Abstract
Studies on named entity recognition (NER) often require a substantial amount of human-annotated training data. This makes technical domain-specific NER from industry data especially challenging as labelled data are scarce. Despite English as the surface language, technical jargon and writing conventions used in technical documents render the low-resource language challenges where techniques such as transfer learning hardly work. Relieving labour intensive annotations using automatic labelling is thus an important research topic, seeking ways to obtain labelled data quickly and consistently. In this work, we propose an iterative deep learning NER framework using distant supervision for automatic labelling of domain-specific datasets. The framework is applied to mineral exploration reports and produced a large BIO-annotated dataset with six geological categories. This quality-labelled dataset, OzROCK, is made publicly available to support future research on technical domain NER. Experimental results demonstrated the effectiveness of this approach, further confirmed by domain experts. The generalisation ability is verified by applying the framework to two other datasets: one for disease names and the other for chemical names. Overall, our approach can effectively reduce annotation efforts by identifying a much smaller subset, that is challenging for automatic labelling thus requires attention from human experts.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The extraction of named entities from free text has been a core task of information extraction (IE). Identifying salient information units, specifically a single token or a sequence of tokens, as entities, and their types is the critical first step of IE. Named entity recognition (NER) [23] refers to the IE techniques that identify and classify entities with predefined generic semantic types such as person, location, and organisation. The term domain named entity recognition (DNER), on the other hand, is used to emphasise domain-specific names, such as medical or biological in DrugNER [29] and BioNER [38]. In this study, named entities in the geological domain are of interest.
Recent years have witnessed success of deep learning architectures in advancing the state of the art of NER. These deep learning solutions require little or no feature engineering, addressing a common problem traditional NER systems suffered from. However, deep learning models require large amounts of training data reliably annotated with labels for the entities [9]. It has been shown that such NER systems would perform at an unsatisfactory level, if there is insufficient labelled data [43]. Deep model learning and evaluation depend heavily on the reliability of the annotations [20].
For general purpose NER, labelled English text data are abundant. Annotated benchmark datasets and off-the-shelf tools are available. For example, CoNLL-2003 English benchmark dataset [28] is a collection of documents from Reuters news articles, annotated with four entity types: persons, organisations, locations and miscellaneous names.Footnote 1 It contains around 300,000 tokens of 22,137 sentences. OntoNotes5.0 [39] is an annotated corpus comprising 2.9 million words in topics of news, phone conversations, weblogs, broadcast, talk shows in three languages (English, Chinese and Arabic) with structural information (syntax and predicate argument structure) and shallow semantics (word sense linked to an ontology and co-reference).Footnote 2 Off-the-shelf standard NER tools are able to recognise named entities of a restricted list of predefined entity types, such as location, person and organisation names, money, date and time. Widely used tools include NLTK [2], SpaCy [16], Stanford Named Entity Recogniser [10] and AllenNLP [12, 24].
However, when it comes to real-world applications for domain-specific text, e.g. geological exploration reports, they face the low-resource data problem similar to machine translation between rare languages. There is no benchmark-annotated dataset relevant to this domain, and it is not possible to find the right pivot language that allows us to take advantages of existing high-resource NER tools. In geological domain, mineral and rock names are more important and far common than person or organisation names.
Our domain-specific dataset is a collection of Western Australian mineral exploration reports (WAMEX).Footnote 3 The Department of Mines, Industry Regulation and Safety (DMIRS) and Geological Survey and Resource Strategy Division (GSRSD) promote mineral exploration investments through publishing geoscientific data to the exploration industry. Based on statutory requirements, mineral explorers report their exploration activities and submit collected data to DMIRS. After a period of confidentiality, these reports and data are made publicly available to avoid repeated work and to reduce risks associated with exploration. For mineral explorers, past exploration reports are an important resource to understand mineral deposits and their geological and depositional environment in which they form. This may include identifying the rocks that host the mineral deposit, their geological age and the key genetic processes that may be revealed in stratigraphic formations. The six primary entity groups of interest are, namely ROCK (rock types), MINERAL (minerals), TIMESCALE (geological time scales), STRAT (stratigraphic unit names), LOCATION (locations in Western Australia) and ORE_DEPOSIT (economically important elements or minerals that are concentrated in rocks as well as mineral deposits). Presently, we are interested in six primary entity groups, but they can be extended to incorporate other groups such as major geological structures in the area, which could be used as conduits for mineralising fluids; and geochemical anomalies that may constrain the source of the mineralisation.
Among the different types of techniques for addressing the low-resource issue in the literature, distant supervision [30] is the most readily applicable to this study, as compared to transfer learning [42], rule learning [34] and co-training [3]. This is because of the geological terminologies reasonably readily available to us. Thus, in this paper, we propose a deep learning-based distant supervision technique for automatic bootstrapping of labelled geological text data. A customised geological dictionary is created for initial data annotation, following the standard BIO notation [26]. The annotated data are then used for training NER models for labelling seen and unseen documents. Four architecturally different models are evaluated, which employ RNNs (BiLSTM) and CNNs for word and character embeddings with CRF and softmax for label decoding. Based on the experimental results, character-level and word-level BiLSTM model and also word-level BiLSTM model performed higher than other two models and generated similar results for our sequence tagging task.
This solution relies heavily on the coverage of the domain vocabulary, which incorporates as much domain named entities as possible. Experiments were conducted with randomly initialised, uniformly sampled word embeddings to train models based on end-to-end NER architectures. The quality of our auto-labelled entities in the geological domain has been assessed and confirmed by domain experts. The Auto-Labelled Set is evaluated on the Evaluation Set, which is manually annotated by the domain experts. To further demonstrate the effectiveness of our proposed approach for auto-labelling of domain-specific data, further experiments were conducted using two datasets outside the geology domain: one for disease names and other for chemical names. Then the results are discussed in terms of the ability to detect unseen entities in text and the influence of dictionary coverage. In summary, the main contribution of the paper is twofold:
-
A deep learning-based framework for generating high-quality labelled data for domain-specific and low-resource named entity recognition;
-
OzROCKFootnote 4 - An annotated dataset, which is released to public to support information extraction for the mineral exploration domain.
The paper is organised as follows. Related work is discussed in Sect. 2. Section 3 explains the end-to-end deep learning-based distant supervision framework. Then the experimental results are presented in Sect. 4. The paper concludes in Sect. 5 with an outlook to future work.
2 Related work
2.1 Auto-labelling approaches
Learning an effective NER model for augmenting the labelled dataset is proposed to reduce the manual annotation effort in creating the training dataset [11, 30]. The main methods for auto-labelling include transfer learning, rule learning, co-training and distant supervision to address the challenge.
Transfer learning To train a deep learning model with a small amount of labelled data, transfer learning [25, 42] or retraining models that have been trained for different tasks that have enough training data in the same domain are used by ignoring unlabelled data available for the domain.
Rule learning Rule-based information extraction [4, 41] is important to industry practitioners, because of the need for a massive training dataset for machine learning techniques and the ability to trace errors. Using a small labelled dataset and a large unlabelled dataset, Snuba [34] iteratively generates rules to help in assigning of labels to the unlabelled dataset.
Co-training This method takes advantage of both labelled and unlabelled data to train two independent models on two separate views of the data [3, 36]. Two learning algorithms are trained separately and then predict on unlabelled data to enlarge the training set of the other model.
Distant supervision Distant supervision makes use of information present in knowledge bases or domain vocabularies. This approach is chosen in our study because the geological terminologies are well defined and consistent. We can integrate and make good use of well curated, existing databases for various kinds of geological named entities. Shang et al. [30, 37] designed frameworks for noisy labels, generated using a dictionary only. A fuzzy LSTM+CRF model with modified IOBES [27] labelling scheme is introduced to tackle the multi-label tokens. AutoNER and AutoBioNER frameworks were proposed with tie or break labelling scheme for dictionary-based noisy labels. Their models are refined by unknown category phrases mined from the documents and corpus-aware dictionary tailoring was done for categorised entities. Deep learning-based auto-labelling is experimented using a domain dictionary only on the domain-specific biomedical and laptop review datasets. GeoDeepDive [44] supports data mining and knowledge base creation in the geosciences and biosciences. It provides a digital library with toolkits that acquire and manage articles. GeoDeepDive provides a corpus with user-prescribed keywords and user-developed rules to retrieve the data from matching publications in their digital library. SwellShark [11] is a distant supervised model in the biomedical domain. No human-annotated dataset is used in SwellShark, but it employs domain vocabulary, expert involvement in designing effective regular expressions and special case tuning.
To enrich the vocabulary, word embeddings trained on the same domain [8] or phrase detection approaches [30] can help extract more entities. Category labelling of unknown entities can be assisted by external resources [45].
2.2 Geological NER literature
A geological application of NER using CRFs was developed by Sobhana et al. [32]. They used a geology-related dataset of scientific reports and articles on the geology of the Indian subcontinent. The data were manually annotated with the tags for names of countries, states, water bodies, minerals, people and organisations. A number of studies are reported for KG creation from geological data in Chinese text. Wang et al. [35] extracted generic and geology terms from geology dictionaries. They created a KG from the terms and their co-occurrences, and visualised it to give a view of the corpus. CNNs is used to classify the paragraphs and extract information about copper deposit in the Sichuan Province in China from geoscience text data [31]. They extracted the keywords based on the four categories: terminology, technical methods, data processing methods and descriptive words. The frequency statistics are conducted on words and paragraphs to extract the key content words. They also created a KG from the key terms and their co-occurrences. Zhu et al. [45] also proposed a design to construct a KG towards geological data. Entity extraction was done by Chinese word segmentation, frequency statistics, category labelling using online dictionary (Baike.com) and keyword extraction. A geological domain dictionary for institutional names, place names and person names, a modern Chinese dictionary and a customised dictionary are used for type identification.
3 Proposed approach
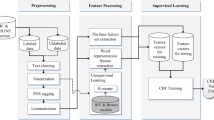
As the main purpose of the framework is to reduce the annotation load, firstly, we create dictionary-labelled data using dictionary matching. Then this dictionary-labelled data are divided into training, validation and test sets to train four different sequence labelling architectures. The best performing architecture is selected out of the four models. Then the model learns from the dictionary-labelled data and predicts the labels to create the model-predicted data. Next, these two types of labels are combined and the Auto-Labelled Set is constructed as a training set. This auto-labelled dataset is applied to train the chosen sequence labelling model. An ensemble approach can be taken here. In other words, instead of choosing the best model, the output of four models can be compared and aggregated to construct the auto-labelled dataset. For the practicality of this framework, we opt for selecting one or two best performing models. Finally, the Evaluation Set, which is manually annotated by experts, is used for evaluating the proposed framework.
This section introduces the proposed deep learning-based distant supervision approach for automatic annotation for domain-specific data. Figure 1 provides a system overview for the auto-labelling process. The system employs the combination of dictionary-based labelling and deep learning-based sequence labelling to create auto-labelled data. The domain experts participate in the dictionary construction and result validation. The proposed framework aims to iteratively enhance the annotation quality, until the annotated dataset is deemed satisfactory by the human experts.
3.1 Dictionary matching
Existing entity types in standard NER tools are insufficient and not fine-grained in extracting the geological entities. As a result, many geological terms are either missed or tagged incorrectly. For example, a stratigraphic unit name or a mining location is misclassified as a person or an organisation. Therefore, customised types need to be defined and used for extracting domain entities of interest.
A geological dictionary of domain vocabulary is created with the help of domain experts to provide initial labelling for entities through dictionary matching. This domain dictionary is a list of entity names that each belongs to a specific geological entity type. This dictionary is used to create a training dataset for our entity recognition and annotation task.
We retrieve vocabularies for minerals, rocks, ores and deposits, geological time scales, stratigraphic unit names and locations in Western Australia (WA). See Table 1 for the details of the domain dictionary. The six categories of entities are collected, and the total of 16,300 terms are included in the dictionary. The sources of these terminologies are Explanatory Notes System database,Footnote 5 Mindat open database,Footnote 6 the GeoNames geographical database,Footnote 7 Australian Stratigraphic Units Database,Footnote 8 and Wikipedia.Footnote 9
Entities are first annotated through dictionary lookup. Multiple labels are assigned for STRAT, ORE_DEPOSIT, MINERAL, ROCK, TIMESCALE and LOCATION columns in Table 2. Although multiple labels can be associated with a single token, we decided to allow one label for each token. For example, the token iron can be an entity with a single token or a part of a phrase (multiple tokens). When iron is a single-token entity, it is tagged as MINERAL. When iron is in a phrase iron mica, its tag is MINERAL, but in iron gravel, it is ROCK, in iron ore, it is ORE_DEPOSIT.
During the dictionary-based annotation process, which involves a single token and nested tagging situations, two resolution rules are followed to ensure a single label for each token. When nested tags are found for a token, the longest phrase is prioritised as an entity type, over a single token or shorter phrases. When multiple tags are found for the same token, the last tag is selected.
Table 2 shows, for the final label in Tag column, a single label for each token is assigned. For example, the nested tagging occurs for banded iron formation as ROCK and iron as MINERAL. The system favours the longer phrase banded iron formation over the single-word entity iron. Also Hamersley Group as STRAT is selected over Hamersley as LOCATION. Gold deposits is also recognised as a type of ore and deposit, but gold as MINERAL is not selected, because the longer the phrases, the higher the priority.
Automatic NER models are typically trained using a standard labelling scheme. One of the most popular schemes, simple yet effective, is the BIO notation [26] and used for many entity tagging frameworks [15, 22]. The BIO notation marks a word: B for a beginning of an entity, I for inside an entity, and O means others. The BIO scheme is adopted to annotate the text using our domain dictionary. The dictionary-labelled data are then used as labelled data for training, validating and testing the neural network-based sequence labelling models for the next stage.
3.2 Selecting a sequence labelling architecture
This section describes how four different neural network models are trained with our dictionary-labelled data. The performances of the models are evaluated to select the highest performing model out of the four models. The ultimate aim is to annotate more text data that were not in the training set, in order to grow the labelled dataset.
Deep learning-based NER models learn to make predictions by training on example inputs and their expected labels. The NER task is mostly formulated as a sequence labelling task, which involves the algorithmic assignment of a categorical label to each word in a sequence of observed words. In general, a NER task [20] includes three components: distributed representations for an input, context encoding for capturing the context dependencies and label decoding for converting predicted scores into target labels. The input is a sequence of words or characters, and the output is a sequence of labels for the words.
Distributed representations (embeddings) are prepared for the input sequence to represent each word or each character as low-dimensional real-valued dense vectors. These employ randomly initialised character-level or word-level embeddings or pretrained word embeddings that trained over large collections of text through unsupervised algorithms. Word-level models [6, 17] use the representations that are based on words. A sequence of words is given to create a word embedding for each word and predict a label for each word. Character or word embeddings can be learned from an end-to-end neural network-based model [22]. A sentence is then represented as a sequence of characters. The sequence of characters is given to create an embedding for each character and predicts labels for the sequence. Character-level representations are created with convolutional neural networks (CNNs) [22, 24] and recurrent neural networks (RNNs) [1, 15, 18] such as BiLSTM [14]. Hybrid representations [5, 17, 40] incorporate word-level or character-level embeddings with additional information such as character capitalisation, spelling and/or affix features.
Context encoding can employ CNNs, RNNs, language models or transformer models for learning from the input representation and capturing contextual dependencies. In this paper, we chose models that employ a long short-term memory (LSTM), which is a form of RNNs. LSTM has three gated units: input, output and forget, which is able to control the passing of information along the sequence in order to improve the modelling of long-range dependencies [13]. A stacked bidirectional LSTM (BiLSTM) model [14] uses a forward LSTM network for past states and a backward LSTM network for future states for a given time step to transform word features into named entity tag scores.
Label decoding component predicts labels for tokens in the input sequence. Sequence labelling algorithms are often probabilistic and rely on statistical inference to find the best sequence. Most sequence labelling methods employ a Conditional Random Field (CRF) [19] or softmax. Models employ CRFs as the tag decoder, on top of BiLSTM [17, 24] or on top of CNNs [1]. A softmax layer can also be used as the tag decoder [6].
Four different sequence labelling models are experimented on the dictionary-labelled data, which were annotated using the domain dictionary as described in Sect. 3.1. The purpose of using these models is to compare different neural networks and their combinations on the geological NER task. Word-level (WL) BiLSTM is demonstrated to lead the state of the art in NER [5, 22]. We therefore selected WL BiLSTM as the core architecture and attempted to compare with character-level (CL) CNN alone and combination with character-level models as shown in Table 3. The models are CL CNN, CL CNN+WL BiLSTM, WL BiLSTM and CL+WL BiLSTM. Our experiments are by no means exhaustive, one may also notice that transformer models and models working on word pieces can be considered as well. The best performing architectures are then chosen based on the NER task performance and employed for predicting the labels for the data. Figure 2 shows a schematic diagram of CL+WL BiLSTM model.
3.3 Auto-labelling
Once the experts are satisfied with the dictionary coverage and the models are selected based on the \(F_1\) performance, the models are trained on the Auto-Labelled Set, which are created by combining the dictionary-based labels with the model-predicted labels. Joining the dictionary-based labels with the model-predicted labels refines our dataset. The following rules are applied in the label selection:
-
1.
when either one of the dictionary-based label or model-predicted label is available, that existing label is selected.
-
2.
when nested tags exist, the longest phrase is prioritised.
-
3.
when conflicted tags exist for the same entity, a dictionary-based tag is preferred.
Now the chosen NER model is ready to annotate more data automatically with the six entity types: ROCK (rock types), MINERAL (mineral types), TIMESCALE (geological time scale), STRAT (stratigraphic unit names), LOCATION (location names in Western Australia) and ORE_DEPOSIT (elements or minerals that are concentrated in rocks or in mineral deposits). Finally, the model is evaluated on the Evaluation Set, which was manually annotated by the domain experts using Redcoat [33], a Web-based annotation tool for labelling data for entity typing.
3.4 Domain expert verification
The two types of annotations are created, respectively, by dictionary matching and by model prediction. Their annotation outputs match most of the time, conflicts can also happen. We collect the labelled sentences from those that contain conflicts between the two kinds of annotations. From our experiments, the number of conflicted sentences is around 100 in 10,000 sentences. Domain experts annotate these conflicted sentences manually. The results of dictionary tags, predicted tags are then compared with the tags from the experts.
As a result of the verification, shown in Table 4, the ores and deposits category was newly created to include the mineral ore and deposit names that affected the number of missed entities. (11.61% was missed by dictionary matching, and 10.42% was missed by the model prediction.) A total of 335 entities were annotated by the domain experts. After introducing the new category ORE_DEPOSIT, 21 missed entities were labelled and the percentage of missed entities was reduced to 5% in dictionary-based labels and 3.3% in model-predicted labels. Updating categories as a result of the verification process serves as a positive knowledge elicitation tool to prompt the domain experts to rethink through the dictionary coverage. Manual verification is an important process in fine-tuning the category labels, but the labelling effort is minimised because only the conflicted subset needs to be verified. The domain experts may suggest to enrich the dictionary in order to cover the Missed entities, remove unimportant categories or combine similar categories. Once the domain dictionary is updated, the auto-labelling process needs to be performed again. Until the annotated dataset is satisfactory to the domain experts, the auto-labelling is an iterative process that injects the changes of the domain vocabulary.
3.5 Evaluation
Precision (P), recall (R) and \(F_1\) metrics are used for NER evaluation [5, 15, 22, 30]. The main metric is \(F_1\), which is a balanced score between P and R.
where C is the number of correctly predicted entities, N the number of predicted entities and T the number of ground-truth entities.
Our dictionary-labelled test set is used when comparing the sequence labelling models. We need the models to be able to identify as many entities as dictionary can label and predict more. The chosen models are then further evaluated on the manually annotated Evaluation Set as ground truth.
4 Experiments
Experiments include (1) annotating geological named entities using the domain vocabulary, (2) comparing different deep learning architectures, (3) applying the best performing architectures for the NER task and (4) evaluating outcomes. As our main results, a geological annotated dataset is generated and a NER model is trained and ready to annotate seen and further unseen data in mineral exploration text. To further evaluate the proposed approach, we conduct experiments on two more low-resource, domain-specific, manually labelled datasets. NCBI corpus [7] is selected for recognising disease names, and BC5CDR corpus [21] is selected for recognising chemical names.
4.1 Dataset description
WAMEX dataset contains reports for geological exploration of mineral resources in Western Australia. The reports were converted from PDF format to text, so spelling mistakes or joined tokens do exist. There are also sentences in incorrect structure, due to the fact that data were pulled from tables or figures in the original documents. We selected 34,000 sentences that contain geological entities out of WAMEX dataset for the experiment. In total, 32,000 sentences are automatically annotated using the geological domain dictionary and the sequence labelling model. The remaining 2000 sentences are manually annotated by the domain experts and kept as the Evaluation Set in order to evaluate how well our auto-labelling framework can perform against the manually curated annotation.
4.2 Annotation using a domain dictionary
The 32,000 sentences are annotated automatically using dictionary lookup using the BIO annotation scheme. Table 1 contains the domain dictionary details with entity categories and the size of each category. Dictionary-based labelling results are presented in Table 8 together with other data, which will be explained next. Training, validation and test sets are created from this dictionary-labelled sentences. These datasets are used to train and validate the different neural network-based NER architectures. Therefore, we can compare the deep learning models on automatic labelling.
4.3 Auto-labelling with NER models
The dictionary-labelled data are used to train, validate and test the performance of the four deep learning-based models. Distributed representations of words are created from words and/or characters in the input sequence and Context encoder and Label decoder predict a sequence of labels for the input. The four architectures and their \(F_1\) scores for each entity category are presented in Table 5 for performance comparison. The experiment shows that the larger the training dataset, the better the models are performed, as more data expose the models to larger variety of examples. So the \(F_1\) on the dataset of 32,000 sentences is higher than on the dataset of 18,000 sentences. CL+WL BiLSTM model performed the highest with \(F_1\) score of 97.35 on 18k data, but WL BiLSTM model performed the highest with \(F_1\) score of 97.85 on 32k data. The score differences of the models were 0.23 on 32k to 0.37 on 18k data. Therefore, both models were selected to be evaluated on the Evaluation Set.
The training process was run five times for each model, and then, the highest \(F_1\) scores for each model are selected to show the performance. Also the best scored, trained models are saved for further annotation of seen and unseen text. Each process used random splits of training, validation and test sets in order to train the models, by using the below hyper-parameters. The dimension of word embeddings is 64 and character embeddings is 30. We experimented with the input embeddings that are randomly initialised for each training with uniform samples from \([-\sqrt{3/\mathrm{dim}}, +\sqrt{3/\mathrm{dim}}]\), where dim is the dimension of the embeddings. For character-level CNNs, the window size of 3 is used with 30 filters. 100 hidden units are given for both forward and backward LSTMs. The number of epochs is 60, and batch size is 32. We applied dropout rate of 0.5 both before and after the context encoding layer.
4.4 Evaluation
4.4.1 Geological NER evaluation
Using the dictionary-annotated dataset of 32,000 sentences as the training data, the chosen two models identified the entities and predicted their labels on Evaluation Set. Performance results of the models are shown in Table 6, where WL BiLSTM has \(F_1\) score of 78.53 and CL+WL BiLSTM has \(F_1\) of 77.59. The deep learning models performed lower than the dictionary matching.
To improve the training dataset, the model was applied on the 32,000 sentences. Then the predicted labels of the model is combined with the dictionary-based labels and constructed the Auto-Labelled Set as the final training dataset. The proposed framework performed with \(F_1\) score of 82.19 as shown in Table 7 on Evaluation Set. The labelling performance is improved by 3.66% on WL BiLSTM model. The process of using the expert verification to improve the domain vocabulary can be repeated till the result is improved to the satisfaction.
A total of 296 more entity mentions were annotated in the Auto-Labelled Set as a result of combining process. Table 8 shows the statistics of the datasets. The table contains number of entities for each category in each dataset that are created during this experiment. Auto-Labelled Set contains 102 unique new entities, which do not exist in the domain vocabulary. For example, metaliferrous rock was not labelled by dictionary lookup, due to it not being in the dictionary, but the trained model tagged it as ROCK correctly. Even with its misspelling (metalliferous is correct), the model recognised it by learning that the word rock is often used in the entities of ROCK type. More misspelled entities were annotated correctly, including De Gray Group (De Grey Group is correct) as STRAT, grabbroic rock (gabbroic rock is correct) as ROCK, felspathic siltstone (feldspathic siltstone is correct) as ROCK and Hamerlsey Group (Hamersley Group is correct) as STRAT. Moreover, many new entities are discovered, although they do not exist in the dictionary such as ultramafic komatiitic rocks, metabasaltic rocks, mafic sequence, ultramafic dykes are ROCK types, while eolian deposit is ORE_DEPOSIT, Bangemall formation and Moogie metamorphic suite are STRAT types. In addition, the new entities like Mount Burges, Mount Menzies, Mount Gibson, Emu Lake, Collier Basin, Shaley Basin, Dome Pit, Sovereign mine, King mine, Mount Hill and Padbury Basin are identified as LOCATION, so did misspelled Port Hedtand (correctly Port Hedland). The model learned that mount, lake, basin, pit, mine and port are generally found in location names and labelled them accordingly.
The result shows the trend that the entities of a single token or fewer tokens such as ORE_DEPOSIT, LOCATION, MINERAL and TIMESCALE are easier to predict for the model, while STRAT and ROCK types are harder to predict. As shown in the dictionary in Table 1, the most entity categories contain no more than 4 tokens, while ROCK and STRAT types can contain up to 8 and 9 tokens, respectively. However, we found out that the entity length is not the main case of the poor performance here.
In fact, the main reason of the stratigraphic unit names perform poorly is that they include many different location and rock names in their often multiple-token names. For example, Wallaby Conglomerate is STRAT type, but the model labelled Wallaby as LOCATION and Conglomerate as ROCK. The model is not wrong in case for each token, but we prefer longer entities as they represent the fine-grained data units. This mix of different entities in a single entity confuses the model sometimes and the model can label them as separate entities of different categories, which then affects the model performance for the involved categories. Therefore, when the different categories do not share the same tokens often, the deep learning model can learn effectively, perform well and improve the dictionary-based annotation.
OzROCK Dataset As a result of the proposed framework, OzROCK dataset is created for NER in the mineral exploration domain. It contains 34,000 labelled sentences, which are divided into Auto-Labelled Set (32,000 auto-labelled sentences) for training and Evaluation Set (2,000 manually labelled sentences) for evaluation. Note that this data contain errors in spelling and structure, due to documents were converted from PDF to text format.
Importantly, the learned sequence labelling model allows us to label unseen text. Therefore, the labelled dataset can be updated with new reports and keeps growing. With a high-quality domain dictionary, our approach reduces the load of manual annotation of large amounts of data. The proposed approach can be applied to data sets of any size and the framework may be applicable for other domain-specific NER systems.
4.4.2 Experiments and discussion on other domains
In order to demonstrate the generalisation ability of our approach, we experimented on two more datasets from different domains. The National Center for Biotechnology Information (NCBI) corpus [7] and the BioCreative V Chemical Disease Relation (BC5CDR) corpus [21] are selected for auto-labelling and recognising unseen entities. NCBI corpus is used for recognising disease names, while BC5CDR corpus is used for recognising chemical names.
Our auto-labelling framework is applied to both datasets, and their performances are compared against dictionary matching and WL BiLSTM neural network model. The results have shown around 5% increase in F1 score for disease names and 8% increase for chemical names.
NCBI corpus is fully annotated for disease names to serve as a research resource for biomedical NER. It contains 793 biomedical literature abstracts with 6,892 disease mentions and 790 unique disease concepts. The corpus is separated for training (593 abstracts), development (100 abstracts) and testing (100 abstracts) sets. A vocabulary of disease names is prepared from the training set only and used for generating the domain dictionary. The created dictionary contains 1,690 disease names including the abbreviated versions. The development set contains 199 unseen entities, and the test set contains 235 unseen entities that do not exist in the domain dictionary nor in the training set. The neural network model performed with F1 score of 74.39 for DISEASE entities on the test set as shown in Table 9. The proposed auto-labelling approach improved the NER performance by 5.38% in comparison to dictionary matching.
In the test set, auto-labelling detected 95 new entities including Waardenburg syndrome, Stuve–Wiedemann syndrome, hepatic copper accumulation, hereditary non-polyposis colorectal cancer, insulin-dependent diabetes mellitus, malformation of the eye, sporadic T-cell prolymphocytic leukaemia and hereditary ovarian cancer. Common tokens that helped detecting disease names are deficiency, disease, cancer, leukaemia, syndrome, tumour, dysplasia, disorder, carcinoma, malignancy, abnormality, anomaly, malformation and defect.
BC5CDR corpus was first released in the BioCreative V Chemical Disease Relation task and is annotated for diseases and chemicals for biomedical NER. Our approach is tested on the chemical entities using this dataset. It has 1,500 articles containing 15,935 CHEMICAL mentions. We used 1000 articles for the model training and a test set of 500 articles to evaluate the proposed auto-labelling framework. The domain dictionary contains 2,203 chemical names that are labelled in the training set. The test set contains 718 unseen entities that do not exist in the dictionary nor in the training set. The framework performed with F1 score of 79.05 on the test set as shown in Table 10. Although the auto-labelling and model prediction are scored close, the recall score is improved by the proposed approach. The proposed approach improved the labelling performance by 8.18% on dictionary matching.
In the test set, auto-labelling detected 265 new entities including ammonium, quinine, 4-aminopyridine, remoxipride, pyrrolidine, hydroquinone, fungizone, lorazepam, nitric oxide NADPH, S-312, ICRF-187, hepatitis B surface antigen, 5-fluorouracil, 5-hydroxyindoleacetic acid, all-trans-retinoic acid, ATRA, vitamin A, 3-methoxy-4-hydroxyphenethyleneglycol, mefenamic acid, platinum and D-glucarates. The main context words associated with chemical entities include induced, treatment, administration, association, related, toxicity, occur, effect and concentration.
Discussion. The experiments show that dictionary matching performs well in terms of the precision, but the recall is often low. On the other hand, the neural network model is able to improve the recall. By combining their annotations, the proposed auto-labelling approach is often able to improve overall performance of NER. The auto-labelling improvement is due to the common tokens that are used in defining the entities and the surrounding words of the entities that are learned by the model and then combining the labels from both model prediction and dictionary matching.
In order to explore the influence of domain dictionary coverage, an experiment is performed with 50%, 60%, 70%, 80%, 90% and 100% of the domain dictionary. Figure 3 shows the performance comparisons on the NCBI and BC5CDR corpora with different percentages of vocabulary coverage. Domain coverage 50% means 50% of the domain dictionary is removed and the training dataset does not contain labels for those removed entities. For example, NCBI contains 1,690 disease entities in the vocabulary as 100% coverage of the training dataset, but only 845 entities are used for training when 50% coverage is applied. Figure 3 shows that the importance of dictionary coverage is corpus and domain-specific. For NCBI, the increase in the dictionary coverage plays a significant role in model performance. However, for BC5CDR, the neural network model injected a significant boost in performance (from 29% to 62% even with only 50% of dictionary terms). This warrants future research in measuring the “learnability” of a domain.
In summary, the proposed auto-labelling approach is able to learn and label seen and unseen entities based on the domain dictionary. Our approach can help build annotated datasets efficiently for domain-specific named entities, by utilising vocabulary in existing established databases in low-resource domains.
5 Conclusion and future work
In this research, we developed a framework and demonstrated its effectiveness of automatically labelling named entities in a low-resource, domain-specific real-world dataset. An automatic NER approach is proposed by adopting dictionary matching and deep learning-based sequence labelling, with the potential of integrating domain expert validation. One major advantage of this work is its convenience in labelling named entities solely using a domain vocabulary. Therefore, a large amount of labelled data can be created efficiently.
The annotated dataset OzROCK was created and has been made publicly available. We evaluated the outcomes of the proposed approach on the manually annotated dataset, which was labelled by the geological domain experts. Identifying domain-specific salient entities and annotating them automatically have proved to be effective and significantly reduces the costly and subjective manual annotation work. Our approach effectively identifies the subset of the data that is challenging to automatically annotate and thus needs expert assistance for annotation. We further applied our auto-labelling framework on the disease corpus and chemical corpus, and compared the performance against dictionary matching and WL BiLSTM neural network model. The results have shown approximately around 5% increase in F1 score for disease names and 8% increase for chemical names.
Moreover, a large number of seen and unseen documents can be efficiently labelled using the deep learning-based models trained over OzROCK dataset. This is a step towards building a knowledge graph in the mineral exploration domain to allow efficient storage and retrieval of information.
Currently, a single tag is allowed for each token in the text. Future work may include an inclusion of multiple labels for each entity.
In conclusion, the deep learning-based NER approach incorporated with a domain dictionary showed significant potential for automatically identifying and labelling domain-specific entities. Automatic labelling of unstructured domain-specific text is an important step for knowledge discovery in low-resource domains. This research is a part of our ongoing work to extract mineralisation knowledge from geological exploration reports and similar documents.
Notes
References
Akbik A, Blythe D, Vollgraf R ( 2018) Contextual string embeddings for sequence labeling. In: Proceedings of the 27th international conference on computational linguistics. pp. 1638–1649
Bird S, Klein E, Loper E (2009) Natural language processing with Python: analyzing text with the natural language toolkit. O’Reilly Media Inc., Sebastopol
Blum A, Mitchell T (1998) Combining labeled and unlabeled data with co-training. In: Proceedings of the eleventh annual conference on Computational learning theory’. ACM 92–100
Chiticariu L, Li Y, Reiss F (2013) Rule-based information extraction is dead! long live rule-based information extraction systems. In: Proceedings of the 2013 conference on empirical methods in natural language processing, pp. 827–832
Chiu JP, Nichols E (2016) Named entity recognition with bidirectional lstm-cnns. Trans Assoc Comput Linguist 4:357–370
Devlin J, Chang M.-W, Lee K, Toutanova K (2018) Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805
Doğan RI, Leaman R, Lu Z (2014) Ncbi disease corpus: a resource for disease name recognition and concept normalization. J Biomed Inf 47:1–10
Enkhsaikhan M, Liu W, Holden E.-J, Duuring P (2018) Towards geological knowledge discovery using vector-based semantic similarity. In: International conference on advanced data mining and applications. Springer, pp. 224–237
Feng X, Feng X, Qin B, Feng Z, Liu T (2018) Improving low resource named entity recognition using cross-lingual knowledge transfer. In: Proceedings of the 27th international joint conference on artificial intelligence. AAAI Press, pp. 4071–4077
Finkel JR, Grenager T, Manning C (2005) Incorporating non-local information into information extraction systems by Gibbs sampling. In: Proceedings of the 43rd annual meeting on association for computational linguistics. Association for Computational Linguistics 363–370
Fries J, Wu S, Ratner A, Ré C (2017) Swellshark: a generative model for biomedical named entity recognition without labeled data. arXiv preprint arXiv:1704.06360
Gardner M, Grus J, Neumann M, Tafjord O, Dasigi P, Liu NF, Peters M, Schmitz M, Zettlemoyer LS (2017) Allennlp: a deep semantic natural language processing platform
Gers FA, Schmidhuber JA, Cummins FA (2000) Learning to forget: continual prediction with lstm. Neural Comput. 12(10):2451–2471. https://doi.org/10.1162/089976600300015015
Graves A, Mohamed A-R, Hinton G (2013) Speech recognition with deep recurrent neural networks. In: 2013 IEEE international conference on acoustics, speech and signal processing’. IEEE 6645–6649
Guillaume L, Miguel B, Sandeep S, Kazuya K, Chris D (2016) Neural architectures for named entity recognition. In: Proceedings of NAACL-HLT
Honnibal M ( 2017) ‘Spacy’. https://explosion.ai/blog/introducing-spacy
Huang Z, Xu W , Yu K ( 2015) Bidirectional lstm-crf models for sequence tagging. arXiv preprint arXiv:1508.01991
Kuru O, Can OA , Yuret D (2016) Charner: character-level named entity recognition. In: Proceedings of COLING 2016, the 26th international conference on computational linguistics: Technical Papers’, pp. 911–921
Lafferty J, McCallum A , Pereira FC ( 2001) Conditional random fields: probabilistic models for segmenting and labeling sequence data
Li J, Sun A, Han J , Li C ( 2018) A survey on deep learning for named entity recognition. arXiv preprint arXiv:1812.09449
Li J, Sun Y, Johnson RJ, Sciaky D, Wei C-H, Leaman R, Davis AP, Mattingly CJ, Wiegers TC, Lu Z (2016) Biocreative v cdr task corpus: a resource for chemical disease relation extraction. Database
Ma X, Hovy E ( 2016) End-to-end sequence labeling via bi-directional lstm-cnns-crf. In: Proceedings of the 54th annual meeting of the association for computational linguistics (Volume 1: Long Papers), Vol. 1, pp. 1064–1074
Nadeau D, Sekine S (2007) A survey of named entity recognition and classification. Lingvisticae Investigationes 30(1):3–26
Peters ME, Neumann M, Iyyer M, Gardner M, Clark C, Lee K , Zettlemoyer L ( 2018) Deep contextualized word representations. arXiv preprint arXiv:1802.05365
Qu L, Ferraro G, Zhou L, Hou W, Baldwin T ( 2016) Named entity recognition for novel types by transfer learning. In: Proceedings of the 2016 conference on empirical methods in natural language processing. pp. 899–905
Ramshaw LA, Marcus MP (1995) Text chunking using transformation-based learning. CoRR arxiv: cmp-lg/9505040
Ramshaw LA, Marcus MP (1999) Text chunking using transformation-based learning. In: Natural language processing using very large corpora. Springer, pp. 157–176
Sang EFTK , De Meulder F ( 2003) Introduction to the conll-2003 shared task:language-independent named entity recognition, CoNLL-2003
Segura-Bedmar I, Martínez P, Segura-Bedmar M (2008) Drug name recognition and classification in biomedical texts: a case study outlining approaches underpinning automated systems. Drug Discov Today 13(17–18):816–823
Shang J, Liu L, Gu X, Ren X, Ren T , Han J (2018) Learning named entity tagger using domain-specific dictionary. In: Proceedings of the 2018 conference on empirical methods in natural language processing. pp. 2054–2064
Shi L, Jianping C, Jie X (2018) Prospecting information extraction by text mining based on convolutional neural networks-a case study of the lala copper deposit, china. IEEE Access 6:52286–52297
Sobhana N, Mitra P, Ghosh S (2010) Conditional random field based named entity recognition in geological text. Int J Comput Appl 975:8887
Stewart M, Liu W, Cardell-Oliver R ( 2019) Redcoat: a collaborative annotation tool for hierarchical entity typing. In: Proceedings of the 2019 Conference on empirical methods in natural language processing and the 9th international joint conference on natural language processing (EMNLP-IJCNLP): system demonstrations. pp. 193–198
Varma P, Ré C (2018) Snuba: automating weak supervision to label training data. Proceedings of the VLDB Endowment 12(3):223–236
Wang C, Ma X, Chen J, Chen J (2018) Information extraction and knowledge graph construction from geoscience literature. Comput Geosci 112:112–120
Wang R, Liu W, McDonald C ( 2016) Featureless domain-specific term extraction with minimal labelled data. In: Proceedings of the Australasian language technology association workshop 2016. pp. 103–112
Wang X, Zhang Y, Li Q, Ren X, Shang J, Han J ( 2019) Distantly supervised biomedical named entity recognition with dictionary expansion. In: 2019 IEEE International conference on bioinformatics and biomedicine (BIBM), IEEE, pp. 496–503
Wang X, Zhang Y, Ren X, Zhang Y, Zitnik M, Shang J, Langlotz C, Han J (2019) Cross-type biomedical named entity recognition with deep multi-task learning. Bioinformatics 35(10):1745–1752
Weischedel R, Palmer M, Marcus M, Hovy E, Pradhan S, Ramshaw L, Xue N, Taylor A, Kaufman J, Franchini M et al (2013) Ontonotes release 5.0 ldc2013t19. Linguistic Data Consortium, Philadelphia
Yadav V, Sharp R, Bethard S (2018) Deep affix features improve neural named entity recognizers. In: Proceedings of the seventh joint conference on lexical and computational semantics. pp. 167–172
Yang LC, Tan IK, Selvaretnam B, Howg EK , Kar LH ( 2019) Text: traffic entity extraction from twitter. In: Proceedings of the 2019 5th international conference on computing and data engineering. pp. 53–59
Yang Z, Salakhutdinov R, Cohen WW ( 2017) Transfer learning for sequence tagging with hierarchical recurrent networks. arXiv preprint arXiv:1703.06345
Zhang B, Pan X, Wang T, Vaswani A, Ji H, Knight K, Marcu D (2016) Name tagging for low-resource incident languages based on expectation-driven learning. In: Proceedings of the 2016 conference of the North American chapter of the association for computational linguistics: human language technologies’, pp. 249–259
Zhang C, Govindaraju V, Borchardt J, Foltz T, Ré C, Peters S (2013) Geodeepdive: statistical inference using familiar data-processing languages. In: Proceedings of the 2013 ACM SIGMOD international conference on management of data. ACM, pp. 993–996
Zhu Y, Zhou W, Xu Y, Liu J, Tan Y (2017) (2017) Intelligent learning for knowledge graph towards geological data. Scientific Programming
Acknowledgements
We thank the Geological Survey and Resource Strategy Division (GSRSD) of the Department of Mines, Industry Regulation and Safety in Western Australia for assistance with accessing the WAMEX dataset and the GSRSD Explanatory Notes System database. Paul Duuring publishes with permission from the Executive Director of the Geological Survey of Western Australia.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Enkhsaikhan, M., Liu, W., Holden, EJ. et al. Auto-labelling entities in low-resource text: a geological case study. Knowl Inf Syst 63, 695–715 (2021). https://doi.org/10.1007/s10115-020-01532-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10115-020-01532-6