Abstract
The Asian rice gall midge [Orseolia oryzae (Wood-Mason)] is an important rice pest causing an annual average yield loss of about US $80 million in India. Rice varieties possess several discrete resistance (R) genes conferring resistance against the pest in two distinct ways, i.e., with (HR+ type) or without (HR− type) the expression of hypersensitive reaction (HR). The aim of the present work is to understand the molecular basis of compatible and incompatible (HR− type) rice gall midge interactions between the rice variety Kavya and the two gall midge biotypes: the virulent GMB4M and the avirulent GMB1 using transcriptional microarray gene expression analysis. A large number of differentially expressed genes (602genes in incompatible interaction and 1,330 genes in compatible interaction with at least twofold changes, p value <0.05) was obtained from the microarray analysis that could be grouped into six clusters based on their induction during both or either of the interactions. MapMan software was used for functional characterization of these genes into 13 categories (BINs). Real-time polymerase chain reaction validation of 26 genes selected through the analysis revealed four genes viz. NADPH oxidase, AtrbohF, cinnamoyl-CoA reductase, and von Willebrand factor type A domain containing protein coding genes to be significantly upregulated during the incompatible interaction. But most of the signature genes related to HR+ type resistance like salicylic acid pathway-related genes and disease resistance protein coding genes were downregulated. On the other hand, during the compatible interaction, genes related to primary metabolism and nutrient transport were upregulated and genes for defense and signaling were downregulated. We propose a hypothesis that HR− type of resistance in the rice variety Kavya against gall midge could be due to the constitutive expression of an R gene and a case of extreme resistance which is devoid of cell death. Compatible interaction, however, modulated a large number of differentially expressed transcripts to reprogram cell organization, cell remodeling, and relocation of nutrients through transport to support insect growth.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Asian rice gall midge, Orseolia oryzae (Wood-Mason), is an important rice pest. This insect is essentially a wet-season pest and can cause severe damage under favorable weather with high humidity and moderate temperatures, resulting in an estimated annual yield loss amounting US$ 80 million in India (Bentur et al. 2003) or about US$ 500 million in Asia (Herdt 1991). The genetics of rice gall midge interaction is well-characterized. Eleven resistance (R) genes (designated Gm1 through Gm11) have been identified, so far, from different rice varieties (Himabindu et al. 2010). Seven distinct gall midge biotypes, differing in their virulence against these R genes have been reported (Bentur et al. 2003; Vijaya Lakshmi et al. 2006). The interaction between the plant–insect is on gene-for-gene basis and defense response is triggered by initial feeding of the maggot within 48 h. Two distinct interactions are noted when gall midge attempts to feed on a rice plant. In compatible interaction, the rice variety is susceptible and the virulent insect establishes in the plant and a gall is formed. The insect is able to complete its development and emerge from the plant as an adult fly. In incompatible interaction, the rice variety is resistant and the avirulent insect fails to establish and is killed within the plant. Incompatibility is manifested in two distinct ways, i.e., with or without the expression of hypersensitive reaction (HR; HR+ or HR− type; Bentur and Kalode 1996). The HR+ type resistance response of the plant manifests in tissue necrosis at the feeding site and the insect mortality. In contrast, insect mortality in HR− type response is without tissue necrosis. Rice varieties carrying gall midge resistance genes such as Gm1, Gm8, and few other uncharacterized genes show HR− type response, while those carrying other known R genes exhibit HR+ type of response. Efforts are being made to identify and clone some of the gall midge resistance genes from the rice plant. However, the molecular basis of rice gall midge interactions is still to be deciphered. The rice variety Kavya, carrying Gm1gene, is resistant to four out of seven gall midge biotypes found in India and shows HR− type of resistance (Biradar et al. 2004). Development and cultivation of the gall midge-resistant rice varieties has been the main approach to mitigate the pest inflicted production loss. However, cultivation of resistant rice varieties with a single resistance gene has often led to evolution of new virulent biotypes (Vijaya Lakshmi et al. 2006). Pyramiding of two distinct and effective genes in a single cultivar is the strategy being adopted to counter this phenomenon.
Studies carried out so far on molecular basis of insect–plant interactions have shown induction of resistance through a complex network of signal perception, transduction, and induction of a cascade of downstream genes involved in the secondary metabolism and defense. Plant defense against insect herbivores has been generally reported to be distinct from that noted against plant pathogens (Gatehouse 2002). Response of plants against leaf-feeding insects has been found, akin to wounding, to be associated with jasmonic acid pathway while defense against plant sap sucking insects like aphids and the sedentary gall forming insects has been found associated with salicylic acid pathway (Walling 2000; Gatehouse 2002). Expression of HR as a manifestation of plant resistance is widely documented against pathogenic fungi (Alexander et al. 1993), bacteria (Poncianoa et al. 2006), and viruses (Pfitzner and Goodman 1987). Involvement of HR in insect–plant interaction is also not uncommon (Grover 1995). Genotypic variations in HR expression have also been reported in resistant germplasm (Bentur and Kalode 1996; Hoglund et al. 2005). Thus, plant resistance to pathogen or to a specific group of insects need not always to be HR mediated. HR-independent resistance (HR− type) has been reported in many plant–pest interactions. These include barley resistance against all races of powdery mildew, Erysiphe graminis sp hordei, conferred by the mlo gene (Freialdenhoven et al. 1994; Knogge 1996), the potato Rx gene-mediated resistance to potato virus X (Kohm et al. 1993) and wheat resistance to black rust pathogen Puccinia triticina conferred by Lr34/Yr18 genes (Hulbert et al. 2007). The phenotype resulting from the dnd1 mutation in Arabidopsis indicated that cell death was separate and independent of the resistance mechanisms leading to suppression of the pathogen. The phenotype of the dnd1 mutant of Arabidopsis was loss of HR without loss of disease resistance (Yu et al. 1998). Also consistent with the distinction of HR and disease resistance were the effects of humidity and temperature that could inhibit the HR response without compromising resistance (Baulcombe et al. 1994; Hammond-Kosack et al. 1996). The cell death response may be to reinforce or stimulate the induction of defenses (Dangl et al. 1996; Alvarez et al. 1998) by activation of local and systemic acquired resistance (Staskawicz et al. 1995; Ryals et al. 1996).
Earlier, to gain insight into the molecular basis of gall midge resistance in rice, we have analyzed differentially expressed genes in the rice variety Suraksha with Gm11 gene expressing HR+ type resistance using suppression subtractive hybridization library (Rawat et al. 2010; Rawat 2012). In continuation, we have now examined rice gall midge interactions in the rice variety Kavya adopting a transcriptome profiling strategy using microarray analysis. We observed that the signature genes typically involved in HR+ type resistance were downregulated in this incompatible interaction. Thus, this study reports a different mechanism of resistance. This study also covered compatible interaction which involved upregulation of the genes related to nutrient transport and metabolism.
Material and methods
Plant and insect material
The experimental material consisted of the rice (Oryza sativa L.) variety Kavya grown under greenhouse conditions at the Directorate of Rice Research, Hyderabad, India and the two gall midge biotypes (GMB1 and GMB4M) maintained on the appropriate differential rice cultivars and in physical isolation from each other, as described in detail by Vijaya Lakshmi et al. (2006) and Himabindu (2009). GMB1 is avirulent while the GMB4M is virulent on Kavya. Uninfested Kavya plants were used as the control.
Experimental treatments and RNA extraction
The Kavya seedlings infested with GMB4M or GMB1 represented tissue samples for compatible or incompatible interactions, respectively. Seedlings under the same conditions but without infestation were used as uninfested control. Fifteen-day-old seedlings of the test cultivars were exposed to newly emerged female and male adult insects (25 female and 10 male adults on each tray) according to the standard procedure (Himabindu 2009; Rawat et al. 2010). Each experiment contained three independent sets of seedlings that were treated in the same way and at different times in different chambers as biological replicates. Larval hatching time was monitored with additional infested plants under the same growth chamber. Tissue samples were collected at four time points (24, 48, 96, and 120 h after larval hatching) from 20 plants and pooled for each replication. Only those plants with 10–15 larvae per plant were chosen for tissue collection. The time chosen for sample collection was based on the pest biology, 24 h is critical to determine whether a larva can live or die on a specific plant. Only the tissue at the feeding site (about 1.5–2 cm of the meristematic stem tissue) was collected for RNA extraction.
A portion of the seedlings in each experiment was kept for phenotypic confirmation later. Basal part of the plants, above the root, were cut and dissected to remove live and dead maggots and meristematic tissue from the feeding site was collected and immersed in RNALater (Ambion, USA). Total RNA was extracted from meristematic stem tissues by using TRIzol (Invitrogen, USA) according to the procedure provided by the manufacturer. The RNA samples were further purified with DNAase (Qiagen, GmbH, Germany) through an RNeasy Plant Mini kit according to the manufacturer’s instruction (Qiagen, GmbH, Germany).
Microarray analyses
Transcriptome analysis was performed using Affymetrix GeneChip Rice Genome Arrays (Affymetrix), and three biological replicates were analyzed. RNA quality was verified using an Agilent Bioanalyzer (Agilent Technologies) and spectrophotometric analysis (NanoDrop ND-1000; NanoDrop Technologies) to determine concentration and the A260/A280 and A260/A230 ratios. Preparation of labeled copy RNA from 5 μg of total RNA, target hybridization, as well as washing, staining, and scanning of the arrays were carried out by Ocimum Biosolutions (Hyderabad, India) as described in the Affymetrix GeneChip Expression Analysis Technical Manual (http://media.affymetrix.com/support/downloads/manuals/expression_analysis_technical_manual.pdf).
Microarray normalization and data analysis
The hybridization data was analyzed using GeneChip Operating Software (GCOS 1.4) (http://media.affymetrix.com/support/downloads/manuals/expression_analysis_technical_manual.pdf). The scanned images were examined for any visible defects. Satisfactory images were analyzed to generate raw data saved as CEL files using the default settings of GCOS 1.4 from Affymetrix. A global scaling factor of 500, a normalization value of 1, and default parameter settings for the rice genome array were used. The detection calls (present, absent, or marginal) for the probe sets were made by GCOS. CEL files generated by GCOS were imported into Avadis 4.3 (Strand Life Sciences, India) for further analysis. Raw intensity data were initially normalized using the robust multichip average algorithm allowing gene expression levels with statistical significance (p < 0.05) above background level were recorded as present. For calculating the expression index of probe sets, we used the perfect match/mismatch model and setup “absent” by truncating the low-expression values to the tenth percentile of the probe set signals. The expression values were log2 transformed after calculating the expression index. The signal log ratio is the change in the expression level of a transcript between the control and experimental samples, and log2 (ratio) ≥1 (twofold change) or log2 (ratio) ≤ −1 (0.5-fold change). A linear model approach has been used for identifying differentially expressed genes for a specific threshold fold change (FC) and an adjusted p value. During analysis, the log fold change was set to 1 (FC = 2) and the adjusted p value was set 0.05. The analysis was performed using “Limma” software from R-package (http://www.r-project.org). The locus information corresponding to the Affymetrix probe set IDs was obtained from the National Science Foundation rice nucleotide array project (https://www.ricearray.org) through the rice genome server of The Institute for Genome Research (TIGR; version 5.0). Functional analyses of the differentially expressed genes were done with the MapMan software based on the Wilcoxon Rank Sum Test.
Data submission and accession number
The microarray data has been deposited in NCBI’s Gene Expression Omnibus (https://www.ncbi.nim.nih.gov/geo) and is accessible through GEO series accession number GSE22538.
Functional annotation and metabolic pathway analysis using MapMan software
Functional annotation and metabolic pathway analysis were performed using MapMan (Thimm et al. 2004; Usadel et al. 2005). MapMan was also used to identify the functional categories associated with a set of sequences (e.g., differentially expressed) and to find the metabolic pathways or other cellular functions up- or downregulated in microarray experiments. Functional classification in the mapping file (Osa_affy_150909) that structures the rice gene from an Affymetrix into distinct metabolic and cellular processes from the MapMan program was used. Differentially expressed rice genes were functionally annotated by performing Basic Local Alignment Search Tool alignment against the TIGR database. MapMan software was employed to show the differences in gene expression in different cellular and metabolic process. Ratios were expressed in a log2 scale for importing into the software and changes in expression were displayed via a false color code.
Real-time PCR validation
The microarray results were validated via real-time polymerase chain reaction (PCR) with two types of samples: the same RNA samples that were used for microarray analysis and newly prepared RNA samples from independent infestations. Real-time PCR conducted with RNA samples prepared from independent infestations with two time points (24 and 120 h after gall midge infestation). Three micrograms of RNA was used for first-strand cDNA synthesis using the SuperScript® III RT enzyme (Invitrogen, USA) following the manufacturer’s guidelines. Real-time PCR was performed using Applied Biosystems 7500 real-time PCR System with the SYBR green chemistry (Applied Biosystems, USA) according to the manufacturer’s instructions.
Twenty-six genes having greater than twofold change when compared to control were further validated using quantitative PCR (qRT). Gene-specific primers for real-time PCR were designed using Primer Express® Software (Applied Biosystems; Table 1). Rice ubiquitin gene, OsUbq (GenBank accession no. AK059694) was used as the endogenous control. Total real-time PCR volume of 20 μl contained 10 μl SYBR Green I PCR Master Mix (Applied Biosystems, USA), 500 nM each of forward and reverse primers and 20 ng of the cDNA samples. To calculate mean relative expression levels, cDNAs from three independent biological samples in three technical replications each were used. PCR was initiated with a pre-incubation at 50°C for 2 min and denaturation at 95°C for 10 min followed by 40 cycles of denaturation at 95°C for 15 s and annealing and extension at 60°C for 1 min. The real-time PCR efficiencies for the reactions were >95 %. A melting curve analysis was performed to determine the specificity of the reaction. Fluorescence data produced sigmoid-shaped amplification plots when plotted against the number of cycles. After normalization, quantity of each mRNA was calculated from the threshold points located in the log-linear range. The data were analyzed using the 7500 Sequence Detection Software (Applied Biosystems, USA) with default baseline and threshold.
Results
Gene expression profiling through microarray analysis
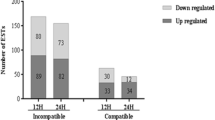
Of the 51,279 probe sets contained in the Affymatrix Rice GeneChip, 50,382 hybridization signals were detected in our study. Total number of probe sets for analysis was reduced to 24,150 transcripts after removing probe sets with ambiguous signals and those that were not called “present” in at least two replicates. Among these transcripts, 602 and 1,330 genes recorded at least twofold changes in expression levels at a p value <0.05 in paired t tests (either up or down) between uninfested control vs. GMB1-infested plant samples representing incompatible interaction (ESM Table 1, S1) or between uninfested control vs. GMB4M-infested samples representing compatible interaction (ESM Table 1, S2), respectively. Thus, number of genes that were either up- or downregulated during compatible interaction was about two times greater than that during incompatible interaction. We also identified 236 upregulated and 224 downregulated genes common to both the interactions. However, only 553 and 44 genes showed upregulation only in compatible or only in incompatible interactions, respectively. In contrast, 317 and 98 genes were specifically downregulated only during these interactions, respectively (Fig. 1). Further, these up- or downregulated genes obtained from the microarray data were grouped into six expression clusters using Euclidean distance as distance measure and complete linkage algorithm (Fig. 2). Genes were analyzed for their putative function using rice database (rapdb.dna.affrc.go.jp). Out of 236 commonly upregulated genes in both the interactions, cluster I contained 126 genes with specific function and remaining 110 genes were from hypothetical, expressed protein, unknown function, or transposons class. Genes in this cluster were those involved in various cellular processes including metabolism, transport, and regulation of gene expression and/or signal transduction. Similarly, out of 224 commonly downregulated genes, cluster II contained 110 genes that were mostly related to oxidative burst and pathogenesis-related proteins, phenylalanine ammonia lyase, cytochrome P450—the signature genes known to play important role in HR+ type resistance. Out of 553 upregulated genes in compatible interaction, cluster III contained 385 genes that were mostly related to stress response, including various heat shock proteins, primary metabolism and nutrient transporters and those involved in regulation and/or signal transduction. Out of 317 downregulated genes of compatible interaction, cluster IV contained 156 genes which were mostly related to disease resistance proteins, Myb transcription factor, and AP2 domain containing proteins. Out of 44 upregulated genes of incompatible interaction, cluster V had 29 genes pertaining to respiratory burst protein, late embryonic abundant protein, NADPH oxidase, AtrbohF, cinnamoyl CoA reductase, and TINY ethylene responsive transcription factor. Out of 98 downregulated genes of incompatible interaction, cluster VI contained 49 genes those were related to hypersensitive-related proteins, flavonol synthase, thionin, OsWAK transcription factors, and transport-related genes.
Venn diagram representing common and specifically up- and downregulated genes in the rice variety Kavya during compatible (after infestation with the rice gall midge biotype, GMB4M) and incompatible (after infestation with the rice gall midge biotype, GMB1) interactions. Numbers represent genes upregulated in incompatible (A), compatible (B), common to both (E) interactions and genes downregulated in incompatible (C), compatible (E), common to both (rice gall) interactions
Cluster display of differentially expressed genes during compatible and incompatible interactions of the rice variety Kavya with gall midge biotypes GMB1 or GMB4M. Differentially expressed genes were grouped into six clusters: cluster I (upregulated) and cluster II (downregulated) in both the interactions; cluster III (upregulated) and cluster IV (downregulated) specific to the compatible interaction and, cluster V (upregulated) and cluster VI (downregulated) specific to the incompatible interaction. Numbers correspond to differentially expressed genes in comparison with uninfested control. Genes were clustered using Euclidean distance and complete linkage algorithm. The cutoff values for grouping of up- and downregulated genes were kept at ≥4- or ≤4-folds, respectively
Differentially responsive genes
The differentially expressed candidates were evaluated for their biological relevance through Gene Ontology (http://www.geneontology.org/; ESM Table 2) and KEGG Pathway analysis (http://www.genome.jp; ESM Table 3). Fisher’s exact test was carried out to determine the significance of the GO term. We further analyzed the genes from each cluster that appear to be the major targets of differential regulation during compatible and incompatible interactions using MapMan software (Thimm et al. 2004; Usadel et al. 2005) with O. sativa mapping file. Mapping file functionally classified all the genes of Affymetrix rice chip in 36 major categories (BINs) and several subBINs. Out of 36 major BINs, 13 BINs involved in these interactions are described in detail.
Expression of genes common to both the interactions
Two functional categories “Transcription factors” (BIN 27) and “Development” (BIN 33) were found to be commonly upregulated (ESM Table 1, S3). However, compatible interaction induced larger number of genes compared to incompatible interaction. The main type of transcription factors commonly induced in both the interactions were AP2/EREBP, MADS box transcription factor family, basic helix–loop–helix family, and Homeobox transcription factor family. But GRAS transcription factor family, WRKY domain transcription factor family, growth-regulating factor and histone deacetylase 2b in this BIN were induced only in compatible interaction. In “Development BIN” (BIN 33) genes coding for nodulin-like protein 5NG4, protein RCN1, protein MTN3 and MARD1 were commonly upregulated. However, genes of this BIN coding for embryogenesis-associated protein EMB8 in compatible interaction and late embryogenesis abundant protein D-34 in incompatible interaction were specifically upregulated. One gene encoding senescence associated protein DH was found to be commonly downregulated in both the interactions.
Five functional categories such as secondary metabolism (BIN 16), hormone signaling (BIN 17), defense-related genes (BIN 20), redox (BIN 21), and signal transduction (BIN 30) were found to be commonly downregulated in both the interactions (ESM Table 1, S4). In secondary metabolism (BIN 16), phenylpropanoids, chalcones, dihydroflavonols were prominent. However, few genes such as flavonol synthase/flavanone-3-hydroxylase and dihydroflavonol-4-reductase were downregulated only in incompatible interaction while isoflavone reductase was downregulated only in compatible interaction. Plant hormone signaling-related pathway genes (BIN 17) such as jasmonate synthesis-related gene lipooxygenase, gibberelin synthesis-related genes ent kaurene synthase, salicylic synthesis-related benzoate carboxyl methyltransferase, and phenylalanine ammonia lyase were commonly downregulated in both the interactions. Significantly, most of the genes in defense-related category (BIN 20) were found to be commonly downregulated in the interactions like those encoding for pathogenesis-related proteins (PR1β and PR10α), xylanase inhibitors, germin-like protein subfamily 1, hypersensitive-induced response protein. However, few genes of this BIN such as those coding for nucleotide-binding site–leucine-rich repeats (NBS–LRR) disease-resistance protein, von Willebrand factor type A domain containing protein and NADPH oxidase were upregulated in both the interactions. Genes of redox BIN (BIN 21) like catalase and l-ascorbate oxidase and l-ascorbate peroxidases were commonly downregulated. Genes related to signal transduction (BIN 30) such as receptor kinase DUF26, wall associated kinase, Ca+2 signaling receptors were commonly downregulated. But few genes encoding for leucine-rich repeat III and G-protein receptor of the BIN were upregulated in both the interactions.
Expression of genes specific to the compatible interaction
Out of 13 BINs, genes present in the six BINs such as cell wall metabolism (BIN 10), primary metabolism (BIN 13), DNA synthesis and chromatin remodeling (BIN 28), protein synthesis and turnover (BIN 29), cell organization and division (BIN 31), and transporters (BIN 34) were upregulated in compatible interaction (ESM Table 1, S5). However, none of the BINs was found to be downregulated. In BIN 10, genes related to cell wall degradation like pectate lyases and polygalacturonases and cell wall modification-related genes were upregulated significantly in compatible interaction. Large number primary metabolism (BIN 13) genes involved in glycolysis, TCA cycle, nucleotide, and lipid metabolism were upregulated in the compatible interaction. Genes such as replication factor, superfamily II DNA helicase, DNA glycosylase, DNA repair protein RAD51, and exonuclease of DNA synthesis and chromatin remodeling BIN (BIN 28) were upregulated during this interaction. In protein synthesis and turnover BIN (BIN 29) genes of different sub-BINs like protein activation, protein synthesis, protein targeting, post-translational modification, and protein degradation were upregulated in compatible interaction. In cell organization and division BIN (BIN 31), genes related to cell division such as microtubule-associated protein MAP65-1a, meiotic recombination protein DMC1, and genes related to cell cycle such as cyclin-A2, CYCD6, cyclin delta-3, and cyclin-dependent protein kinase were upregulated. A large number of genes from transporters category (BIN 34) such as amino acids transporters, metabolite transporters, peptides and oligopeptides, ABC transporters, auxin hydrogen symporter, antiporter/drug transporter, and phosphatidylinositol transporter were upregulated in compatible interaction. Further, defense-related (BIN 20) abiotic stress responsive genes (BIN 20.2) such as heat shock proteins and chaperones were also upregulated.
Expression of genes specific to the incompatible interaction
Genes specifically modulated in incompatible interaction, were relatively few in number (44 upregulated and 98 downregulated). Among the upregulated genes, biotic stress and respiratory burst (BIN 20.1.1) genes, AtrbohF, cytochrome P450 protein encoding genes, respiratory burst oxidase and cinnamoyl-CoA reductase were present (ESM Table 1, S6). Protein synthesis and degradation (BIN 29)-related genes such as cysteine proteinase 2 and serine carboxypeptidase 1 were also upregulated. From primary metabolism category (BIN 11), phospholipase D beta 1, lipid-binding protein, and aldose 1-epimerase of lipid metabolism were upregulated. BINs for secondary metabolism (BIN 16), salicylic acid (SA) hormone signaling (BIN 17), and defense- (BIN 20) and transport-related (BIN 34) genes were noticeably downregulated in the incompatible interaction.
Validation of selected transcripts through real-time PCR
Microarray results of 26 differentially expressed genes from both the interaction were validated using real-time PCR (Tables 1 and 2). The real-time PCR results were generally in agreement with microarray data as shown in ESM Fig. S1. Correlation value was 0.25 and 0.22 between microarray data and real-time PCR data at 24 and 120 h time point, respectively, which were significant at p ≤ 0.05.
Genes modulated at 24 h after infestation
Out of 26 genes, 10 genes in compatible interaction and four genes in incompatible interaction were upregulated with high fold values (Fig. 3). Out of 10 upregulated genes in compatible interaction, six genes were from BIN 20 viz., xylanase inhibitor, PAZ domain containing protein, respiratory burst oxidase, cytochrome P450 protein, hypersensitive-induced response protein, and hsp20 alpha 2 proteins; the other four genes were coding for GDSL lipase (BIN 13), cytokinine dehydrogenase (BIN 17), PPR repeat containing protein (BIN 27), and annexin (BIN 31). The four genes upregulated in the incompatible interaction were von Willebrand factor A containing protein (BIN 20, 9.5-fold), MADS transcription factor (BIN 27, 2.5-fold), peptidyl prolyl cis/trans isomerase (BIN 31, fivefold), and transmembrane amino acid transporter (BIN 34, eightfold; Fig. 3). Four genes upregulated in both the interactions with similar fold values were for cinnamoyl-CoA reductase (BIN 20), thionin (BIN 20), CCT/B-box Zn finger protein (BIN 27) and aquaporin protein (BIN 34). Four genes were found to be downregulated in both the interactions such as flavonol synthase/flavanone 3-hydroxylase (BIN 16), ent-kaurene synthase (BIN 17), OsPR10α (BIN 20) and Lipooxygenase (BIN 29). The remaining four genes such as Aldose-1-epimerase (BIN 13), glycosyl hydrolase 16 (BIN 13), hsp20 alpha 1 protein (BIN 20), and auxin response factor (BIN 27) were not significantly modulated in either of the interactions.
Genes modulated at 120 h after infestation
At 120 h after gall midge infestation, 15 genes were downregulated in both the interactions (Fig. 4). Nine genes were upregulated in compatible interaction. Among these, three genes such as transmembrane amino acid transporter (18-fold, BIN 34), MADS transcription factor (15-fold, BIN 27) and thionin (8.5-fold, BIN 20) had very high fold values while the remaining six genes were induced with 2- to 2.5-fold value in compatible interaction. Two genes coding for von Willebrand factor A containing protein (BIN 20, 5.5-fold) and cinnamoyl-CoA reductase (BIN 20, sevenfold) were upregulated in incompatible interaction.
Discussion
Plants have developed diverse mechanisms of biotic stress resistance mediated through either jasmonic acid or SA pathways and with or without expression of HR. Most of but not all, R-gene-triggered resistance is accompanied with HR. Insect resistance with HR-like symptoms have also been observed in cases such as wheat (Triticum aestivum) resistance to the Hessian fly (Mayetiola destructor; Grover 1995), peach (Prunus persica) resistance to the aphid Myzus persicae (Sauge et al. 1998), rice (O. sativa) against the gall midge O. oryzae (Bentur et al. 2003), willow (Salix viminalis) tree varieties against the gall midge (Dasineura marginemtorquens; Hoglund et al. 2005) and bean (Phaseolus vulgaris) resistance to the bean-pod weevil (Apion godmani; Garza et al. 2001). Cell death that occurs during HR+ type resistance is reported to be controlled by the oxidative burst (Atkinson et al. 1996; Heath 2000), production of NO (Delledonne et al. 2002), and the interaction between some of these different signaling pathways (Delledonne et al. 2002). Despite the observation that several resistance responses (RR) are accompanied by HR, the signal transduction requirement for the RR and HR can be different depending on the host–pathogen combination (Greenberg and Yao 2004). Host cell death occurs in most of the cases, but not in all the reported interactions between plant and pathogens. Non-cell death or HR− type resistance are rare (Yu et al. 1998), though. However, the rice gall midge interactions involve both HR+ and HR− types of resistance.
The present study revealed unique features of HR− type of resistance in the rice variety Kavya against the gall midge at the molecular level. HR− type of resistance as studied in potato carrying Rx gene against potato virus X revealed that resistance conferred by Rx gene against potato virus X is likely due to the involvement of NBS–LRR protein (Bendahmane et al. 1999) as noted for other R genes that conferred HR+ type of resistance. Upregulation of phenylpropanoid pathway and most of the typical R gene-mediated pathways were also observed in microarray study conducted on wheat expressing HR− type resistance conferred by Lr34/Yr18 gene against the rust P. triticum (Hulbert et al. 2007; Bolton et al. 2008). In both the cases, defense-related genes were also induced including those encoding various pathogenesis-related (PR) proteins. The response conditioned by these genes is reminiscent of some components of HR+ type resistance but without the accompanying localized cell death. Such type of phenotype was also observed in Salix–Dasineura interaction which clearly suggested that hypersensitivity and symptomless resistance (HR− type) are two different mechanisms (Hoglund et al. 2005).
In contrast, our microarray studies revealed downregulation of secondary metabolism (BIN 16), hormone signaling (BIN 17) mainly SA pathway (BIN 17.8) and defense-related genes (BIN 20) and these results have been further validated in real-time PCR studies involving HR− type resistance in the rice variety Kavya against GMB1. Thus, HR− type resistance in the rice variety Kavya did not merely suppress cell death but also suppressed other defense-related genes as in case of compatible interaction. This is a novel and unique observation for any plant–pest interaction reported in the literature. It is important to note that SA-mediated pathway, phenylpropanoid pathway, secondary metabolism, and defense-related genes such as PAL, NBS–LRR, and OsPR10α were induced in rice variety Suraksha expressing HR+ type of resistance against GMB4 biotype of gall midge (Rawat et al. 2010). We have also documented that OsPR10α was upregulated only in HR+ type resistance in Suraksha possessing Gm11 gene but not in HR− type resistance in the rice variety Kavya with gall midge resistance gene Gm1 (Rawat et al. 2010).
Based on the above information, we propose a hypothesis to understand the HR− type resistance mechanism in Kavya against rice gall midge. The HR− type of resistance could be due to the expression of a “constitutive R gene” and could be a result of “extreme resistance” which does not require invoking of a cell death mechanism. The base of this hypothesis is explained based on our microarray and real-time PCR results.
In the present study, we found evidence for strong transcriptional changes in the compatible interaction and a weaker response in the incompatible interaction as 870 and 142 transcripts, respectively, were specifically modulated while 460 transcripts were commonly induced (Fig. 1, ESM Table S1) in these interactions. Thus, the transcriptome of the rice variety Kavya responded weakly to the avirulent biotype of gall midge. This weak transcriptome response is consistent with recent findings in tomato (Solanum lycopersicum) and powdery mildew fungus interaction (Li et al. 2006; Fung et al. 2008). This suggests that transcripts involved in incompatible interaction could be constitutively expressing at high or low levels in this variety and only weakly modulated in response to the gall midge attack. Thus, Gm1 conferred resistance in the rice variety Kavya could be constitutive, unlike Gm11 conferred resistance in Suraksha which was distinctly induced upon pest infestation.
Further, it is also hypothesized that, in some host–pathogen interactions, a phenomenon called “extreme resistance” induces sufficient R protein signaling to arrest the pathogen without cell death (Bendahmane et al. 1999). Thus, some RR are extremely robust to arrest the pathogen without cell death. Torres et al. (2002) considered HR as the function of a weak or strong R gene. The weak R gene initiates cell death by triggering molecules related to hypersensitivity followed by R signaling. The response of the strong R gene, despite less production or depletion of ROS, was sufficient enough to trigger the R signaling. This concept is in line with findings of Bendahmane et al. (1999), demonstrating that HR occurrence and magnitude can be correlated directly to R and Avr protein levels. We suggest that Gm1 conferred resistance that could be a mode of “extreme resistance” which is able to kill the insect without requiring cell death in the host as R proteins constitutively expressed (due to Gm1 resistance gene) are sufficient to kill the insect without requiring HR expression.
In our microarray study, four genes were significantly upregulated in incompatible interaction and these were NADPH oxidase, AtrbohF, von Willebrand domain containing protein (vWA), and cinnamoyl CoA reductase (ESM Table S3, S6). The NADPH oxidase complex is composed of RbohF/RbohD genes and is also known as the respiratory burst oxidase (RBO). Several recent reports demonstrate that members of the Rboh family mediate the production of apoplastic ROS during the defense responses, as well as in response to abiotic stress and developmental cues (Torres et al. 2005; Torres et al. 2006). ROS produced by AtrbohD and AtrbohF are negative regulators of the unrestricted cell death in lesion mimic mutant lsd1 of Arabidopsis (Torres et al. 2005).
vWA domain containing protein and the cinnamoyl-CoA reductase were the only other genes which have shown relatively higher fold of transcript upregulation in incompatible interaction when compared with the compatible interaction at 120 h in real-time PCR validation. The vWA domain containing protein is a well-studied domain involved in cell adhesion, in extracellular matrix proteins, and in integrin receptors. Higher expression of vWA has been reported in resistance interaction between rice and parasitic plant Striga hermonthica, during the fourth and 11th day (∼30-fold) of interaction through microarray analysis (Swarbrick et al. 2008). In the present study, a higher upregulation of vWA gene in incompatible interaction at both the time point suggests involvement of this gene in rice gall midge interaction. Cinnamoyl CoA reductase 1 (OsCCR1) is involved in lignin biosynthesis (Kawasaki et al. 2006). Lignin is polymerized through peroxidase activity of NADPH oxidase by using H2O2 in the cell wall and provides an undegradable mechanical barrier to most of the pathogens (Kawasaki et al. 2006). CCR gene is also reported to be involved in production of monolignols which have antimicrobial activities (Keen and Littlefield 1979). In the absence of H2O2, the conversion of monolignol to lignin is reduced dramatically. H2O2, a molecule for ROS generation, was found to be depleted in HR− type (symptomless) resistance in Salix–Dasineura interaction whereas levels of H2O2 were found to be more in HR+ genotypes (Hoglund et al. 2005). Therefore, we propose that low levels of H2O2 in HR− type of resistance prevent the polymerization of monolignols to lignin. As a result, monolignols can get accumulated due to NADPH oxidase and CCR1 gene activities, in the incompatible interaction and this may prove to be detrimental for gall midge larva. However, the exact role of altered lignin biosynthesis pathway in HR− type of resistance mechanism is still to be investigated.
The present study provides the first ever details of a novel molecular basis of relatively durable gall midge resistance of the HR− type conferred by Gm1, a gene that has been widely deployed through breeding to develop gall midge-resistant rice varieties. Another important implication of the current study is that it provides a better understanding of the mechanism of resistance conferred by Gm1 and its suitability in gene pyramiding along with HR+ type resistance conferred by genes such as Gm11. We suggest that HR− type resistance in the rice variety Kavya against GMB1 has a novel mechanism that could be explained by the proposed hypotheses. The hypothesis suggests that HR− type resistance could be due to the involvement of a “constitutive R gene” as revealed by low profile of transcriptome induction by Gm1 gene. This could also be a case of “extreme resistance mechanism” which is devoid of cell death. It is also noteworthy that resistance conferred by Gm1 gene in the rice variety Kavya is more durable than that conferred by other gall midge-resistant genes which exhibit inducible HR+ type resistance (Bentur et al. 2008). Nevertheless, this novel mechanism of resistance needs further characterization.
In the present study, transcriptome profiling of compatible interaction of Kavya with GMB4M suggested that the virulent gall midge elicits a range of physiological and morphological changes in the rice plant. These changes are understandably brought about by the activation of susceptibility-related transcription regulators, which in turn upregulate expression of genes involved in the metabolism, cell organization, transport of nutrients while downregulating genes involved in defense. The virulent larva, thus orchestrates the plant genetic machinery so as to accommodate its growth. This generalized picture emerged through the present microarray study and real-time PCR validation. However, upregulation of stress-related genes such as respiratory burst oxidase (24-fold), thionin (8.5-fold), and heat shock proteins (fivefold) was observed at 24 h in compatible interaction. These results indicate that Kavya mounted an elevated stress response during early hours of virulent gall midge infestation. This response subsides during the course, resulting in susceptibility. Similarly in the wheat–Hessian fly interaction, virulent larva is initially exposed to the stress response of the host during compatible interaction (Anderson and Harris 2006; Harris et al. 2006; Liu et al. 2010). Downregulation of the genes related to biotic stress defense pathway (BIN 20.1; Fig. 5a and b) including NB-ARC, PR genes, xylanase inhibitors and disease resistance proteins and genes related to secondary metabolism (BIN 16) especially phenylpropanoid pathway (BIN 16.2) genes such as PAL and anthranilate N-benzoyltransferase protein suggests that the virulent larvae efficiently suppressed the plant defense (Fig. 6a and b). The present study also provides evidence for increased and improved food quality that came from the strong upregulation of genes involved in primary metabolism (Fig. 7a and b) as also observed in the wheat–Hessian fly compatible interaction (Liu et al. 2007). Evidence for increased nutrient flow comes from the strong upregulation of genes that encode various transporters (BIN 34), e.g., sugar transporters, peptide transporter PTR2, a phosphate carrier protein, aquaporin, and amino acid transporter-like protein. Our study also revealed induction of transmembrane amino acid transporter gene up to 18-fold during later hours of compatible interaction (as revealed by real-time PCR validation). Earlier studies on rice gall midge interaction reported increased concentrations of amino acids in the host that favored gall midge multiplication (Regupathy and Subramanian 1972) and probably hints at the role of amino acids in the susceptibility of plant. In this regard, transmembrane amino acid transporter may help virulent larva to get better access to food/nutrients. In contrast to the wheat–Hessian fly studies, genes related to DNA structure, synthesis and repair (BIN 28), and cell organization (BIN 31) such as cell division (BIN 31.2) and cell cycle (BIN 31.3) were significantly (p < 0.05) upregulated in the compatible interaction (Fig. 6a and b). Thus to support maggot survival, growth, and gall formation, the entire plant cell machinery is geared up to enhance/augment vegetative growth in the compatible interaction. Transcriptional reprogramming in compatible interaction of rice gall midge featured here suggested that susceptibility is not a default phenotype but is the result of coherent interplay of a large number of genes. Manipulating any of the key genes would result in resistance against the pest.
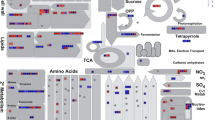
MapMan-based visualization of the transcripts involved in “secondary metabolism” in the rice variety Kavya after infestation with gall midge biotypes (a) GMB1 and (b) GMB4M. Functional subBINs (small squares) shown in red or blue indicate their upregulation or downregulation, respectively. Red rectangles represent commonly downregulated pathways of phenylpropanoids and flavonoids. Color key log2 scale
MapMan-based visualization of the transcripts involved in “cellular response” in the rice variety Kavya after infestation with gall midge biotypes (a) GMB1 and (b) GMB4M. Functional subBINs (small squares) shown in red or blue indicate their upregulation or downregulation, respectively. Red rectangles represent differentially expressed pathways of cell division and cell cycle. Color key log2 scale
MapMan-based visualization of the transcripts involved in “glycolysis and TCA cycle” in the rice variety Kavya after infestation with gall midge biotypes (a) GMB1 and (b) GMB4M. Functional subBINs (small squares) shown in red or blue indicate their upregulation or downregulation, respectively. Red rectangles represent differentially expressed genes of glycolysis pathway. Color key log2 scale
Further, to validate the hypothesis that Gm1-conferred resistance is due to constitutive expression of a R gene. We propose proteomics and metabolomics approach and induce mutation in the Kavya variety aimed at loss of resistance which will help to provide a better insight into the novel mechanism of resistance.
References
Alexander D, Goodman RM, Gut-Rella M, Glascock C, Weymann K, Friedrich L, Maddox D, Ahl-Goy P, Luntz T, Ward E (1993) Increased tolerance to 2 oomycete pathogens in transgenic tobacco expressing pathogenesis related protein-1α. Proc Natl Acad Sci USA 90:7327–7331
Alvarez ME, Pennell RI, Meijer PJ, Ishikawa A, Dixon RA, Lamb C (1998) Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92:773–784
Anderson KG, Harris MO (2006) Does R gene resistance allow wheat to prevent plant growth effects associated with Hessian fly (Diptera: Cecidomyiidae) attack? J Econ Entomol 99:1842–1853
Atkinson MM, Mildland SL, Sims JJ, Keen NT (1996) Syringolide triggers Ca2+ influx, K+ efflux, and extracellular alkalization in soybean cells carrying the disease-resistance gene rpG4. Plant Physiol 112:297–302
Baulcombe DC, Gilbert JE, Goulden M, Köhm BA, Santa Cruz S (1994) Molecular biology of resistance to potato virus X in potato. Biochem Soc Symp 60:207–218
Bendahmane A, Kanyuka K, Baulcombe DC (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11:781–791
Bentur JS, Kalode MB (1996) Hypersensitive reaction and induced resistance in rice against Asian rice gall midge, Orseolia oryzae. Entomol Exp Appl 78:77–81
Bentur JS, Pasalu IC, Sarma NP, Prasada Rao U, Mishra B (2003) Gall midge resistance in rice. Directorate of Rice Research, Hyderabad
Bentur JS, Cheralu C, Rao PRM (2008) Monitoring virulence in Asian rice gall midge populations in India. Entomol Exp Appl 129:96–106
Biradar SK, Sundaram RM, Thirumurugan T, Bentur JS, Amudhan S, Shenoy VV, Mishra B, Bennet J, Sarma NP (2004) Identification of flanking SSR markers for a major rice gall midge resistance gene Gm1 and their validation. Theor Appl Genet 10:1468–1472
Bolton MD, Kolmer JA, Xu WW, Garvin DF (2008) Lr-34 mediated leaf rust resistance in wheat transcript profiling reveals a high energetic supported by transient recruitment of multiple metabolic pathways. Mol Plant Microbe Interact 21:1515–1527
Dangl JL, Dietrich RA, Richberg MH (1996) Death don’t have no mercy: cell death programs in plant–microbe interactions. Plant Cell 8:1793–1807
Delledonne M, Murgia I, Ederle D, Sbicego PF, Biondani A, Polverari A, Lamb C (2002) Reactive oxygen intermediates modulate nitric oxide signaling in the plant hypersensitive disease response. Plant Physiol Biochem 40:605–610
Freialdenhoven A, Scherag B, Hollricher K, Collinge DB, Thordal-Christensen H, Schulze-Leferd P (1994) Nar-1 and Nar-2, two loci required for Mla 12 specified race specific resistance to powdery mildew in Barley. Plant cell 10:947–956
Fung RWM, Gonzalo M, Fekete C, Kovacs LG, He Y, Marsh E, McIntyre LM, Schachtman DP, Qiu W (2008) Powdery mildew induces defense-oriented reprogramming of the transcriptome in a susceptible but not in a resistant grapevine. Plant Physiol 146:236–249
Garza R, Vera J, Cardona C, Barcenas N, Singh SP (2001) Hypersensitive response of beans to Apion godmani (Coleoptera: Curculionidae). J Econ Entomol 94:958–962
Gatehouse JA (2002) Plant resistance towards insect herbivores: a dynamic interaction. New Phytol 156:145–169
Greenberg JT, Yao N (2004) The role and regulation of programmed cell death in plant–pathogen interactions. Cellular Microbiol 6:201–211
Grover PBJ (1995) Hypersensitive response of wheat to Hessian fly. Entomol Exp Appl 74:283–294
Hammond-Kosack KE, Silverman P, Raskin I, Jones JDG (1996) Race-specific elicitors of Cladosporium fulvum induce changes in cell morphology and the synthesis of ethylene and salicylic acid in tomato cells carrying the corresponding Cf disease resistance gene. Plant Physiol 110:1381–1394
Harris MO, Freeman TP, Rohfritsch O, Anderson KG, Payne SA, Moore JA (2006) Virulent Hessian fly (Diptera: Cecidomyiidae) larvae induce a nutritive tissue during compatible interactions with wheat. Ann Entomol Soc Am 99:305–316
Heath MC (2000) Hypersensitive response-related death. Plant Mol Biol 44:321–334
Herdt RW (1991) Research priorities for rice biotechnology. In: Khush GS, Toenniessen GH (eds) Rice biotechnology. CABI and International Rice Research Institute, Manila (Philippines), pp 19–54
Himabindu K (2009) Identification, tagging and mapping of rice gall midge resistance genes using microsatellite markers. Dissertation, Acharya Nagarjuna University
Himabindu K, Suneetha K, Sama VSAK, Bentur JS (2010) A new rice gall midge resistance gene in the breeding line CR57-MR1523, mapping with flanking markers and development of NILs. Euphytica 174:179–187
Hoglund S, Larsson S, Wingsle G (2005) Both hypersensitive and non-hypersensitive responses are associated with resistance in Salix viminalis against the gall midge Dasineura marginemtorquens. J Exp Bot 56:3215–3222
Hulbert SH, Bai J, Fellers JP, Pacheco MG, Bowden RL (2007) Gene expression patterns in near isogenic lines for wheat rust resistance gene Lr34/Yr18. Phytopathol 97:1083–1093
Kawasaki T, Koita H, Nakatsubo T, Hasegawa K, Wakabayashi K (2006) Cinnamoyl-CoA reductase a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc Natl Acad Sci USA 103:230–235
Keen NT, Littlefield LJ (1979) The possible association of phytoalexins with resistance gene expression in flax to Melamspora lini. Physiol Plant Pathol 14:265–280
Knogge W (1996) Fungal infection of plants. Plant Cell 8:1711–1722
Kohm BA, Goulden MG, Gilbert JE, Kavanagh TA, Baulcombe DC (1993) A potato virus X resistance gene mediates an induced, nonspecific resistance in protoplasts. Plant Cell 5:913–920
Li C, Bai Y, Jacobsen E, Visser R, Lindhout P, Bonnema G (2006) Tomato defense to the powdery mildew fungus: differences in expression of genes in susceptible, monogenic- and polygenic-resistance responses are mainly in timing. Plant Mol Biol 62:127–140
Liu X, Bai J, Huang L, Zhu L, Liu X et al (2007) Gene expression of different wheat genotypes during attack by virulent and avirulent Hessian fly (Mayetiola destructor) larvae. J Chem Ecol 33:2171–2194
Liu X, Williams CE, Nemacheck JA, Wang H, Subramanyam S, Zheng C, Chen MS (2010) Reactive oxygen species are involved in plant defense against a gall midge. Plant Physiol 152:985–999
Pfitzner UM, Goodman HM (1987) Isolation and characterization of cDNA clones encoding pathogenesis-related proteins from tobacco mosaic virus infected tobacco plants. Nucleic Acids Res 15:4449–4465
Poncianoa G, Yoshikawa M, Lee JL, Ronald PC, Whalena MC (2006) Pathogenesis-related gene expression in rice is correlated with developmentally controlled Xa21-mediated resistance against Xanthomonas oryzae pv. oryzae. Physiol Mol Plant Pathol 69:131–139
Rawat N (2012) Identification and characterization of differentially expressed genes in rice involved in rice gall midge interactions. Dissertation, Osmania University
Rawat N, Sinha DK, Rajendrakumar P, Srivastava P, Neeraja CN, Sundaram RM, Nair S, Bentur JS (2010) Role of pathogenesis related genes in rice gall midge interactions. Curr Sci 99:1361–1368
Regupathy A, Subramanian A (1972) Effect of different doses of fertilizers on the mineral metabolism of IR8 rice in relation to its susceptibility to gall fly Pachydiplosis oryzae, Wood-Manson, and leaf roller, Cnaphalocrosis medinalis Guenee. Oryza 9:81–85
Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD (1996) Systemic acquired resistance. Plant Cell 8:1809–1819
Sauge MH, Kervella J, Pascal T (1998) Settling behaviour and reproductive potential of the green peach aphid Myzus persicae on peach varieties and a related wild Prunus. Entomol Exp Appl 89:233–242
Staskawicz BJ, Ausubel FM, Baker BJ, Ellis JG, Jones JDG (1995) Molecular genetics of plant disease resistance. Science 268:661–667
Swarbrick PJ, Huang K, Liu G, Slate J, Press MC, Scholes JD (2008) Global patterns of gene expression in rice cultivars undergoing a susceptible or resistant interaction with the parasitic plant Striga hermonthica. New Phytol 179:515–529
Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37:914–939
Torres MA, Dangl JL, Jones JD (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci USA 99:517–522
Torres MA, Jones JD, Dangl JL (2005) Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat Genet 37:1130–1134
Torres MA, Jones JDG, Dangl JL (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol 141:373–378
Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138:1195–1204
Vijaya Lakshmi P, Amudhan S, Himabindu K, Cheralu C, Bentur JS (2006) A new biotype of the Asian rice gall midge Orseolia oryzae (Diptera: Cecidomyiidae) characterized from the Warangal population in Andhra Pradesh, India. Int J Trop Sci 26:207–211
Walling LL (2000) The myriad plant responses to herbivores. J Plant Growth Regul 19:195–216
Yu IC, Parker J, Bent AF (1998) Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd 1 mutant. Proc Natl Acad Sci USA 95:7819–7824
Acknowledgments
We thank the Project Director, Directorate of Rice Research, Hyderabad and the Director, International Centre for Genetic Engineering and Biotechnology, New Delhi for the facilities and encouragement. This work was supported by a research grant (NFBSFARA/PCN/AP01/2006-07) from the National Fund of the National Agricultural Innovative Project of the Indian Council of Agricultural Research, New Delhi, to JSB.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary materials
Below is the link to the electronic supplementary material.
10142_2012_275_MOESM3_ESM.xls
List of different rice pathways, downregulated in the Kavya–GMB4M interaction. Shaded area in the sheet represents the significant pathways list (p < 0.05; XLS 54 kb)
Rights and permissions
About this article
Cite this article
Rawat, N., Chiruvuri Naga, N., Raman Meenakshi, S. et al. A novel mechanism of gall midge resistance in the rice variety Kavya revealed by microarray analysis. Funct Integr Genomics 12, 249–264 (2012). https://doi.org/10.1007/s10142-012-0275-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-012-0275-2