necessary for interacts with itself and ZmXHS2
Our previous study indicated that ZmXHS5 interacts with itself and ZmXHS2 (Supplementary Fig. 3). To identify the protein domains of ZmXHS5 responsible for the interaction, we generated two truncation mutants of ZmXHS5 in pGADT7 (Fig. 4a): lacking the XH domain (ZmXHS5-1), the XH domain alone (ZmXHS5-2). We checked the interaction of these truncated ZmXHS5 mutants with full-length ZmXHS5 and ZmXHS2 using the yeast two-hybrid assay described above. ZmXHS5-2 was able to interact with ZmXHS5 and ZmXHS2, because co-transformation of the two pairs enabled yeast cell to grow in the absence of Ade and His (Fig. 4b). However, ZmXHS5-1 did not interact with ZmXHS5 and ZmXHS2. These results indicated that the XH domain of ZmXHS5 is necessary for ZmXHS5–ZmXHS5 and ZmXHS5–ZmXHS2 interactions.
The XH domain mediates ZmXHS5–ZmXHS5 and ZmXHS5–ZmXHS2 interactions a Schematic structure of the full-length and truncated ZmXHS5 proteins used for yeast two-hybrid assays. ZmXHS5-1, truncated ZmXHS5 protein lacking the XH domain; ZmXHS5-2, XH domain alone; b interaction analyses of truncated ZmXHS5 proteins with ZmXHS5 and ZmXHS2 in yeast AH109 cells. The paired AD and BD fusion constructs were co-transformed into yeast. Positive clones selected on –Leu/–Trp were spotted on –Ade/–Leu/–Trp/-Ade medium
cis-Element analysis
By searching the PLACE database, promoter regions (2kp range B73 genomic DNA sequences upstream of translation start site) of ZmXHS genes were analyzed. Amounts up to 58 putative cis-elements more than 6 bp length were identified (Supplementary Table 2). In this study, a series of cis-elements were found that involved in abiotic stress responsive, phytohormone, pollen-specific and quantitative, seed development and germination, light and circadian rhythms regulation, Ca2+-responsive, root hair cell-specific, and CO2-responsive transcriptional activation.
Among these cis-elements, five kinds were distributed in all the ten ZmXHS genes, i.e., the dehydration-stress responsive element MYBCORE and MYCCONSENSUSAT (Solano et al. 1995; Abe et al. 2003), Ca2+-responsive element CGCGBOXAT (Yang and Poovaiah 2002) and EECCRCAH1 (Guo et al. 2010), ABA-responsive element DPBFCOREDCDC3 (Kim et al. 1997; Finkelstein and Lynch 2000; Lopez-Molina and Chua 2000), light-responsive element EBOXBNNAPA and INRNTPSADB (Stalberg et al. 1996; Hartmann et al. 2005), SA-responsive element GT1CONSENSUS (Villain et al. 1996; Le Gourrierec et al. 1999). Another widely distributed cis-element ABRERATCAL (Kaplan et al. 2006), CO2-responsive element was present in nine of the ten ZmXHS genes. Pollen-specific and quantitative element QELEMENTZMZM13 (Hamilton et al. 1998), phytochrome regulation element REBETALGLHCB21 (Degenhardt and Tobin 1996), root hair cell-specific element RHERPATEXPA7 (Kim et al. 2006), early responsive to dehydration element MYCATERD1 (Tran et al. 2004), fermentative pathway responsive element ANAERO1CONSENSUS (Bratić et al. 2009), also exist in most of the ZmXHS gene promoters.
By controlling efficiency of the promoters, cis-elements played essential function in the regulation of gene expression. Studies on cis-elements could provide vital foundation for further functional research of the ZmXHS gene family.
Expression profiles of ZmXHS family in different tissues and organs
In order to identify the spatial and temporal specific expression patterns of ZmXHS genes, we explored microarray data which record the gene expression levels of 60 tissues varying developmental stages of the maize (Sekhon et al. 2011). It can be seen from the heat map that all of the ten detected genes were involved in numerous biological processes, and expressed in almost tissues, but their expression levels were distinct (Fig. 5). ZmXHS2 had higher expression in reproductive organs such as cob, tassel, whole seed (DAP), endosperm (DAP), and embryo (DAP). The transcript level of ZmXHS4 was detected at the higher level in silks, anthers, sheath, outer husk, and different leaves. ZmXHS8a and ZmXHS9 had similar expression pattern but different from that of ZmXHS8b. ZmXHS8b was involved in the development of vegetative organs such as primary root, stem, SAM, sheath, internode, and innermost husk, while ZmXHS8a and ZmXHS9 had higher expression both in reproductive and vegetative organs, i.e., seed, endosperm, embryo, leaf, stem, and husk. ZmXHS5-7 and ZmXHS11 had similar expression pattern, appearing to be lowly expressed among vegetative organs and moderate expressed among reproductive organs (Fig. 5).
Expression profiles of ZmXHS genes under abiotic stresses
Many plant gene families were involved both in stress and development responses. In order to check whether ZmXHS genes were responsive to stresses in seedling stage, quantitative real-time PCR (qRT-PCR) was conducted to analyze all ten genes' transcriptional expression in shoots at the three-leaf stage treated by exogenous ABA, PEG, NaCl, and low temperature (4 °C).
The results indicated that, except for ZmXHS9, the other nine genes were induced in the seedling leaves by at least one of the four stresses applied (Fig. 6). Among them, six genes (ZmXHS1, ZmXHS2, ZmXHS4, ZmXHS7, and ZmXHS11) were obviously induced by PEG stress with distinct patterns. For example, the transcript levels of ZmXHS1, ZmXHS7, and ZmXHS11 were increased at the early stage of PEG stress and then decreased, while the expression level of ZmXHS2 and ZmXHS4 increased gradually and reached the highest level at the late stage of the stress. ZmXHS2, ZmXHS5, and ZmXHS6 were induced by salt stress, while ZmXHS1 and ZmXHS8b were suppressed by salt stress at 1 h after treatment. Under ABA stress condition, the expression level of ZmXHS4 increased gradually and peaked at 12 h, while that of the other nine gsenes were no obviously initiated. There was only one gene, ZmXHS8b responsive to low temperature (4 °C) treatment. The expression of ZmXHS8b was evidently induced within 1 h after low temperature treatment.
Expression levels of maize XHS genes under each treatment, including ABA, cold, NaCl and PEG stress, based on real-time quantitative PCR. The X-axes are treatment time points and the Y-axes are scales of relative expression level. The transcript level at time 0 h (untreated) was used as the calibrator and was given as 1
Expression profiles of ZmXHS genes under biotic stresses
To discover the ZmXHS (only seven genes were detected) genes involved in biotic stresses, we detected differentially expressed genes with microarray data. Data sets of three experiments under the pathogen treatments of Fusarium moniliforme, Sphacelotheca reiliana, and Colletotrichum graminicola infection have been analyzed. As shown in Fig. 7, ZmXHS2 and ZmXHS11 were downregulated after the infection of the three pathogens. ZmXHS5 and ZmXHS8b were suppressed by C. graminicola and S. reiliana, respectively. ZmXHS7 was suppressed by F. moniliforme and C. graminicola while induced by S. reiliana treatment. ZmXHS1 was upregulated while ZmXHS9 was downregulated by C. graminicola (Fig. 7).
Expression levels of maize XHS genes under biotic stresses. CK1: Samples from uninfected control plants were taken at the same time points of T1. T1 Samples from infected leaves were taken at 36 h post infection. CK2 Samples from uninfected control plants were taken at the same time points of T2. T2 Samples from infected leaves were taken at 96 h post infection
Discussion
X1-homologous genes (XHS) encode plant specific proteins containing three basic domains (XH, XS, zf-XS) (Qin et al. 2009). In this study, ten genes belonging to the XHS in maize were identified. Among the better analyzed plant XHS families, Arabidopsis has likely 14 XHS, rice has at least 11 expressed XHS (Qin et al. 2009), sorghum at least 19 XHS (http://www.gramene.org/Multi/blastview). The genome structure of the ZmXHS genes showed that most them had six exons, except for ZmXHS8a, 8b, 9 which had three (Fig. 2). Most rice and sorghum XHS genes also had six exons, but most Arabidopsis XHS genes had seven or eight exons (Supplementary Fig. 4). Five of ten proteins, ZmXHS1, ZmXHS2, ZmXHS4, ZmXHS5 and ZmXHS6 contained the XH domain which was necessary for proteins interaction. In Arabidopsis, the XH domain of FDM1 was necessary for FDM1–FDM1 and FDM1–IDN2 interactions (Xie et al. 2012a). In our study, the XH domain of ZmXHS5 was necessary for interacts with both itself and ZmXHS2 (Fig. 4).
Increasing evidence indicated that some gene families had vital roles both in development and response to stress (Qin et al. 2009). Examples were the genes from CBL-interacting protein kinase (CIPK) and F-BOX family and many transcription factor families such as NAC, MYB, and bZIP (Urao et al. 1993; Shi et al. 1999; Liu et al. 2000; Durfee et al. 2003; Hu et al. 2006; Kim 2006).
The members of XHS gene family in rice showed tissue-specific expression patterns. All the 11 OXHS genes were observably expressed in floral organs, and some were expressed in a wide range of different organs in rice (Qin et al. 2009). Like that in rice, the transcript levels of the ten ZmXHS genes were all had different expression in reproductive and vegetative organs (Fig. 5). The XS domain containing proteins are involved in a wide range of processes such as viral defense and stress response (Qin et al. 2009). In Arabidopsis, SGS3 was required for post-transcriptional gene silencing, natural virus resistance and sgs3 mutant shown enhanced susceptibility to CMV (Mourrain et al. 2000). In tomato, SISGS3 was specifically required for the RNA-silencing defense against geminiviruses (Glick et al. 2008). In our work, most of ZmXHS were up or down regulated by the pathogen treatments of F. moniliforme, S. reiliana, and C. graminicola. In rice, nine OXHS genes were responsive to at least one of the abiotic stresses including drought, salt, cold, and abscisic acid treatment. Over-expression of the gene OXHS2 in rice resulted in reduced tolerance to salt and drought stresses (Qin et al. 2009). In our study, except ZmXHS9, the other nine genes were induced in the seedling leaves by at least one of the four stresses PEG cold, NaCl, or ABA (Fig. 6). In cluster ІII of phylogenetic tree (Supplementary Fig. 5), ZmXHS2 was the closest homologue to OXHS2, and the both proteins have strong similarity in domain composition, suggested that ZmXHS2 might be an orthologue of OXHS2 in maize. Both genes were induced by drought and salt treatment (Fig. 6) (Qin et al. 2009). These results indicated that ZmXHS2 might have the similar function to OXHS2.
Abbreviations
- ABA:
-
Abscisic acid
- bZIP:
-
Basic leucine zipper
- DAP:
-
Day after pollination
- ORF:
-
Open reading frame
- PEG:
-
Polyethylene glycol
- RDR6:
-
RNA-dependent RNA polymeraes 6
- XH:
-
X1 homologue
- XHS:
-
XH and XS domain
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
Ausin I, Mockler TC, Chory J, Jacobsen SE (2009) IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat Struct Mol Biol 16:1325–1327
Bailey TL, Boden M, Buske FA, Martin F, Grant CE, Clernenti L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:202–208
Bateman A (2002) The SGS3 protein involved in PTGS finds a family. BMC Bioinforma 3:21
Bratić AM, Majić DB, Samardžić JT, Maksimović VR (2009) Functional analysis of the buckwheat metallothionein promoter: tissue specificity pattern and up-regulation under complex stress stimuli. J Plant Physiol 166:996–1000
Degenhardt J, Tobin EM (1996) A DNA binding activity for one of two closely defined phytochrome regulatory lements in an Lhcb promoter is more abundant in etiolated than in green plants. Plant Cell 8:31–41
Durfee T, Roe JL, Sessions RA, Inouye C, Serikawa K, Feldmann KA, Weigel D, Zambryski PC (2003) The F-box-containing protein UFO and AGAMOUS participate in antagonistic pathways governing early petal development in Arabidopsis. Proc Natl Acad Sci U S A 100:8571–8576
Elmayan T, Adenot X, Gissot L, Lauressergues D, Gy I, Vaucheret H (2009) A neomorphic sgs3 allele stabilizing miRNA cleavage products reveals that SGS3 acts as a homodimer. FEBS J 276:835–844
Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12:599–609
Ghareeb H, Becker A, Iven T, Feussner I, Schirawski J (2011) Sporisorium reilianum infection changes inflorescence and branching architectures of maize. Plant Physiol 156:2037–2052
Glick E, Zrachya A, Levy Y, Mett A, Gidoni D, Belausov E, Citovsky V, Gafni Y (2008) Interaction with host SGS3 is required for suppression of RNA silencing by tomato yellow leaf curl virus V2 protein. Proc Natl Acad Sci U S A 105:157–161
Guo AY, Zhu QH, Chen X, Luo JC (2007) GSDS: a gene structure display server. Yi Chuan 29:1023–1026
Guo L, Yu Y, Xia X, Yin W (2010) Identification and functional characterisation of the promoter of the calcium sensor gene CBL1 from the xerophyte Ammopiptanthus mongolicus. BMC Plant Biol 10:18
Hamilton DA, Schwarz YH, Mascarenhas JP (1998) A monocot pollen-specific promoter contains separable pollen-specific and quantitative elements. Plant Mol Biol 38:663–669
Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B (2005) Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol Biol 57:155–171
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27:297–300
Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103:12987–12992
Kaplan B, Davydov O, Knight H, Galon Y, Knight MR, Fluhr R, Fromm H (2006) Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell 18:2733–2748
Kim DW, Lee SH, Choi SB, Won SK, Heo YK, Cho M, Park YI, Cho HT (2006) Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 18:2958–2970
Kim SY (2006) The role of ABF family bZIP class transcription factors in stress response. Physiol Plant 126:519–527
Kim SY, Chung HJ, Thomas TL (1997) Isolation of a novel class of bZIP transcription factors that interact with ABA-responsive and embryo-specification elements in the Dc3 promoter using a modified yeast one-hybrid system. Plant J 11:1237–1251
Kozak M (1999) Initiation of translation in prokaryotes and eukaryotes. Gene 234:187–208
Le Gourrierec J, Li YF, Zhou DX (1999) Transcriptional activation by Arabidopsis GT-1 may be through interaction with TFIIA-TBP-TATA complex. Plant J 18:663–668
Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci U S A 97:3730–3734
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C (T)) Method. Methods 25:402–408
Lopez-Molina L, Chua NH (2000) A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol 41:541–547
Lupas A, Van Dyke M, Stock J (1991) Predicting coiled coils from protein sequences. Science 252:1162–1164
Mourrain P, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, Rémoué K, Sanial M, Vo TA, Vaucheret H (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101:533–542
Muangsan N, Beclin C, Vaucheret H, Robertson D (2004) Geminivirus VIGS of endogenous genes requires SGS2/SDE1 and SGS3 and defines a new branch in the genetic pathway for silencing in plants. Plant J 38:1004–1014
Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18:2368–2379
Qin YH, Ye HY, Tang N, Xiong LZ (2009) Systematic identification of X1-homologous genes reveals a family involved in stress responses in rice. Plant Mol Biol 71:483–496
Sekhon RS, Childs KL, Santoro N, Foster CE, Buell CR, de Leon N, Kaeppler SM (2012) Transcriptional and metabolic analysis of senescence induced by preventing pollination in maize. Plant Physiol 159:1730–1744
Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM (2011) Genome-wide atlas of transcription through maize development. Plant J 66:553–563
Shi JR, Kim KN, Ritz O, Albrecht V, Gupta R, Harter K, Luan S, Kudla J (1999) Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 11:2393–2405
Solano R, Nieto C, Avila J, Cañas L, Diaz I, Paz-Ares J (1995) Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYB.Ph3) from Petunia hybrida. EMBO J 14:1773–1784
Sonnhammer EL, Eddy SR, Durbin R (1997) Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28:405–420
Stalberg K, Ellerstom M, Ezcurra I, Ablov S, Rask L (1996) Disruption of an overlapping E-box/ABRE motif abolished high transcription of the napA storage-protein promoter in transgenic Brassica napus seeds. Planta 199:515–519
Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress promoter. Plant Cell 16:2481–2498
Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5:1529–1539
Villain P, Mache R, Zhou DX (1996) The mechanism of GT element-mediated cell type-specific transcriptional control. J Biol Chem 271:32593–32598
Voll LM, Horst RJ, Voitsik AM, Zajic D, Samans B, Pons-Kühnemann J et al (2011) Common motifs in the response of cereal primary metabolism to fungal pathogens are not based on similar transcriptional reprogramming. Front Plant Sci 2:39
Xie M, Ren GD, Costa-Nunes P, Pontes O, Yu B (2012a) A subgroup of SGS3-like proteins act redundantly in RNA-directed DNA methylation. Nucleic Acids Res 40:4422–4431
Xie M, Ren GD, Zhang C, Yu B (2012b) The DNA- and RNA-binding protein FACTOR of DNA METHYLATION 1 requires XH domain-mediated complex formation for its function in RNA-directed DNA methylation. Plant J 72:491–500
Yang T, Poovaiah BW (2002) A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J Biol Chem 277:45049–45058
Yoshikawa M, Peragine A, Park MY, Poethig RS (2005) A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev 19:2164–2175
Zhang D, Trudeau VL (2008) The XS domain of a plant specific SGS3 protein adopts a unique RNA recognition motif (RRM) fold. Cell Cycle 7:2268–2270
Zhang ZB, Li HY, Zhang DF, Liu YH, Fu J, Shi YS, Song YC, Li Y, Wang TY (2012) Characterization and expression analysis of six MADS-box genes in maize (Zea mays L.). J Plant Physiol 169:797–806
Zheng ZM, Xing Y, He XJ, Li W, Hu Y, Yadav SK, Oh J, Zhu JK (2010) An SGS3-like protein functions in RNA-directed DNA methylation and transcriptional gene silencing in Arabidopsis. Plant J 62:92–99
Acknowledgments
We are grateful to editors and reviewers for their helpful comments and the providers who submitted the microarray data to the public expression databases which can be applied freely. This work was supported partly by the Beijing Municipal Science and Technology Commission (No. Z111100066111005 and Z121100001512012); Beijing Nova Program (No. Z121105002512031); Beijing Academy of Agriculture and Forestry Sciences (No. KJCX201102003), and the Youth Foundation (No. QNJJ201303).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, Z., Chen, Y., Zhao, D. et al. X1-homologous genes family as central components in biotic and abiotic stresses response in maize (Zea mays L.). Funct Integr Genomics 14, 101–110 (2014). https://doi.org/10.1007/s10142-013-0343-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-013-0343-2