Abstract
Homeodomain-leucine zipper (HD-Zip) transcription factors regulate developmental processes and stress responses in plants, and they vary widely in gene number and family structure. In this study, 55 predicted maize HD-Zip genes were systematically analyzed with respect to their phylogenetic relationships, molecular evolution, and gene expression in order to understand the functional diversification within the family. Phylogenetic analysis of HD-Zip proteins from Zea mays, Oryza sativa, Arabidopsis thaliana, Vitis vinifera, and Physcomitrella patens showed that they group into four classes. We inferred that the copy numbers of classes I and III genes were relatively conserved in all five species. The 55 maize HD-Zip genes are distributed randomly on the ten chromosomes, with 15 segmental duplication and 4 tandem duplication events, suggesting that segmental duplications were the major contributors in the expansion of the maize HD-Zip gene family. Expression analysis of the 55 maize HD-Zip genes in different tissues and drought conditions revealed differences in the expression levels and patterns between the four classes. Promoter analysis revealed that a number of stress response-, hormone response-, light response-, and development-related cis-acting elements were present in their promoters. Our results provide novel insights into the molecular evolution and gene expression within the HD-Zip gene family in maize, and provide a solid foundation for future functional study of the HD-Zip genes in maize.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Precise and coordinated gene expression is essential for plant development and environmental stress resistance. Transcription factors (TFs) are a group of regulatory proteins that regulate gene expression by binding to specific cis-acting elements in the promoters of target genes (Badis et al. 2009). Although the genes that encode transcription factors account for only a small portion of the genome, TFs are important players in gene expression in regulated networks (Hobert 2008). Recently, it has been reported that numerous TFs control many critical biological processes during plant development and growth; examples are TFs such as TCP, WRKY, MYB, AT-hook and E2F (Van Camp 2005; Ricachenevsky et al. 2010; Walford et al. 2011).
Homeodomain-leucine zipper (HD-Zip) proteins constitute a large family of TFs that are characterized by the presence of a DNA-binding homeodomain (HD) and an adjacent leucine zipper (Zip) motif. HD-Zip proteins are unique to plants (with the exception of charophycean algae), but they are related to HD proteins found in other eukaryotes. To date, all members of the HD-Zip gene family from >50 plant species have been deposited in the Plant Transcription Factor Database (PlantTFDB) (Jin et al. 2014). With the recent development of high throughput DNA sequencing technology, more and more plant genomes are being sequenced. A great deal of experimental work will be required to determine the specific biological function of each of the HD-Zip genes in these plant species. However, a precise annotation of HD-Zip genes is the first step toward fully understanding their roles in plant development.
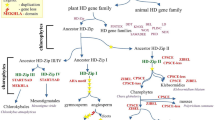
Previous studies have shown that HD-Zip gene family members can be grouped into four different classes, HD-Zip I to IV, based on their additional conserved domains, structures, and physiological functions (Schena and Davis 1994). The different classes of HD-Zip genes in plants regulate a wide array of diverse biological processes, but there seems to be at least some functional conservation within each of these classes (Ariel et al. 2007). HD-Zip I proteins are mainly involved in responses to abiotic stress, auxin, de-etiolation, blue light signaling, and the regulation of organ growth and development (Aoyama et al. 1995; Himmelbach et al. 2002; Johannesson et al. 2003; Wang et al. 2003; Olsson et al. 2004; Ariel et al. 2007). HD-Zip II proteins are involved in responses to illumination conditions, shade avoidance, and auxin signaling (Carabelli et al. 1993; Steindler et al. 1999; Sawa et al. 2002; Sessa et al. 2005). HD-Zip III proteins control the development of the apical meristem, the vascular bundles, and the adaxial domains of lateral organs (Baima et al. 2001; Emery et al. 2003; Ohashi-Ito and Fukuda 2003), and HD-Zip IV proteins play crucial roles in anthocyanin accumulation, epidermal cell differentiation, trichome formation, root development, and cuticle development (Rerie et al. 1994; Di Cristina et al. 1996; Nakamura et al. 2006; Ariel et al. 2007).
To date, functional analysis of the HD-Zip gene family have mainly focused on model plants, such as Arabidopsis and rice. The expansion and functional divergence of this large gene family have not yet been investigated in other land plants. Thus, there is a lack of general understanding of the tempo and mode of HD-Zip gene family evolution in plants. Now, with the completion of the bryophyte Physcomitrella patens (Rensing et al. 2008), the lycophyte Selaginella moellendorffii (Banks et al. 2011), and the Zea mays (Schnable et al. 2009) genome projects, a joint phylogenetic analysis of HD-Zip genes from a bryophyte, a lycophyte, a eudicot (Arabidopsis), and two monocots (rice and maize) will help further our understanding of the expansion and diversification of this large gene family in land plants. Bryophytes are the closest extant relatives of early land plants, which began to diverge ~450 million years ago (Rensing et al. 2008). Lycophytes are early vascular plants with a dominant sporophyte generation (Banks et al. 2011). Compared with lycophytes and bryophytes, the angiosperm plant body possesses more complex organ systems and structures, such as flowers and fruits. One of the major questions in plant evolution concerns the evolution of the body developmental program, which has been modified through time so that the sporophyte became larger and acquired the ability to branch, develop vascular conducting tissues, and produce roots, leaves, seeds, and flowers (Bowman et al. 2007).
As one of the world’s major cereal crops, maize is not only a primary food resource, but is also an important source of industrial materials such as oil and specialty starches. With the maize genome sequence completed, an excellent opportunity is available to conduct whole-genome annotation, evolution, and comparative studies in maize (Schnable et al. 2009). Moreover, previous lines of evidence have already demonstrated that the HD-Zip family may be an excellent candidate to regulate plant development and environmental stress responses. Thus, a systematic analysis of gene expression of maize HD-Zip family members is necessary to fully understand the basic function of this gene family in maize. Previous research have identified 55 HD-Zip genes in maize genome and subsequently the phylogenetic, gene structure, chromosomal location, gene duplication of 55 HD-Zip genes and expression levels of the HD-Zip I genes under drought stress were investigated (Zhao et al. 2011). In this study, a comprehensive analysis of phylogenetic, gene structure, chromosomal locations and gene duplication were conducted to comfirm the above research. Importantly, the molecular evolution, promoter analysis, and gene expression patterns in various tissues and in response to drought stress were also performed. The results provide a biological reference for future studies on the functions of maize HD-Zip genes.
Materials and methods
Identification of HD-Zip proteins in the maize genome
We used several different approaches to identify putative HD-Zip proteins in maize. Initially, protein sequences for maize HD-Zip transcription factors (genome assembly: AGPv3) were retrieved from the Plant Transcription Factor Database 3.0 (available online: http://planttfdb.cbi.pku.edu.cn) (Jin et al. 2014). Genes in the maize genome that were annotated with a Pfam HD domain (PF00046) and leucine zipper motif (PF02183) were retrieved from Phytozome v10.0 (available online: http://www.phytozome.net/eucalyptus.php). In addition, BLAST searches were also performed using Arabidposis HD-Zip proteins against the maize genome to identify any additional HD-Zip members. All but the longest splice variants were removed and redundant sequences were removed using the decrease redundancy tool (available online: http://web.expasy.org/decrease_redundancy). The presence of HD domain and leucine zipper motif in all family proteins; START (PF01852) and MEKHLA (PF08670) domains in HD-Zip III class; and the START (PF01852) domain in HD-Zip IV class were evaluated using Pfam (available online: http://pfam.xfam.org/) (Finn et al. 2014) and SMART (available online: http://smart.embl-heidelberg.de/) (Letunic et al. 2004) database searches. Arabidopsis HD-Zip proteins were downloaded from TAIR 10 (available online: http://www.arabidopsis.org), which contained 48 members. Rice (genome assembly: IRGSP-1.0), Physcomitrella (Genome assembly: Phypa1.1), Selaginella (Genome assembly: phytozomeV10.0/v1.0), and grape (Genome assembly: Genoscope 12X) HD-Zip proteins were downloaded from the Plant Transcription Factor Database (available online: http://planttfdb.cbi.pku.edu.cn) (Jin et al. 2014).
Gene structure and phylogenetic analysis
Genomic sequences of maize HD-Zip genes were downloaded from Phytozome v10.0 (available online: http://www.phytozome.net/eucalyptus.php) and untranslated regions were removed. To show the exon/intron organization for individual HD-Zip genes, coding sequences were aligned to genomic sequences and schematics generated using GSDS (Gene Structure Display Server) (available online: http://gsds.cbi.pku.edu.cn) (Guo et al. 2007).
To determine the phylogenetic relationships of the HD-Zip proteins, multiple alignments were created with the ClustalW program (Chenna et al. 2003) using a Gonnet protein weight matrix, as well as multiple alignment gap opening/extension penalties of 10/0.5 and pairwise gap opening/extension penalties of 10/0.1. Phylogenetic trees were then constructed with MEGA 5.0 software (Tamura et al. 2011) using the NJ method. Bootstrap analysis was performed on 1000 replicates with the pairwise deletion option.
In silico promoter sequence analysis
For detection of putative cis-acting regulatory elements in the promoter sequence, the online search tool of PlantCARE (plant cis-acting regulatory elements, http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was utilized.
Chromosomal localization and test for selection neutrality
Chromosomal location informations of maize HD-Zip genes were retrieved from maize genome (genome assembly: AGPv3) in Phytozome v10.0 (http://planttfdb.cbi.pku.edu.cn). The tandem duplicates were identified according to Hanada et al. (2008) as pairs of HD-Zip genes within 100 kb of each other and having 10 or fewer nonhomologous genes in between. Whole-genome (segmental) duplicated gene pairs were analyzed on the Plant Genome Duplication Database (PGDD) server (available online: http://chibba.agtec.uga.edu/duplication/index/locus) with a display range of 100 kb. The synonymous substitution rates (Ks) between the tandemly or segmental duplicated gene pairs of HD-Zip genes were calculated using the CODEML program in PAML (Yang 2007). The approximate date of the duplication events was calculated using T = Ks/2λ by assuming a clock-like rate (λ) in grasses of 6.5 × 10−9 synonymous mutations per synonymous site per year (Gaut et al. 1996).
Expression of HD-Zip genes in different tissues
Expression patterns of HD-Zip genes in different maize tissues were analyzed using the genome-wide gene expression atlas of the inbred B73 line of maize that was reported previously (Sekhon et al. 2011). Normalized gene expression values expressed as FPKM (fragments per kilobase of exon per million fragments mapped) were transformed using log2(FPKM) for further expression analysis.
Plant material and growth conditions
Maize (Inbred line B73) growth conditions and drought treatments were performed according to Mao (Mao et al. 2015). Briefly, maize seeds were surface-sterilized in 1 % (v/v) Topsin-M (Rotam Crop Sciences Ltd.) for 10 min, then washed in deionized water and germinated on wet filter paper at 28 °C for three days. The germinated seeds were placed in a nutrient solution (0.75 mM K2SO4, 0.1 mM KCl, 0.25 mM KH2PO4, 0.65 mM MgSO4, 0.1 mM EDTA-Fe, 2.0 mM Ca(NO3)2, 1.0 mM MnSO4, 1.0 mM ZnSO4, 0.1 mM CuSO4, 0.005 mM (NH4)6Mo7O24) for hydroponic cultivation for eight days. The hydroponically cultured seedlings were placed on a clean bench and subjected to dehydration (28 °C, relative humidity of 40–60 %). Samples were exposed for 0, 5, and 10 h, and the time points were determined by measuring the relative leaf water content (RLWC), corresponding to the drought-treated leaf samples, which were approximately 98, 70 and 60 %, respectively. Leaf samples from a minimum of 3 seedlings were frozen in liquid nitrogen immediately and then stored at −80 °C prior to RNA isolation. Total RNA was isolated from the leaf samples collected using TRIZOL reagent (Biotopped, China) according to the manufacturer’s instructions. The RNA samples were inspected for their quality and quantity using NanoDrop® spectrophotometer.
Microarray-based gene expression analysis of HD-Zip genes
Total RNA from 6 leaf samples that includes two biological replicates for each of the three time points 0, 5 and 10 h under dehydration was sent to the Shanghai Biotechnology Corporation (Shanghai, China) for microarray analysis. A custom 60 K maize oligo microarray (Agilent Technology) (60-mer oligos) was used. mRNA from leaf samples of drought-treated and control plants were reverse transcribed and labeled with Cy3 and Cy5, respectively. Dye-swap experiments were carried out to control the technique errors, in which each pair of drought-treated and untreated samples was inversely labeled with Cy5 and Cy3. Hybridization and data processing were carried out according to the standard analysis pipeline (Agilent Technology). The microarray data were clustered using Cluster3.0 with Euclidean distances and the hierarchical cluster method of complete linkage clustering. The clustering tree was constructed and viewed in Java Treeview.
RNA isolation and transcript level analysis
In order to validate the microarray results, we performed the quantitative real time-PCR (qRT-PCR) analysis using primers corresponding to maize HD-Zip I genes. The RNA samples was subsequently treated with RNase-free DNase I (Takara, China) to remove genomic DNA contamination. First-strand cDNA was synthesized from 1 µg of total RNA using Recombinant M-MLV reverse transcriptase (Promega, USA). Quantitative real time-PCR (qRT-PCR) was performed in optical 48-well plates using the ABI7300 Thermo-cycler (Applied Biosystems, USA). Reactions were carried out in a 10-μl volume containing 1 μl diluted cDNA, 200 nM gene-specific primers, and 5 μl SYBR Premix Ex Taq II (Takara, China) with the following conditions: 10 min at 95 °C, 40 cycles of 15 s at 95 °C, and 30 s at 60 °C. The specificity of the amplicon for each primer pair was verified by melting curve analysis. The expression of ZmUbi-2 (UniProtKB/TrEMBL; ACC:Q42415) was used as an internal control. The quantification method (2−ΔΔCt) (Livak and Schmittgen 2001) was used and the variation in expression was estimated from the three biological replicates. The primer pairs used in this study are listed in Table S1.
Results
Identification of HD-Zip genes in maize
Fifty-five full-length genes (ZmHDZ1–ZmHDZ55) encoding putative HD-Zip proteins were identified in the maize genome. In order to confirm the reliability of HD-Zip candidate proteins in maize, the amino acid sequences of the 55 proteins were searched for the presence of both HD domain and leucine zipper motif, and additional START or MEKHLA domains using Pfam and SMART programs. The results showed that all of the 55 HD-Zip proteins in maize contained both HD domain and leucine zipper motif, indicating that they probably belong to the HD-Zip family (Table S2). In addition, five HD-Zip proteins also contained START and MEKHLA domains, and fifteen HD-Zip proteins cotained START domain (Table S2). The full-length coding sequences (CDS) of the maize HD-Zip genes ranged from 666 bp (ZmHDZ28) to 2862 bp (ZmHDZ39), with deduced protein lengths of 221–953 amino acids (Table 1).
Phylogenetic and structural analysis
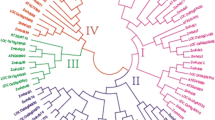
Phylogenetic relationships among the 55 HD-Zip proteins were reconstructed using the neighbor-joining (NJ) method. The resulting phylogenetic tree showed that the maize HD-Zip proteins formed four distinct clades with >76 % bootstrap support, corresponding to classes I, II, III, and IV, and contained 17, 18, 5, and 15 members, respectively (Fig. 1a; Table 1). We subsequently performed an exon–intron structure analysis to support the phylogeny reconstruction. The schematic structures revealed that each HD-Zip gene coding sequence is disrupted by one or more introns. Based on these results, the HD-Zip genes can be divided into four groups as shown in Fig. 1c. Among the four class HD-Zip genes, only class III genes showed well conserved exon–intron structures, where 4 of the five genes have exactly 18 exons. Class III HD-Zip genes contain the largest number of introns, while class I and II genes contain the fewest (Fig. 1c). Pairwise comparisons of the 55 full-length HD-Zip protein sequences revealed that HD-Zip I amino acid sequences are more diverse and HD-Zip III are well conserved. Otherwise, the protein sequence identities within each HD-Zip class are higher than they are between classes (independent-sample t test, P < 0.0001) (Fig. 2). These results are consistence with the previous study (Ariel et al. 2007; Arce et al. 2011).
Predicted maize HD-Zip protein phylogeny, expression profiles, and gene structures. a Neighbor-Joining (NJ) phylogeny of maize HD-Zip proteins, showing subfamily classification and bootstrap values >50. b Expression profile of maize HD-Zip genes, except for ZmHDZ16, ZmHDZ31 and ZmHDZ41, in root (Rt), coleoptile (Cp), stem (St), internode (In), cob (Co), leaves (Le), shoot (Sh), tassel (Ta), silk (Si), husk (Hu), embryo (Em), pericarp (Pe), and seed (Se). Normalized gene expression values are shown in different colors that represent the relative levels of expression indicated on the scale bar. c Position of exons and introns in the maize HD-Zip genes. Introns are shown as lines, exons as boxes
Pairwise sequence identity of full-length maize HD-Zip proteins. A, B, C, and D represent pairwise sequence identities of classes I, II, III, and IV HD-Zip proteins, respectively. AB, AC, and AD represent pairwise sequence identities between classes I and II, I and III, and I and IV HD-Zip proteins; BC and BD represent pairwise sequence identities between classes II and III, and II and IV HD-Zip proteins; and CD represents pairwise sequence identities between classes III and IV HD-Zip proteins. The box plot shows the median (black line), interquartile range (box), and maximum and minimum scores (whiskers) for each data set. Outliers are shown as circles outside of the whiskers
Expansion of the HD-Zip family in maize and rice
It is apparent that the number of HD-Zip genes in maize is larger than in rice. To investigate the extent of lineage-specific expansion of the HD-Zip genes in maize and rice, we performed a joint phylogenetic analysis of the maize and rice HD-Zip proteins. As in the maize-only analysis, the 55 maize and 42 rice HD-Zip proteins grouped into four distinct classes (Fig. 3a). We identified the nodes that led to maize- and rice-specific clades (red circles in Fig. 3a). These nodes indicate the divergence point between maize and rice, and thus represent the most recent common ancestral genes prior to the split. Some HD-Zip genes might have been present in the most recent common ancestor (MRCA) of maize and rice but were later lost in either maize or rice. We found seven clades that contained only maize HD-Zip genes (Fig. 3a, red arrows) and six clades that contained only rice HD-Zip genes (Fig. 3a, black arrows), indicating that gene loss could have occurred in these clades. The number of clades indicated that there were at least 47 ancestral HD-Zip genes before the maize-rice evolutionary split occurred. After split from the MRCA, maize has gained more genes than rice (Fig. 3b).
Phylogenetic analysis (a) and copy number changes (b) of maize and rice HD-Zip proteins. In a, a NJ tree was constructed from a sequence alignment of predicted maize and rice HD-Zip proteins using MEGA 5.0 software. Numbers on branches indicate the bootstrap percentage values calculated from 1000 replicates, and only values >50 % are shown. The nodes that represent the most recent common ancestral genes before the maize and rice split are indicated by red circles (bootstrap support >50 %). Clades that contain only maize or rice HD-Zip proteins are indicated by black and red arrows, respectively. In b, the numbers in circles and rectangles represent the numbers of HD-Zip genes in extant and ancestral species, respectively. Numbers on branches with plus and minus symbols represent the numbers of gene gains and losses, respectively
Duplication mechanisms accounting for the maize HD-Zip family expansion
We examined the distribution of the 55 HD-Zip genes across the 10 maize chromosomes. The distribution of the HD-Zip genes on the chromosomes appears to be random. Interestingly, chromosome 8 was found to harbor only a single HD-Zip gene (Fig. 4). In addition, four pairs of HD-Zip genes (ZmHDZ19/29, ZmHDZ22/23, ZmHDZ22/28, and ZmHDZ23/28) are arranged in tandem repeats at two loci on chromosomes 9 and 5 (Fig. 4; Table 2).
Genomic localization of maize HD-Zip genes. Schematic view of chromosome reorganization by the most recent whole-genome duplication in maize. Regions that are assumed to correspond to homologous genome blocks are shaded gray and connected by lines. Paralogous HD-Zip genes are indicated by red lines within the gray-shaded trapezoids
The maize genome has undergone several rounds of genome duplication, including that from a paleopolyploid ancestor ~70 million years ago (mya) and an additional whole-genome duplication event ~5–12 mya (Schnable et al. 2009). In this study, we identified 15 duplicated HD-Zip gene pairs, including ZmHDZ4/6, ZmHDZ5/11, ZmHDZ5/13, ZmHDZ8/10, ZmHDZ9/12, ZmHDZ19/21, ZmHDZ20/33, ZmHDZ26/33, ZmHDZ30/35, ZmHDZ37/39, ZmHDZ42/48, ZmHDZ42/49, ZmHDZ43/53, ZmHDZ48/52, and ZmHDZ51/53, and each of them are located in a pair of paralogous blocks (Fig. 4; Table S3). It is noteworthy that among the 19 sister pairs identified at the terminal nodes of the phylogenetic tree shown in Fig. 1a, eight gene pairs (ZmHDZ2/3, ZmHDZ14/17, ZmHDZ25/34, ZmHDZ18/27, ZmHDZ38/40, ZmHDZ41/45, and ZmHDZ46/55) (ZmHDZ22 and ZmHDZ28 were found to be involved in tandem duplication) were located just outside the segmentally duplicated regions. However, their gene structure and phylogenetic relationships indicated that they share a similar evolutionary history and are closely related to each other. These results provide strong evidence that gene duplication, especially the segmental duplication, has made an important contribution to the expansion of the maize HD-Zip gene family.
Based on the ratio of nonsynonymous to synonymous substitutions (Ka/Ks), the history of selection acting on coding sequences can be estimated. A pair of sequences will have Ka/Ks < 1 if one sequence has been under purifying selection but the other has been drifting neutrally, while Ka/Ks = 1 if both sequences are drifting neutrally and Ka/Ks > 1 at specific sites that are under positive selection (Juretic et al. 2005). A summary of Ka/Ks ratios for 19 HD-Zip duplicated pairs is shown in Table 2. The result suggests that gene pairs ZmHDZ4/6, ZmHDZ5/11, ZmHDZ5/13, ZmHDZ9/12, ZmHDZ19/21, ZmHDZ20/33, and ZmHDZ26/33 have evolved mainly under the influence of positive selection, and the other gene pairs evolved under purifying selection. Based on the divergence rate of 6.5 × 10−9 synonymous mutations per synonymous site per year as previously proposed for grasses (Gaut et al. 1996), the duplications in these 19 paralogous pairs are estimated to have occurred between 6.92 and 58.46 Mya (Table 2).
Comparation of HD-Zip genes in land plants
Two recently-available plant genomes (the lycophyte, Selaginella moellendorffii and the bryophyte, Physcomitrella patens) allow new insights into the evolution of the HD-Zip gene family in land plants. In this study, we identified 9 and 29 HD-Zip genes from the Selaginella and Physcomitrella genomes, respectively (Table S4). Using the NJ method, we reconstructed the phylogenetic relationships among 207 HD-Zip proteins from Physcomitrella, Arabidopsis, Vitis, rice, and maize. Again, the phylogenetic tree showed that all the HD-Zip proteins grouped into four distinct clades (classes I, II, III and IV) with >74 % bootstrap support (Fig. 5A; Fig. S1). Class I genes generally comprise the largest of the subfamilies in these plant species, while class III has the smallest number of HD-Zip gene members.
Unrooted phylogenetic tree (a) and copy number comparisons (b) of the HD-Zip genes from five land plant species. Numbers at the internal branches leading to the four HD-Zip classes indicate the bootstrap support from 1000 replicates. HD-Zip protein classes I, II, III, and IV are shaded red, blue, green, and yellow, respectively
The marked difference in HD-Zip gene family size among bryophytes, lycophytes, and angiosperms (Fig. 5b; Table S4) suggests that expansion of the HD-Zip gene family occurred after the divergence of the lycophytes and euphyllophytes. Because of the lack of genomic information for gymnosperms and monilophytes, the tempo of HD-Zip gene family evolution in land plants cannot yet be precisely defined. We found highly conserved copy numbers of the class III HD-Zip genes in each of the five species. Similarly, we also observed relatively conserved copy numbers of the class I HD-Zip genes in the five species: there are 16 copies in Physcomitrella, 13 in Vitis, 15 in rice, and 17 each in Arabidopsis and maize. However, pronounced copy number variations for class II and class IV HD-Zip genes were observed in these plant species. In class II, Physcomitrella only contains four genes, while Vitis, Arabidopsis, rice, and maize have 7, 10, 12, and 18 genes, respectively. Similarly, In class IV, Physcomitrella contains only four genes, but Vitis, Arabidopsis, rice, and maize have 8, 16, 11, and 15 genes, respectively (Fig. 5b).
Expression patterns of maize HD-Zip genes in various tissues
Tissue-specific mRNA abundance can be suggestive of a gene’s biological function. HD-Zip proteins have been shown to participate in the transcriptional regulation of a series of biological processes related to plant growth and development. In order to better understand the function of each class of HD-Zip genes in maize, their expression profiles were investigated in 13 different tissues sampled from maize plants grown under non-limiting growth conditions. Using transcriptomic data from maize B73 (Sekhon et al. 2011), an expression heat map was constructed for 55 maize HD-Zip genes in 13 different tissues, except for ZmHDZ16, ZmHDZ31 and ZmHDZ41 (Fig. 1b). The results showed that the expression patterns of the different HD-Zip genes varied greatly. All class III HD-Zip genes showed a relatively high level of expression compared to the other genes examined, possibly indicating the importance of these genes in maize development. Except for ZmHDZ43, −46, −51, −53, −54, and −55, which were constitutively expressed at lower levels in the various tissues, all class IV HD-Zip genes showed selectively higher expression in one or more tissues. Similarly, except for ZmHDZ20, −22, −24, −26, −27, −28, and −30 which were constitutively expressed at lower levels in the various tissues, all class II HD-Zip genes were selectively expressed at higher levels in one or more specific tissues. However, substantially more variation in expression patterns was found among the class I HD-Zip genes, and all class I genes showed higher expression levels in one or some specific tissues. Thus, the class II and class IV maize HD-Zip genes displayed obvious differences in expression patterns and levels; some genes were consistently expressed at lower levels in all tissues, while the others showed higher expression in one or more tissues (Fig. 1b; Table S5).
Expression differences of maize HD-Zip genes under drought stress
To gain further insight into the roles of maize HD-Zip genes in drought tolerance, we investigated the expression profiles of all maize HD-Zip genes in response to drought stress using microarray analysis. We found that the maize HD-Zip genes could be mainly classified into two groups based on their expression patterns. The expression levels of 21 HD-Zip genes were continuously up-regulated (fold-change >1) in response to both drought stress conditions (5 h and 10 h of drought treatments). Among these genes, 16 genes showed two or more times fold change in one or two drought stress conditions (Table S6). In contrast, 29 HD-Zip genes were continuously down-regulated (fold-change <−1) under both drought stress conditions. Among these genes, only 9 genes showed two or more times fold change in one or two drought stress conditions (Table S6). Expression of the remaining five genes, ZmHDZ14, −15, −17, −45, and −54, were either suppressed or induced under one of the drought conditions (Fig. 6a; Table S6). We noticed that most of the class II HD-Zip genes (67 %) were up-regulated, while all class III HD-Zip genes (100 %) and most of the class IV HD-Zip genes (73 %) were down-regulated in response to drought stress. Also, nearly half of the class I HD-Zip genes (47 %) were up-regulated in response to drought stress (Fig. 6b, c). Further, qRT-PCR analysis of the HD-Zip I genes in leaves of different drought-treated maize seedlings were performed to validate their expression in the microarray. The results showed the significantly correlation between RT-qPCR and microarray data under drought stress conditions (5 and 10 h of drought treatments; P < 0.0001) (Fig. S2), suggesting a reliability of microarray data. The results clearly show the functional divergence of HD-Zip genes in response to drought stress in maize seedlings.
Expression profile of maize HD-Zip genes in response to drought stress. a Microarray-based expression analysis of maize HD-Zip genes. The heat map was generated based on the fold-change values in the treated samples when compared with the unstressed control. The color scale for fold-change values is shown at the bottom. The drought-treated leaf samples were collected at two time points, 5 and 10 h, which reflected relative leaf water content (RLWC) of 70 and 60 %, respectively. b The percentage of up-regulated maize HD-Zip genes in each of the four classes. c The percentage of down-regulated maize HD-Zip genes in the four classes
Promoter analysis
In order to compare the promoter sequences of genes and check whether they have similar or different motifs, we analyzed the ~1500 bp promoter regions of the maize HD-Zip genes using PLACE databases. The results showed that a number of stress response-, hormone response-, light response-, and development-related cis-acting elements were present in their promoters (Table S7). The genes belonging to the same subfamily had a diverse profile of cis-acting elements enriched in the promoters, suggesting the possible role of promoter sequences in functional diversification of the HD-Zip genes of the same subfamilies (Table S7). We found that cis-acting element, AuxRR-core (Auxin-responsive element), only distributed in the promoters of some class III and IV HD-Zip genes; GATA-motif (one of light-responsive elements) distributed in the promoters of some class II and IV HD-Zip genes; and as-2-box (one of light-responsive elements) distributed in the promoters of some class I and II HD-Zip genes. The other cis-acting elements are distributed in promoters of four class HD-Zip genes (Table S7). This results combined with the expression analysis indicated that HD-Zip genes may regulate a wide array of biological processes in maize.
Discussion
HD-Zip transcription factors are ubiquitous in plants and carry out essential roles in various aspects of normal plant growth and developmental processes and also in environmental responses (Ariel et al. 2007). HD-Zip proteins belong to a large gene family with >40 members in the rice, Arabidopsis, and maize genomes. A previous study showed that Arabidopsis HD-Zip proteins could be classified into four groups, and that the groups have distinct functions (Ariel et al. 2007). In this study, we found that the 207 HD-Zip proteins from the Physcomitrella, Arabidopsis, Vitis, rice, and maize genomes also clustered into four distinct groups (Fig. 5a; Fig. S1). In addition, the protein sequence identities within each group of maize HD-Zip proteins were higher than they were between the groups (Fig. 2). Thus, in this study, the HD-Zip proteins were grouped into four classes (HD-Zip I, II, III, and IV).
Gene duplication is one of the major evolutionary mechanisms for generating novel genes that help organisms adapt to new and changing environments (Bowers et al. 2003; Gu et al. 2003). Three principal evolutionary patterns are attributed to gene duplications; these include segmental duplication, tandem duplication, and transposition events such as retroposition and replicative transposition (Kong et al. 2007). Among these, segmental duplication occurs most frequently in plants because most plants are diploidized polyploids that retain numerous duplicated chromosomal blocks within their genomes (Cannon et al. 2004). Maize is an important model plant for fundamental research on evolutionary history. Previous studies have shown that the maize genome has undergone several rounds of genome duplication, including that from a paleopolyploid ancestor ~70 million years ago (mya) and an additional whole-genome duplication event ~5–12 mya (Schnable et al. 2009). In this study, we found that among the 55 maize HD-Zip genes, 15 gene pairs (26 genes) were created by segmental duplication event and four gene pairs (five genes) resulted from the tandem duplication event (Fig. 4; Table 2). This indicates that segmental duplication was the major process responsible for the rapid expansion of the maize HD-Zip gene family, and this has also been observed in other plant species (Hu et al. 2012; Chen et al. 2014) and some other maize gene families, such as the genes for mitogen-activated protein kinase and CCCH-type zinc finger proteins (Peng et al. 2012; Liu et al. 2013). In addition, over half of the whole-genome duplication pairs showed evidence of significant overall purifying selection, suggesting that at least some of the retained duplicates are still functional and may provide adaptive advantages (Table 2).
Previous studies have discussed three outcomes of gene duplication: neofunctionalization, subfunctionalization, and pseudogenization (Lynch and Conery 2000; Li et al. 2005). In the neofunctionalization model, one duplicate copy (paralog) accumulates beneficial mutations and acquires a new function, whereas the other duplicate copy retains the original gene function. In the subfunctionalization model, each paralog partitions the ancestral gene function (Li et al. 2005). Studies for 19 duplicated gene pairs showed that most of them exhibited similar expression profiles in various tissues or in response to drought stress (Figs. 1b, 6a), such as ZmHDZ4/6, ZmHDZ22/28, ZmHDZ37/39, ZmHDZ42/49, and ZmHDZ51/53, which indicated that the duplicated genes might have redundant functions. However, divergence was also detected between some duplicated gene pairs, such as ZmHDZ5/11, ZmHDZ19/21, ZmHDZ20/33, and ZmHDZ48/52, suggesting neofunctionalization or subfunctionalization of the duplicated paralogs, which is the major feature of most duplicated genes (Blanc and Wolfe 2004).
The growth and productivity of plants are frequently threatened by environmental stresses such as drought and high salinity. Many stress-related genes are induced to help plants adapt to these environmental stresses. Compared with HD-Zip classes II, III, and IV, class I genes play important roles in abiotic stress tolerance (Ariel et al. 2007). Several specific HD-Zip I family members have been characterized with respect to their roles in regulating drought responses. Previous phylogenetic analysis placed OsHOX22, OsHOX24, ATHB-7, and ATHB-12 in the same subgroup of HD-Zip family I, and they are all involved in the regulation of drought stress (Harris et al. 2011). In maize, 21 of 55 HD-Zip genes were found to be up-regulated (fold-change >1) in response to drought stress, and these genes are mainly from classes I and II (Fig. 6). There is little functional evidence to suggest a role for class II HD-Zip genes in plant growth adaptation responses to water deficit. However, expression studies using microarrays have shown that HAT2 and HAT22 expression is up-regulated by drought in Arabidopsis (Huang et al. 2008). In our study, 12 of 18 class II HD-Zip genes were up-regulated in response to drought, indicating that these genes may play an important role in the drought stress response. In addition, based on their expression patterns in seedlings subjected to drought stress, all class III HD-Zip genes and most of the class IV HD-Zip genes were found to be down-regulated (fold-change <−1) in response to drought stress (Fig. 6). These results highlight the functional difference of maize HD-Zip genes in the drought stress reponse.
Moreover, tissue-specific expression analysis revealed that the expression patterns of four class HD-Zip genes are different. The expression patterns of class III HD-Zip genes relatively conserved while the other classes are varied greatly (Fig. 1b). Further promoter analysis revealed that a number of stress response-, hormone response-, light response-, and development-related cis-acting elements were present in their promoters, but the genes belonging to the same subfamily had a diverse profile of cis-acting elements enriched in the promoters (Table S7). Thus, the different classes of HD-Zip genes in plants regulate a wide array of diverse biological processes. HD-Zip I and HD-Zip II proteins contribute to the plasticity of plant growth and are responsible for modulating plant development in response to environmental stimuli, while HD-Zip III and HD-Zip IV proteins are involved in various developmental events (Ariel et al. 2007; Harris et al. 2011).
In summary, we identified 55 HD-Zip transcription factor genes in maize. Moreover, the maize genome contained many more HD-Zip gene family members than did either Arabidopsis or rice, and it appears that segmental duplication events played important roles during the maize HD-Zip family expansion. In addition, by exploring the currently available genome information in land plants, our comparative evolutionary analysis coupled with gene expression analysis provides new insights into the evolution of the HD-Zip family in maize. The findings presented here give researchers a novel draft about molecular evolution and changes in gene expression in the HD-Zip family in maize and lay a solid foundation for uncovering the biological roles of HD-Zip genes in maize development and the stress response.
References
Aoyama T, Dong CH, Wu Y, Carabelli M, Sessa G, Ruberti I, Morelli G, Chua NH (1995) Ectopic expression of the Arabidopsis transcriptional activator Athb-1 alters leaf cell fate in tobacco. Plant Cell 7:1773–1785
Arce AL, Raineri J, Capella M, Cabello JV, Chan RL (2011) Uncharacterized conserved motifs outside the HD-Zip domain in HD-Zip subfamily I transcription factors; a potential source of functional diversity. BMC Plant Biol 11:42
Ariel FD, Manavella PA, Dezar CA, Chan RL (2007) The true story of the HD-Zip family. Trends Plant Sci 12:419–426
Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, Kuznetsov H, Wang CF, Coburn D, Newburger DE, Morris Q, Hughes TR, Bulyk ML (2009) Diversity and complexity in DNA recognition by transcription factors. Science 324:1720–1723
Baima S, Possenti M, Matteucci A, Wisman E, Altamura MM, Ruberti I, Morelli G (2001) The Arabidopsis ATHB-8 HD-Zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol 126:643–655
Banks JA, Nishiyama T, Hasebe M, Bowman JL, Gribskov M, dePamphilis C, Albert VA, Aono N, Aoyama T (2011) The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332:960–963
Blanc G, Wolfe KH (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16:1679–1691
Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422:433–438
Bowman JL, Floyd SK, Sakakibara K (2007) Green genes-comparative genomics of the green branch of life. Cell 129:229–234
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4:10
Carabelli M, Sessa G, Baima S, Morelli G, Ruberti I (1993) The Arabidopsis Athb-2 and -4 genes are strongly induced by far-red-rich light. Plant J 4:469–479
Chen X, Chen Z, Zhao H, Zhao Y, Cheng B, Xiang Y (2014) Genome-wide analysis of soybean HD-Zip gene family and expression profiling under salinity and drought treatments. PLoS ONE 9:e87156
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31:3497–3500
Di Cristina M, Sessa G, Dolan L, Linstead P, Baima S, Ruberti I, Morelli G (1996) The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J 10:393–402
Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13:1768–1774
Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A (2014) Pfam: clans, web tools and services. Nucleic Acids Res 34:D247–D251
Gaut BS, Morton BR, McCaig BC, Clegg MT (1996) Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci USA 93:10274–10279
Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH (2003) Role of duplicate genes in genetic robustness against null mutations. Nature 421:63–66
Guo AY, Zhu QH, Chen X, Luo JC (2007) GSDS: a gene structure display server. Yi Chuan 29:1023–1026
Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu SH (2008) Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol 148:993–1003
Harris JC, Hrmova M, Lopato S, Langridge P (2011) Modulation of plant growth by HD-Zip class I and II transcription factors in response to environmental stimuli. New Phytol 190:823–837
Himmelbach A, Hoffmann T, Leube M, Höhener B, Grill E (2002) Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J 21:3029–3038
Hobert O (2008) Gene regulation by transcription factors and microRNAs. Science 319:1785–1786
Hu R, Chi X, Chai G, Kong Y, He G, Wang X, Shi D, Zhang D, Zhou G (2012) Genome-wide identification, evolutionary expansion, and expression profile of homeodomain-leucine zipper gene family in poplar (Populus trichocarpa). PLoS ONE 7:e31149
Huang D, Wu W, Abrams SR, Cutler AJ (2008) The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J Exp Bot 59:2991–3007
Jin J, Zhang H, Kong L, Gao G, Luo J (2014) PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res 42:D1182–D1187
Johannesson H, Wang Y, Hanson J, Engström P (2003) The Arabidopsis thaliana homeobox gene ATHB5 is a potential regulator of abscisic acid responsiveness in developing seedlings. Plant Mol Biol 51:719–729
Juretic N, Hoen DR, Huynh ML, Harrison PM, Bureau TE (2005) The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res 15:1292–1297
Kong H, Landherr LL, Frohlich MW, Leebens-Mack J, Ma H, dePamphilis CW (2007) Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J 50:873–885
Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P (2004) SMART 4.0: towards genomic data integration. Nucleic Acids Res 32:D142–D144
Li WH, Yang J, Gu X (2005) Expression divergence between duplicate genes. Trends Genet 21:602–607
Liu Y, Zhang D, Wang L, Li D (2013) Genome-wide analysis of mitogen-activated protein kinase gene family in maize. Plant Mol Biol Rep 31:1446–1460
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155
Mao H, Wang H, Liu S, Li Z, Yang X, Yan J, Li J, Tran LS, Qin F (2015) A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat Commun 6:8326
Nakamura M, Katsumata H, Abe M, Yabe N, Komeda Y, Yamamoto KT, Takahashi T (2006) Characterization of the class IV homeodomain-Leucine Zipper gene family in Arabidopsis. Plant Physiol 141:136–1375
Ohashi-Ito K, Fukuda H (2003) HD-Zip III homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are involved in procambium and xylem cell differentiation. Plant Cell Physiol 44:1350–1358
Olsson AS, Engström P, Söderman E (2004) The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol Biol 55:663–677
Peng X, Zhao Y, Cao J, Zhang W, Jiang H, Li X, Ma Q, Zhu S, Cheng B (2012) CCCH-type zinc finger family in maize: genome-wide identification, classification and expression profiling under abscisic acid and drought treatments. PLoS ONE 7:e40120
Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319:64–69
Rerie WG, Feldmann KA, Marks MD (1994) The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev 8:1388–1399
Ricachenevsky FK, Sperotto RA, Menguer PK, Fett JP (2010) Identification of Fe-excess-induced genes in rice shoots reveals a WRKY transcription factor responsive to Fe, drought and senescence. Mol Biol Rep 37:3735–3745
Sawa S, Ohgishi M, Goda H, Higuchi K, Shimada Y, Yoshida S, Koshiba T (2002) The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J 32:1011–1022
Schena M, Davis RW (1994) Structure of homeobox-leucine zipper genes suggests a model for the evolution of gene families. Proc Natl Acad Sci USA 91:8393–8397
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, Minx P, Reily AD (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115
Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM (2011) Genome-wide atlas of transcription during maize development. Plant J 66:553–563
Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, Becker J, Morelli G, Ruberti I (2005) A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev 19:2811–2815
Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I (1999) Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126:4235–4245
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Van Camp W (2005) Yield enhancement genes: seeds for growth. Curr Opin Biotech 16:147–153
Walford SA, Wu Y, Llewellyn DJ, Dennis ES (2011) GhMYB25-like: a key factor in early cotton fibre development. Plant J 65:785–797
Wang Y, Henriksson E, Söderman E, Henriksson KN, Sundberg E, Engström P (2003) The Arabidopsis homeobox gene, ATHB16, regulates leaf development and the sensitivity to photoperiod in Arabidopsis. Dev Biol 264:228–239
Yang ZH (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Zhao Y, Zhou Y, Jiang H, Li X, Gan D, Peng X, Zhu S, Cheng B (2011) Systematic analysis of sequences and expression patterns of drought-responsive members of the HD-Zip gene family in maize. PLoS ONE 6:e28488
Acknowledgments
We thank all our collaborators who made the work on HD-Zip transcription factors a rewarding experience. We would like to thank the members of the Bioinformatics Center of Northwest A&F University for their useful input. This work was supported by the National Natural Science Foundation of China (31471505).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mao, H., Yu, L., Li, Z. et al. Molecular evolution and gene expression differences within the HD-Zip transcription factor family of Zea mays L.. Genetica 144, 243–257 (2016). https://doi.org/10.1007/s10709-016-9896-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-016-9896-z