Abstract
X1-homologous genes (XHS) encode plant-specific proteins containing three major domains (XH, XS, zf-XS), but their functions are largely unknown. We report the systematic identification and characterization of XHS genes in the rice genome. Eleven putative XHS protein sequences (OXHS1–11) were identified in the sequenced genome of Oryza sativa japonica cv. Nipponbare, and these sequences, along with other plant XHS homologues, were classified into five subgroups based on phylogenetic analysis. Distinct diversification of the XHS proteins occurred between monocotyledon and dicotyledon plants. The OXHS family has diverse exon–intron structures and organizations of putative domains and motifs. The OXHS proteins showed no transactivation activity, and no interaction between the XH domain and the XS domain in yeast. Four representative OXHS proteins were targeted to cytoplasm, which contradicts the previous speculation that XHS proteins are putative transcription factors. All the OXHS genes are predominantly expressed in floral organs, and some are expressed in a wide range of tissues or organs in indica rice Minghui 63. Nine OXHS genes are responsive to at least one of the abiotic stresses including drought, salt, cold, and abscisic acid treatment. Over-expression of one stress-responsive gene OXHS2 in rice resulted in reduced tolerance to salt and drought stresses. These results suggest that the OXHS family may be functionally diversified and some members of this family may play important roles in regulating stress tolerance in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sessile plants have evolved numerous plant-specific genes for growth and development and responses to the ever-changing environment across their life cycles. Although the genomes of plants such as Arabidopsis and rice have been finely sequenced and intensively annotated on the basis of bioinformatics analysis, the biological functions of a large proportion of the genes in these genomes remain to be experimentally elucidated. This is especially true for many plant-specific gene families that are potentially important for both basic biological processes, such as growth and development, and plant-specific responses and adaptation to environmental stresses. Detailed analyses of developmental and phenotypic changes of transgenic or mutant plants and spatio-temporal expression patterns of some genes have greatly contributed to our understanding of plant architecture and elucidation of the developmental programs of the life cycle (Itoh et al. 2005). In addition, it is of fundamental biological importance to understand the functions of the numerous unknown genes for perceiving environmental signals and transmitting the signals to cellular machinery to activate adaptive responses (Mittler 2006).
Increasing evidence suggested that some gene families have very significant roles both in development and response to stress. Examples are the genes from the C-terminal domain phosphatase-like (CPL) and F-BOX families and many transcription factor families such as NAC and MYB. The CPL family is a focal control point of complex processes such as plant stress responses and development with unique and overlapping transcriptional regulatory functions that differentiate the signal output that determines the plant response (Koiwa et al. 2002). F-BOX family genes are required for panicle, leaf, and seed development (Chae et al. 2004; Durfee et al. 2003; Woo et al. 2001) and are regulated by light and temperature stress (Calderon-Villalobos et al. 2007; Jain et al. 2007). NAC transcription factor family genes are involved in the controls of lateral root development (Xie et al. 2000) and the formation of secondary walls in woody tissues in Arabidopsis (Mitsuda et al. 2007; Yamaguchi et al. 2008) and drought and salt stress resistance (He et al. 2005; Hegedus et al. 2003; Hu et al. 2006). MYB family genes have important roles in the formation of root hairs, flower development, and stress responses. For example, the MYB gene family has pleiotropic effects on flower development, epidermal cell size, and trichome branching in addition to trichome and root hair formation in Arabidopsis thaliana (Rotman et al. 2005; Tominaga et al. 2008; Zhu et al. 2004), and many genes of this family also participate in the regulation of freezing, drought, and salt stress tolerance (Dai et al. 2007; Urao et al. 1993).
Gene X1 was first identified in rice, and this gene encodes a protein with putative protein–protein interaction and zinc finger domains (Chen and Bennetzen 1996). The first X1-homologous gene in Arabidopsis was SGS3, which was identified through analysis of the SGS3 mutant impaired in post-transcriptional gene silencing (Mourrain et al. 2000). Through a detailed analysis of the homologous sequences of SGS3, a plant-specific gene family with unknown function was identified and the genes of this family were predicted to encode putative transcription factors (Bateman 2002; Mourrain et al. 2000).However, no experimental evidence has been reported so far to support the speculation that the family members are transcription factors. The predicted protein sequences of X1- or SGS3-homologous genes contain one to three of three novel protein domains: XH, XS, and zf-XS (Bateman 2002), and this family was thus named XHS family. The XS domain contains a completely conserved aspartate and a predicted secondary structure with mixed alpha helix and beta sheet. The XH domain is located at the C-terminal region of XHS proteins and contains one completely conserved glutamate that may be a functionally important site. The zf-XS domain, which usually accompanies a XS domain, is an N-terminal cysteine/histidine zinc binding domain (Bateman 2002).To date, no experimental data have been reported regarding the functions of this family, except for the member SGS3 in Arabidopsis. SGS3 is required for post-transcriptional gene silencing and natural virus resistance (Mourrain et al. 2000), which are needed for endogenous gene silencing from DNA viruses (Glick et al. 2008; Muangsan et al. 2004) and for juvenile development and the production of trans-acting siRNAs in Arabidopsis (Peragine et al. 2004).
Although the sequence characteristics of typical XHS proteins were well addressed (Bateman 2002), no systematic identification and functional insight of this family in a given plant species had been reported. Here, we analyzed the sequences of genes containing XH, XS, and/or zf-XS domains in rice (designated as the OXHS family) and investigated the expression profiles of this family in various tissue or organs and under stress treatments. In addition, we investigated the phenotypic changes of the mutants and/or over-expression transgenic plants of a few genes in relation to development or response to abiotic stresses. Our results suggested that the XHS family may play important roles in response to environmental stresses.
Materials and methods
Identification and sequence analysis of OXHS family
Using the BLAST program, the protein sequences of known rice protein X1 (Q9SBW2) and Arabidopsis protein SGS3 (Q9DLX1) from the protein sequence database ExPASy (http://www.expasy.org/) (Gasteiger et al. 2003) were used to search for homologous proteins in the databases of japonica rice (TIGR: http://www.tigr.org, Yuan et al. 2005; KOME: http://cdna01.dna.affrc.go.jp/cDNA, Kikuchi et al. 2003) and of Arabidopsis (TAIR: http://www.arabidopsis.org, Huala et al. 2001). The pHMM was used to identify new XHS sequences in the rice and Arabidopsis genomes. The HMM profile of XH domain (accession no. PF03469), XS domain (accession no. PF03468), and zf-XS domain (accession no. PF03470) were downloaded from the Pfam database (http://www.sanger.ac.uk/Software/Pfam, Bateman et al. 2004). All hits with expected values less than 1.0 were collected. We used a BLASTN search to determine the chromosome locations of OXHS genes, the rice BAC/PAC clones to which they were mapped, and the intron–exon structures of OXHS genes by mapping cDNAs to genomic sequence.
Multiple sequence alignment of XHS proteins was performed with CLUSTALX (van de Graaff et al. 1982) and refined manually. Phylogenetic tree was reconstructed with the program MrBayes version 3.0 (Ronquist and Huelsenbeck 2003) under the JTT-f model of amino acid substitution and 4-g category model. A total of 400,001 generations were performed with four Markov chains with default heating values and tree sampling every 100 generations. The Markov chain converged after 9,000 generations, and the first 100 sampled trees were discarded. The major consensus tree was deduced from the remaining 901 sampled trees. The tree was edited with TreeView 1.5 (Page 1996).
Isolation of XHS genes and rice transformation
For each OXHS gene, a pair of primers (Supplemental Table 1) was designed to amplify the predicted full-length cDNA from different tissues of Minghui 63 (Oryza sativa L. ssp. indica). Ex-Taq DNA polymerase (Takara) was used in the PCR with the following cycling profile: 94°C for 4 min; 30 cycles of 94°C for 40 s, 55°C for 40 s, and 72°C for 2 or 3 min; and extension at 72°C for 10 min. The amplified products were cloned into pGEM-T vector (Promega) and sequenced from both ends by using BigDye Terminator Sequencing Ready kit (version 2.0 or 3.0) in an ABI PRISM 377 or 3730 sequencer (Applied Biosystems) by the vector-border primers T7 and SP6 (Promega). The sequence-confirmed fragments of OXHS genes were cut by KpnI and/or BamHI from pGEM-T clones and ligated into the transformation vector pCAMBIA1301U under the control of the promoter of maize ubiquitin gene. The Agrobacterium-mediated transformation method was used to introduce the constructs into rice Zhonghua 11 (Oryza sativa L. ssp. japonica) (Hiei et al. 1994).
Plant growth, treatments, and measurements
For transcript level measurement of OXHS genes, rice plants of Minghui 63 were grown under normal growth conditions and 2-week-old seedlings were treated with ABA and abiotic stresses. ABA treatment was conducted by spraying leaves with 0.1 mM followed by sampling at 0, 0.5, 2, 4, and 8 h. Abiotic treatments were conducted essentially according to Saijo (2000). For cold stress, seedlings were transferred to a growth chamber at 4°C and sampled at 0, 0.5, 1, 3, and 6 h after treatment. Drought stress at the seedling stage was applied by exposing 3-week-old intact plants to the air without water supply and plant leaves were sampled at 0, 0.5, 2, 4, and 8 h after treatment. The seedlings were submerged with 200 mM NaCl solution for salt stress and sampled at 0, 15 min, 30 min, 1 h, and 2 h after treatment. For salt or mannitol resistance testing of plants at the seedling stage, the plantlets that germinated on plates with MS medium were transferred to MS medium with 150 mM NaCl or mannitol in autoclaved visible boxes. The lengths of shoot and fresh weight were measured 1 week after transplanting. Free proline content in leaves was determined by following the methods reported by Ohnishi et al. (2005).
Drought stress was applied to transgenic plants growing in field at the panicle development stage. Sixteen plants were grown in two rows for each transgenic family with three replications. The wild type (WT) control was inserted every two transgenic families to minimize the potential variation of soil the field. Relative spikelet fertility or yield, the ratio of the spikelet fertility or yield under stress conditions to that under normal growth conditions, was used to evaluate drought resistance performance at the reproductive stage of rice according to Yue et al. (2006). Drought stress treatment, data collection and statistical analysis were conducted according to Xiao et al. (2009).
Transcript level analysis
Total RNA was extracted from various tissues and organs from the life cycle of Minghui 63 using the TRIZol reagent (Invitrogen) according to the manufacturer’s instructions. The total RNA was treated with amplification-grade DNase I (Invitrogen) for 15 min to degrade possibly contaminated residual genomic DNA. SuperScript II reverse transcriptase (Invitrogen) was used to synthesize first-strand cDNA and about 1/20 of the first-strand cDNA generated from 3 ug total RNA was used as template for PCR in a reaction volume of 50μl with the rTaq DNA polymerase (Takara). Semi-quantitative PCR or RT-PCR was performed in an ABI 9700 Thermocycler (Applied Biosystems) with the following cycling profile: 94°C for 3 min; 25–40 cycles (depending on the expression level) at 94°C for 40 s, 55°C or 60°C for 40 s, and 72°C for 1 min. The RT-PCR product was separated in a 1.2% agarose gel and stained with ethidium bromide for visualization. We used rice Actin1 gene (accession no. AK060893) as the internal control. For each OXHS gene, a pair of primers with an average amplification length of 500 bp was used for the RT-PCR. All experiments were repeated three times with independently reverse-transcribed templates.
Relative quantification of gene expression was performed by real-time PCR on an ABI PRISM 7500 instrument (Applied Biosystems). The primers for real-time PCR were designed by Primer Express Version 2.0 (Applied Biosystems; Supplemental Table 2). Rice Actin1 gene was used as the endogenous control. Real-time PCR was performed in an optical 96-well plate, including 12.5 μl 23 SYBR Green Master mix reagent (Applied Biosystems), 1 μl cDNA samples, and 0.2 mM of each gene-specific primers, in a final volume of 25 μl, using the thermal cycles as follows: 50°C for 2 min, 95°C for 10 min, 40–45 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min. Disassociation curve analysis was performed as follows: 95°C for 15 s, 60°C for 20 s, 95°C for 15 s. The relative expression levels were determined as described previously.
Biochemical assay in yeast
For yeast assay, the full ORFs of OXHS1, OXHS2, OXHS8, and OXHS10 were generated by PCR using the primer pairs (Supplemental Table 3) and fused in frame to the yeast GAL4 DNA-binding domain in PDBLeu vector by restriction-ligation reaction or in pDESTT32 by Gateway recombination reaction. The full ORFs were fused to the yeast GAL4 DNA-activation domain in AD502 or in pDESTT22. For transactivation assay, fusion constructs of OXHS-GAL4 DNA-binding domain were transformed in yeast strain MV203. For protein–protein interactions, one fusion protein of OXHS-GAL4 DNA-binding domain and another fusion protein of OXHS-GAL4 activation domain were co-transformed into the yeast strain MV203. The colony-lift filter assay (X-gal assay) was performed to check the transactivation activity or protein–protein interaction as described by the manufacturer (Invitrogen).
To determine the subcellular localization of the OXHS proteins, the coding regions of OXHS gene amplified using the primer pairs (Supplemental Table 3) was fused in frame to the coding sequence of enhanced green fluorescence protein (EGFP) under the control of a maize ubiquitin promoter in the vector of pCAMBIA1391Xb. The constructs were introduced into onion (Allium cepa) epidermal cells by the bombarding method and fluorescence signal was captured by confocal microscopy.
Results
Identification of XHS family genes in rice and Arabidopsis
To identify all putative genes containing the XH-XS domain in rice, two reported protein sequences containing the XH-XS domain (X1 and SGS3) were used as queries to search the GeneBank database, the rice annotation database at The Institute for Genomic Research (TIGR), and the Knowledge-Based Oryza Molecular Biological Encyclopedia (KOME). After removing the redundant sequences, a total of 11 genes encoding XH-XS-domain-containing proteins, designated as OXHS1–OXHS11 (Table 1), were identified in the japonica rice genome. Some generic information for these OXHS genes are listed in Table 1, including the accession numbers of full-length cDNAs available in KOME and corresponding BAC/PAC clones and their chromosome locations. In the KOME database, the full-length cDNAs were available for all OXHS genes except for OXHS7, OXHS9, and OXHS11. Based on a comparison of the full-length cDNAs and the corresponding genomic sequences for the OXHS genes in GenBank or TIGR, we noticed that the 5′-terminus of the open reading frame (ORF) of OXHS2 (cDNA accession no. AK065750) was annotated incorrectly in KOME, and thus a portion of the XS domain in the predicted protein sequence of OXHS2 in TIGR (LOC_Os01g03570) is absent in the predicted protein sequence of the cDNA. Therefore, we isolated the full-length cDNA of this gene again from Oryza sativa japonica cv. Nipponbare and confirmed that the annotation based on the genomic sequence in TIGR database is correct (not shown). The cDNA sequences for complete ORFs of all the OXHS genes except OXHS11 were also successfully isolated from O. sativa indica cv. Minghui 63 with gene-specific primers (Supplemental Table 1). Comparison of the cDNA sequences of OXHS genes from Minghui 63 and Nipponbare revealed a few single-nucleotide polymorphisms in four OXHS genes that led to a few changes of amino acids (Supplemental Table 4). However, all the amino acid changes occurred in nonconserved regions, except one change in the XH domain of OXHS5. For simplicity, all the subsequent analyses were based on the sequences of OXHS genes from the japonica rice Nipponbare.
Using a similar search against the annotation database of Arabidopsis at The Arabidopsis Information Resource (TAIR), 14 genes encoding XH-XS-domain-containing proteins were identified (Supplemental Fig. 1). Homologous sequences of XHS proteins were also found in other plant species but not in human and other animals, suggesting that XHS family may have evolved specifically in plants.
Phylogenetic analysis of XHS family
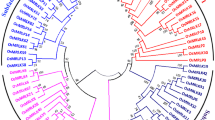
To study the evolutionary relationships of XHS family genes in plants, 35 putative XHS protein sequences, including 11 from rice, 14 from Arabidopsis, and 10 from other flowering plants, were collected for phylogenetic analysis. The results indicate that the XHS family is evolutionarily diversified. The collected XHS protein sequences can be classified into five subgroups (S1–S5) in the unrooted phylogenetic tree (Fig. 1). Nearly all XHS proteins from monocotyledon species (e.g., rice, maize, and wheat) are in subgroups S1 and S4, except that OXHS5 and OXHS3 are grouped into S5 and S3, respectively. Interestingly, all XHS proteins from dicotyledon plants (e.g., Arabidopsis and Medicago truncatula) are in subgroups S2, S3, and S5. Subgroup S4 contained only three rice proteins. These results suggest that plant XHS genes are derived from a common ancestor but differentiated separately in monocotyledon and dicotyledon plants. It is noteworthy that OXHS8 in subgroup S3 is the closest homologue to At5g23570, which is known as the SGS3 protein required for post-transcriptional gene silencing and natural virus resistance (Mourrain et al. 2000). Moreover, both OXHS8 and SGS3 contain only the XS domain, and not the XH domain present in most of the other XHS proteins. Such a strong similarity in both sequence and domain composition suggest that OXHS8 might be an orthologue of SGS3 in rice.
Phylogenetic tree of XHS proteins in plants. Phylogenetic analysis was based on the full XHS proteins sequences of putative 35 XHS proteins from different plants including Oryza sativa, Arabidopsis thaliana, Boechera drummondii, Sorghum bicolor, Medicago truncatula, Triticum monococcum, and Zea mays
Diverse domain composition and gene structure in OXHS family
The OXHS protein sequences were further analyzed for putative domain compositions. Based on a Pfam database search, three putative domains including XH (PF03469), XS (PF03468), and zf-XS (PF03470) that have been identified in the X1 and SGS3 proteins were detected in the OXHS family. The 11 OXHS proteins can be classified into three types according to the composition of the putative domains detected (Fig. 2). The protein sequences in type I, including OXHS1, OXHS2, OXHS3, OXHS4, and OXHS6, contain the XH domain, XS domain, and zf-XS domain (except OXHS2). Four OXHS proteins (OXHS5, OXHS7, OXHS9, and OXHS10) each containing only the XH domain were classified as type II. Type III OXHS proteins (OXHS8 and OXHS11) contain only the XS domain, although OXHS11 has an additional zf-XS domain.
Diverse domain composition and gene structure in the OXHS family. The boxes and lines represent exons and introns, respectively. The double slash (//) and line bar represent 3 kb and 200 bp, respectively. In the boxes, the black regions are UTR regions and other regions represent different putative domains as illustrated. The OXHS family can be classified into three types (I–III) based on the domain compositions
Sequence alignment of the putative domains of the XHS family in rice (Fig. 3) and Arabidopsis (Supplemental Fig. 2) revealed a few highly conserved amino acids or motifs in each domain. Among the three domains, zf-XS is the most conserved according to the sequence alignment. The zf-XS domain contains two highly conserved cysteine and histidine residues and is a C2H2 type zinc finger. In addition, some conserved leucine and arginine residues are also important to the zf-XS domain, which has been supposed to bind to the nucleic acid region (Bateman 2002). Except for SGS3 and OXHS8, the other XS domain proteins are highly conserved, especially for a completely conserved aspartate that was suggested to be an enzymatically active site (Bateman 2002). In addition to one completely conserved glutamate in the C-terminal region that could be part of an active site or other functionally important region, the XH domain also contains many conserved glutamate, aspartate, and proline residues, which may suggest that the XH domain is an activation domain with enriched acidic amino acids or proline, although these amino acids may not necessarily be important for function (Schwechheimer et al. 1998). The lengths of the zf-XS (50 AA), XS (110–120 AA), and XH (130–140 AA) domains are also quite conserved among most of the XHS proteins in rice and Arabidopsis. Nevertheless, OXHS8 and OXHS9 have a shortened XS (73 AA) and XH (78 AA) domain, respectively (Fig. 3). In addition, the exon–intron structures of the OXHS genes and splicing patterns had particular associations with the domain compositions of OXHS proteins. For example, the genes for type I proteins have six or seven exons with very similar splicing patterns and lengths of corresponding exons (Fig. 2).
Transactivation and subcellular location assays of OXHS proteins
The OXHS family had yet to be functionally characterized. The first reported OXHS gene (X1) was suggested to encode a transcription factor (Chen and Bennetzen 1996), although Bateman (2002) was suspicious of this inference. Meanwhile, Bateman (2002) predicted that the XS and XH domains might interact. Thus, far, however, there was no experimental support for these speculations.
To address the potential functions of OXHS proteins, we selected three proteins (OXHS1, OXHS8, and OXHS10, representing the three types of domain compositions shown in Fig. 2) to check if they have transactivation activity. The transactivation assay was performed in the yeast strain MV203 transformed with constructs of the full-length sequences of the three genes fused to the DNA-binding domain of GAL4 and the reporter construct corresponding to GAL4. None of the three OXHS proteins showed transactivation activity in the yeast strain (data not shown). Yeast two-hybrid assay suggested that none of the three selected proteins interacted with the other two proteins, and the XH or XS domain alone did not interact with the other domain or itself either (data not shown).
Most plant transcription factors contain a nuclear localization signal (NLS) characterized by a core peptide enriched in arginine (R) and lysine (K; (Boulikas 1994). By using the PredictNLS tool (http://www.predictprotein.org/cgi/var/nair/resonline.pl), we found that only OXHS1 had one potential NLS sequence with 47 amino acids (KKKKDFNINNLIQHASGVGAASNRQAKDK) in the N terminal. The subcellular locations of the OXHS proteins were predicted by MultiLoc (http://www.osc.edu/supercomputing/software/apps/multicoil.shtml); OXHS1, OXHS2, OXHS5, OXHS7, OXHS8, and OXHS11 were predicted to be possible nuclear proteins, whereas the others were predicted to be located in cytoplasm. To check the authenticity of the prediction, we chose four representative sequences (OXHS1, OXHS2, OXHS8, and OXHS10) to make OXHS-GFP fusion constructs and transformed them into onion epithelial cells. The results showed that all the three fusion proteins were located in cytoplasm (Fig. 4), which differed from the predicted nuclear location for OXHS1, OXHS2 and OXHS8.
Expression profiles of OXHS family in different tissues and organs
RT-PCR was performed to check the relative transcript levels of OXHS genes in different types of tissues or organs using gene-specific primers. Transcripts were detected for all the OXHS genes except OXHS11. No transcript was detected—not even with long PCR cycles—for OXHS11 in all the tissues checked, and no EST was found for this gene in the rice EST database either. This result suggests that OXHS11 may either be a pseudogene or specifically expressed in limited tissues or organs that were not included in this experiment. The other 10 OXHS genes can be classified into three types according to their expression patterns (Fig. 5). The first type of genes (including OXHS1, OXHS6, OXHS8, and OXHS10) were expressed in most of the tissues or organs investigated. The expression patterns of these genes have strong similarity, especially OXHS1 and OXHS10, which both show high transcript levels in young panicles, stamen, pistil, root, and callus. Expression patterns of OXHS6 and OXHS8 are similar to OXHS1 and OXHS10, respectively, although the four genes showed relatively weak expression in callus or stamen. The second type of genes (including OXHS2, OXHS3, OXHS4, and OXHS9) were expressed only in young panicles, stamen, and pistil. The third type of genes (including OXHS5 and OXHS7) were expressed specifically in pistil (OXHS7) or stamen (OXHS5). OXHS2 to OXHS9 showed higher transcript levels in reproductive tissues or organs than in vegetative tissues. Transcript was not detected in root and flag leaf for OXHS2 to OXHS10 (Fig. 5). These results suggest that most of the genes in the OXHS family are involved in reproductive development. We also checked the expression profiles of OXHS family genes in the expression profiling database of the rice genome based on gene chip analysis for more than 30 tissues or organs (Wang et al. submitted), and the results (Supplemental Fig. 3) agreed well with the RT-PCR results.
Semi-quantitative PCR analysis of the OXHS family in ten different tissues or organs of Minghui 63. Based on the expression levels, the OXHS genes can be roughly classified into three types (I–III). The rice Actin1 gene was used as the internal control. The numbers after the gene names denote PCR cycles
Expression profiles of OXHS genes under abiotic stresses
Many plant-specific gene families are involved both in development and stress responses. To check whether any genes of the OXHS family are responsive to stresses, real-time PCR was used to quantify the relative transcript levels of the OXHS genes under a few abiotic stresses. The results showed that, except OXHS5 and OXHS11, the other nine genes were induced in the seedling leaves by at least one of the stresses applied, including drought, salt, cold, and abscisic acid (ABA) treatment (Fig. 6). Among them, six genes (OXHS1, OXHS2, OXHS3, OXHS4, OXHS9, and OXHS10) were evidently induced by drought stress with distinct patterns. For example, the transcript levels of OXHS2, OXHS3, OXHS4, and OXHS9 were increased at the early stage of drought stress and then decreased, while the expression level of OXHS10 increased gradually and peaked at the late stage of the stress. OXHS2 and OXHS3 were induced by salt stress, whereas OXHS8 and OXHS10 were suppressed by salt stress. The expressions of OXHS2, OXHS3, OXHS7, and OXHS9 were evidently induced within 3 h after cold (4°C) treatment. We noticed that the expression levels of OXHS2 and OXHS3 fluctuated, increasing within 3 h, being reduced to the control (0 h) level at 6 h, and then increasing more significantly at 12 h. Such a fluctuation was not due to circadian effect because these genes showed a rather constant expression level in a day-night cycle under normal growth conditions (data not shown). In the ABA treatment, OXHS1, OXHS2, OXHS6, OXHS8, and OXHS10 were significantly induced within 3 h after treatment, and OXHS2 showed a slight increase of transcript expression.
Real-time PCR analysis of OXHS genes in response to abiotic stresses at the seedling stage in Minghui 63. Four-leaf-stage Minghui 63 was treated with drought, salt, cold, and ABA and leaves were sampled at different times as indicated. The RNA used for OXHS2 and OXHS3 was from a different batch of stress-treated samples. The error bars are based on three replications
OXHS2 and OXHS3 showed a similar fluctuation of transcript level in the ABA treatment as observed in the cold treatment. In fact, the stress-induced expression patterns of OXHS2 and OXHS3 were very similar to each other in all the four stress treatments. More interestingly, these two genes also showed similar expression profiles in different tissues or organs (Fig. 5). Further analysis of the cis-acting DNA elements in the promoter region (1 kb upstream of transcriptional start site) of OXHS2 and OXHS3 revealed a few stress-related cis-elements (e.g., ABRELATERD1, ACGTATERD1, ACGTTBOX) as well as a few tissue-specific cis-elements (e.g., GTGANTG10 and POLLEN1LELAT52 involved in pollen-specific expression). In general, the putative cis-elements detected in the OXHS genes were closely related with the expression specificity of tissues or organs and the stress responsiveness.
Over-expression of OXHS2 in rice resulted in reduced salt and drought tolerance
As quite a few OXHS genes are responsive to stress treatments, we further tested whether these genes are involved in stress tolerance. For this purpose, one of the representative OXHS genes, OXHS2, which was strongly induced by salt and drought stresses (Fig. 6) and encodes a protein without self-activation in yeast and targeted to cytoplasm in onion epidermal cells (Fig. 4) was over-expressed in japonica rice Zhonghua 11 under the control of a ubiquitin promoter (Fig. 7a). Real-time PCR showed that 8 of 15 T0 transgenetic plants showed over-expression of the transgene (Fig. 7b). Salt stress tolerance was tested by growing three independent OXHS2-overexpressed T1 lines (U1, U5, and U24), one non-overexpression T1 line (U7), and WT on MS medium containing 150 mM NaCl. On the MS medium without salt stress, the OXHS2-transgenic and WT plants showed no significant difference in shoot length and fresh weight (t-test, P > 0.05; Fig. 7c). Interestingly, the over-expressed lines showed obviously reduced growth performance under the salt stress (Fig. 7d). The values of shoot length and fresh weight of the OXHS2-overexpression lines were significantly lower than that of U7 and WT (t-test, P < 0.01, Fig. 7e, f), although the root length showed no significant difference (Fig. 7d). We measured the proline content in the leaves of the plants before and after the stress, and result showed that the stress-induced increase of proline content in the over-expression plants were significantly lower than that in U7 and WT (t-test, P < 0.05; Fig. 7g). These results suggested that the salt tolerance of the OXHS2-overexpressed plants has been impaired.
Identification and salt tolerance testing of OXHS2 over-expressed plants in rice. a Schematic diagram of OXHS2 overexpression construct. Ubi, Maize ubiquitin gene promoter; Hpt, hygromycin resistance gene. b Real-Time PCR of part of T0 transgenic plants and WT.c, d Growth performance of WT and OXHS2 transgenic plants under normal condition (c) and salt stress with 150 mM NaCl for 7 days (d). e, f Shoot length (E) and fresh weight (F) of WT and OXHS2 transgenic plants under normal condition and salt stress for 7 days. g Content of free proline in the OXHS2 transgenic plants compared to the WT plants under normal conditions and salt stress. U1, U5 and U24 are OXHS2-overexpressing lines; U7, a negative control of transgenic plant; WT, wild type. Error bars are based on three replicates. Significant difference was determined by t test (* P < 0.05; ** P < 0.01)
Drought resistance of three independent OXHS2-overexpression lines (U1, U5, and U24) at T1 generation was also tested at panicle development stage (2 weeks before flowering) in the field at Hainan Island, China. The over-expression lines had significantly (t-test, P < 0.05) lower relative yield and fertility, two commonly used parameters for evaluating drought resistance at reproductive stage (Yue et al. 2006; Xiao et al. 2009), than that of the transgenic control line (U7) and WT (Fig. 8; Supplemental Fig. 4). No significant difference was observed between the over-expression lines and the controls under normal growth (data no shown). This result suggests that over-expression of OXHS2 in rice has negative effect on drought tolerance. Nevertheless, no significant difference was detected between the OXHS2 over-expression lines and WT for the tolerance to cold and ABA treatments, suggesting that OXHS2 gene might be specifically involved in salt and drought tolerance in rice.
Relative yield (a) and fertility (b) of over-expressed T1 families of OXHS2 under the field conditions after drought stress applied at panicle development stage. Relative spikelet fertility or yield is the ratio of the spikelet fertility or yield under stress conditions to that under normal growth conditions. Data represent mean ± SD (n = 12) with t test (* P < 0.05; ** P < 0.01)
Discussion
So far, no XHS homologue sequences from human, other animals, or bacteria have been deposited in the public databases, which suggests that XHS family genes may have evolved specifically in plants (Fraser et al. 2005). Using a sequence similarity search we identified 35 putative XHS protein sequences in plants, and these proteins can be divided into five subgroups (Fig. 1). Most of the XHS sequences from monocot and dicot plants are grouped separately, suggesting that XHS proteins derived from several common ancestors before the monocot-dicot divergence and the evolution of XHS proteins may be independent in monocot plants (such as rice and maize) and dicot plants (Arabidopsis and M. truncatula). Such an evolutionary feature has been also observed for other plant-specific gene families, such as SPL (Xie et al. 2006). In general, the OXHS genes from the same subgroup have similar gene structure. For example, OXHS1, OXHS2, OXHS3, and OXHS4 in S1 and OXHS7 and OXHS10 in S4 have highly similar exon–intron structures, and both OXHS7 and OXHS10 have only the XH domain. Likewise, OXHS8 has the closest relationship with SGS3 and they both contain only the XS domain.
The XHS family is a plant-specific gene family encoding for proteins characterized by three domains: zf-XS, XS, and XH (Bateman 2002). Proteins with different domain constitutions may have distinct functions. The OXHS family can be roughly divided into three subgroups based on the domain constitutions. The zf-XS, XS, and XH domains are specific to XHS family and are highly conserved in plants. The regions beyond the three domains are variable, and no conserved motif was detected by using MEME program. These regions may contribute to the functional diversification of the proteins with the same constitution of conserved domains.
The first XHS protein (X1) was predicted to be a transcription factor (Chen and Bennetzen 1996) Analysis of the common sequence characteristics of the rice XHS proteins may provide some clues as to whether they are transcription factors. A typical plant transcription factor consists of a DNA-binding domain, an oligomerization site, a transcription regulation domain, and a NLS (Liu et al. 1999; Washburn et al. 1997). The DNA-binding domains of plant transcription factors, usually containing many basic amino acid residues, interact with DNA by binding to specific cis-acting elements, and such protein-DNA interactions determine the specificity or classification of transcription factors (Huang et al. 1996; Marmorstein and Fitzgerald 2003; Spittau et al. 2007; Yamasaki et al. 2008). Using motif composition analysis, we found that the C-terminal of OXHS proteins in type I (except OXHS2) and OXHS11 contain the zf-XS domain that may interact with DNA (Bateman 2002), but this possibility is challenged by the fact that some zinc finger domains mediating protein interactions are found in non-transcription factors, such as ubiquitin ligases (Imai et al. 2003; Joazeiro and Weissman 2000).
Many plant transcription factors can form hetero- and/or homo-oligomers to modulate DNA-binding specificity and affinity (Guiltinan and Miller 1994) and nuclear localization (Sainz et al. 1997a, b). Oligomers are stabilized either by hydrophobic interactions between coiled-coils and β sheets or by reactions between hydrophilic residues (Lupas and Gruber 2005; Yu 2002). The X1 protein (OXHS1) is predicted by NCoil (Lupas et al. 1991) to contain coiled-coils, which suggest that it may oligomerize (Bateman 2002). Some X1-like proteins were found to contain coiled-coil domain proteins belonging to one of five coiled-coil protein families, some of which were predicted to be involved in signal transduction in plants, while others were unknown (Rose et al. 2005). These features suggest XHS proteins may be able to form oligomers. Using the coiled-coil prediction program MultiCoil (Wolf et al. 1997), we found that nine OXHS proteins had a coiled-coil motif located between XS and XH domains, in front of the XH domain, or behind the XS domain (Fig. 2). The coiled-coil regions contain many repeated leucine residues (Supplemental Fig. 5), which may be favorable for the stableness of the coiled-coils (Lupas et al. 1991).
Transcription factors generally have distinct actions because of the divergence in their regulation domains (Yanagisawa and Sheen 1998). Regulation domains, and hence transcription factors, function as either repressors or activators, depending on whether they inhibit or stimulate the transcription of target genes (Liu et al. 1999). Activation domains of plant transcription factors often exhibit sequence divergence; their functions depend on interactions between individual residues, and single strategically placed residues determine activation (Sainz et al. 1997a, b). For example, leucine 253 in the activation domain of the maize Myb-like transcription factor C1 is also involved in the transcription activation and modification of other amino acids in the domain (Sainz et al. 1997a, b). We tested the transactivation activity of three representative OXHS proteins in yeast and none of them showed the activity, although such activity in yeast is not a necessary requirement for all transcription factors. Nevertheless, whether the OXHS proteins have transcriptional suppression activity remains to be tested. The XS and XH domains contain a completely conserved aspartate and a glutamate site, respectively, and both sites might be enzymatically active or functionally important (Bateman 2002). Bateman suggested that XH and XS domains may interact, but our results suggested XS and XH domains could not interact in yeast.
Nuclear-localization is one of the most obvious features for transcription factors, but our results suggested that the four representative OXHS-GFP fusion proteins tested were located in the cytoplasm of onion epidermal cells. However, it cannot be excluded that the native OXHS proteins might function as co-factors in regulation are exported to nucleus by interacting with a nuclear targeting partner, as reported for a bHLH protein (Goldfarb and Lewandowska 1994).
The transcript levels of OXHS genes in different tissues and organs are quite diverse. All the genes are expressed in floral organs and a few of them are specifically expressed in floral organs (Fig. 5). These spatio-temporal expression profiles provided useful clues to further dissect the function of these genes in development. Noticeably, majority (9/11) of the OXHS genes are responsive to at least one of the abiotic stresses applied. To further confirm that the OXHS family is involved in stress tolerance, the OXHS2 gene was over-expressed in rice, and the transgenic rice indeed showed reduced salt tolerance at the and drought tolerance. Even though the molecular functions of OXHS proteins remain unclear, which is mainly due to the limited reports on XHS family and unavailability of mutants for most of the OXHS genes, the facts that most OXHS genes are responsive to abiotic stresses and over-expression of OXHS2 resulted in alteration of stress tolerance suggest that some members of this family may play important roles in stress tolerance. Meanwhile, some of these genes may be also involved in development control of rice as quite a few genes showed tissue- or organ-specific expression.
In conclusion, our data suggest that XH-XS-domain-containing proteins constitute a novel plant-specific family with diverse functions both in development and/or stress resistance. These results provide a general view of the organization and functions of this gene family in rice, and further elucidation of the detail molecular mechanisms of individual OXHS genes will greatly expand our understanding of plant development and stress responses.
Abbreviations
- ABA:
-
Abscisic acid
- XH:
-
X1 homologue
- XHS:
-
XH and XS domain
- PCR:
-
Polymerase chain reaction
- RT:
-
Reverse transcription
References
Bateman A (2002) The SGS3 protein involved in PTGS finds a family. BMC Bioinformatics 3:21
Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR (2004) The Pfam protein families database. Nucleic Acids Res 32:D138–D141
Boulikas T (1994) Putative nuclear localization signals (NLS) in protein transcription factors. J Cell Biochem 55:32–58
Calderon-Villalobos LI, Nill C, Marrocco K, Kretsch T, Schwechheimer C (2007) The evolutionarily conserved Arabidopsis thaliana F-box protein AtFBP7 is required for efficient translation during temperature stress. Gene 392:106–116
Chae HJ, Park JM, Lee GY, Park HR, Chae SW, Jeong GS, Kim HM, Kim SB, Yoo SK, Kim HR (2004) Yuk-Hap-Tang induces apoptosis by intervening mn-SOD in human cervical carcinoma HeLa cells. Am J Chin Med 32:883–895
Chen M, Bennetzen JL (1996) Sequence composition and organization in the Sh2/A1-homologous region of rice. Plant Mol Biol 32:999–1001
Dai X, Xu Y, Ma Q, Xu W, Wang T, Xue Y, Chong K (2007) Overexpression of an R1R2R3 MYB gene, OsMYB3R–2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol 143:1739–1751
Durfee T, Roe JL, Sessions RA, Inouye C, Serikawa K, Feldmann KA, Weigel D, Zambryski PC (2003) The F-box-containing protein UFO and AGAMOUS participate in antagonistic pathways governing early petal development in Arabidopsis. Proc Natl Acad Sci U S A 100:8571–8576
Fraser CM, Rider LW, Chapple C (2005) An expression and bioinformatics analysis of the Arabidopsis serine carboxypeptidase-like gene family. Plant Physiol 138:1136–1148
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788
Glick E, Zrachya A, Levy Y, Mett A, Gidoni D, Belausov E, Citovsky V, Gafni Y (2008) Interaction with host SGS3 is required for suppression of RNA silencing by tomato yellow leaf curl virus V2 protein. Proc Natl Acad Sci U S A 105:157–161
Goldfarb AN, Lewandowska K (1994) Nuclear redirection of a cytoplasmic helix-loop-helix protein via heterodimerization with a nuclear localizing partner. Exp Cell Res 214:481–485
Guiltinan MJ, Miller L (1994) Molecular characterization of the DNA-binding and dimerization domains of the bZIP transcription factor, EmBP-1. Plant Mol Biol 26:1041–1053
He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44:903–916
Hegedus D, Yu M, Baldwin D, Gruber M, Sharpe A, Parkin I, Whitwill S, Lydiate D (2003) Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol Biol 53:383–397
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci U S A 103:12987–12992
Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W, Mueller LA, Bhattacharyya D, Bhaya D, Sobral BW, Beavis W, Meinke DW, Town CD, Somerville C, Rhee SY (2001) The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res 29:102–105
Huang H, Tudor M, Su T, Zhang Y, Hu Y, Ma H (1996) DNA binding properties of two Arabidopsis MADS domain proteins: binding consensus and dimer formation. Plant Cell 8:81–94
Imai N, Matsuda N, Tanaka K, Nakano A, Matsumoto S, Kang W (2003) Ubiquitin ligase activities of Bombyx mori nucleopolyhedrovirus RING finger proteins. J Virol 77:923–930
Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46:23–47
Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP (2007) F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143:1467–1483
Joazeiro CA, Weissman AM (2000) RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549–552
Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, Hotta I, Kojima K, Namiki T, Ohneda E, Yahagi W, Suzuki K, Li CJ, Ohtsuki K, Shishiki T, Otomo Y, Murakami K, Iida Y, Sugano S, Fujimura T, Suzuki Y, Tsunoda Y, Kurosaki T, Kodama T, Masuda H, Kobayashi M, Xie Q, Lu M, Narikawa R, Sugiyama A, Mizuno K, Yokomizo S, Niikura J, Ikeda R, Ishibiki J, Kawamata M, Yoshimura A, Miura J, Kusumegi T, Oka M, Ryu R, Ueda M, Matsubara K, Kawai J, Carninci P, Adachi J, Aizawa K, Arakawa T, Fukuda S, Hara A, Hashizume W, Hayatsu N, Imotani K, Ishii Y, Itoh M, Kagawa I, Kondo S, Konno H, Miyazaki A, Osato N, Ota Y, Saito R, Sasaki D, Sato K, Shibata K, Shinagawa A, Shiraki T, Yoshino M, Hayashizaki Y, Yasunishi A (2003) Collection, mapping, and annotation of over 28, 000 cDNA clones from japonica rice. Science 301:376–379
Koiwa H, Barb AW, Xiong L, Li F, McCully MG, Lee BH, Sokolchik I, Zhu J, Gong Z, Reddy M, Sharkhuu A, Manabe Y, Yokoi S, Zhu JK, Bressan RA, Hasegawa PM (2002) C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. Proc Natl Acad Sci U S A 99:10893–10898
Liu L, White MJ, MacRae TH (1999) Transcription factors and their genes in higher plants functional domains, evolution and regulation. Eur J Biochem 262:247–257
Lupas AN, Gruber M (2005) The structure of alpha-helical coiled coils. Adv Protein Chem 70:37–78
Lupas A, Van Dyke M, Stock J (1991) Predicting coiled coils from protein sequences. Science 252:1162–1164
Marmorstein R, Fitzgerald MX (2003) Modulation of DNA-binding domains for sequence-specific DNA recognition. Gene 304:1–12
Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19:270–280
Mittler R (2006) Abiotic stress, the field environment and stress combination. Trends Plant Sci 11:15–19
Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, Remoue K, Sanial M, Vo TA, Vaucheret H (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101:533–542
Muangsan N, Beclin C, Vaucheret H, Robertson D (2004) Geminivirus VIGS of endogenous genes requires SGS2/SDE1 and SGS3 and defines a new branch in the genetic pathway for silencing in plants. Plant J 38:1004–1014
Ohnishi T, Sugahara S, Yamada T, Kikuchi K, Yoshiba Y, Hirano HY, Tsutsumi N (2005) OsNAC6, a member of the NAC gene family, is induced by various stresses in rice. Genes Genet Syst 80:135–139
Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18:2368–2379
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Rose A, Schraegle SJ, Stahlberg EA, Meier I (2005) Coiled-coil protein composition of 22 proteomes–differences and common themes in subcellular infrastructure and traffic control. BMC Evol Biol 5:66
Rotman N, Durbarry A, Wardle A, Yang WC, Chaboud A, Faure JE, Berger F, Twell D (2005) A novel class of MYB factors controls sperm-cell formation in plants. Curr Biol 15:244–248
Saijo Y (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23:319–327
Sainz MB, Goff SA, Chandler VL (1997a) Extensive mutagenesis of a transcriptional activation domain identifies single hydrophobic and acidic amino acids important for activation in vivo. Mol Cell Biol 17:115–122
Sainz MB, Grotewold E, Chandler VL (1997b) Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell 9:611–625
Schwechheimer C, Smith C, Bevan MW (1998) The activities of acidic and glutamine-rich transcriptional activation domains in plant cells: design of modular transcription factors for high-level expression. Plant Mol Biol 36:195–204
Spittau B, Wang Z, Boinska D, Krieglstein K (2007) Functional domains of the TGF-beta-inducible transcription factor Tieg3 and detection of two putative nuclear localization signals within the zinc finger DNA-binding domain. J Cell Biochem 101:712–722
Tominaga R, Iwata M, Sano R, Inoue K, Okada K, Wada T (2008) Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development 135:1335–1345
Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5:1529–1539
Van de Graaff WB, Thompson WL, Sunshine I, Fretthold D, Leickly F, Dayton H (1982) Adsorbent and cathartic inhibition of enteral drug absorption. J Pharmacol Exp Ther 221:656–663
Washburn KB, Davis EA, Ackerman S (1997) Coactivators and TAFs of transcription activation in wheat. Plant Mol Biol 35:1037–1043
Wolf E, Kim PS, Berger B (1997) MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci 6:1179–1189
Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13:1779–1790
Xiao B, Chen X, Xiang C, Tang N, Zhang Q, Xiong L (2009) Evaluation of seven function-known candidate genes for their effects on improving drought resistance of transgenic rice under field conditions. Mol Plant 2:73–83
Xie K, Wu C, Xiong L (2006) Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol 142:280–293
Yamaguchi M, Kubo M, Fukuda H, Demura T (2008) Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J 55:652–664
Yamasaki K, Kigawa T, Inoue M, Watanabe S, Tateno M, Seki M, Shinozaki K, Yokoyama S (2008) Structures and evolutionary origins of plant-specific transcription factor DNA-binding domains. Plant Physiol Biochem 46:394–401
Yanagisawa S, Sheen J (1998) Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell 10:75–89
Yu YB (2002) Coiled-coils: stability, specificity, and drug delivery potential. Adv Drug Deliv Rev 54:1113–1129
Yuan Q, Ouyang S, Wang A, Zhu W, Maiti R, Lin H, Hamilton J, Haas B, Sultana R, Cheung F, Wortman J, Buell CR (2005) The institute for genomic research Osa1 rice genome annotation database. Plant Physiol 138:18–26
Yue B, Xue W, Xiong L, Yu X, Luo L, Cui K, Jin D, Xing Y, Zhang Q (2006) Genetic basis of drought resistance at reproductive stage in rice: separation of drought tolerance from drought avoidance. Genetics 172:1213–1228
Zhu QH, Ramm K, Shivakkumar R, Dennis ES, Upadhyaya NM (2004) The ANTHER INDEHISCENCE1 gene encoding a single MYB domain protein is involved in anther development in rice. Plant Physiol 135:1514–1525
Acknowledgments
This work was supported by grants from the National Special Key Project of China on Functional Genomics of Major Plants and Animals, the National Natural Science Foundation of China, and the Ministry of Education of China.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Qin, Y., Ye, H., Tang, N. et al. Systematic identification of X1-homologous genes reveals a family involved in stress responses in rice. Plant Mol Biol 71, 483 (2009). https://doi.org/10.1007/s11103-009-9535-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11103-009-9535-5