Abstract
This study employs the widely used Large Language Model (LLM), BERT, to implement Named Entity Recognition (NER) on the CORD-19 biomedical literature corpus. By fine-tuning the pre-trained BERT on the CORD-NER dataset, the model gains the ability to comprehend the context and semantics of biomedical named entities. The refined model is then utilized on the CORD-19 to extract more contextually relevant and updated named entities. However, fine-tuning large datasets with LLMs poses a challenge. To counter this, two distinct sampling methodologies are proposed to apply on each dataset. First, for the NER task on the CORD-19, a Latent Dirichlet Allocation (LDA) topic modeling technique is employed. This maintains the sentence structure while concentrating on related content. Second, a straightforward greedy method is deployed to gather the most informative data of 25 entity types from the CORD-NER dataset. The study realizes its goals by demonstrating the content comprehension capability of BERT-based models without the necessity of supercomputers, and converting the document-level corpus into a source for NER data, enhancing data accessibility. The outcomes of this research can shed light on the potential progression of more sophisticated NLP applications across various sectors, including knowledge graph creation, ontology learning, and conversational AI.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The advent of Large Language Models (LLMs) such as BERT and GPT has heralded a new era in the field of Natural Language Processing (NLP), with groundbreaking achievements demonstrated in tasks such as Named Entity Recognition (NER) [1, 2], text classification [3], text summarization [4], and sentiment analysis [5]. Our research pivots on the most consequential biomedical subject of recent times, Covid-19 The challenges presented by the CORD-19 dataset [14], a considerable corpus with a complex document-level structure, can often impede direct access. However, various innovative information systems have been devised to address this, capable of transmuting literature into data that machines can interpret. These include sophisticated methods such as Named Entity Recognition (NER) [6], knowledge graph (KG) [7], and text summarization [4]. The effective implementation of such systems can help democratize access to crucial information, thereby accelerating scientific advancements and improving public health response strategies.
Our research is primarily focused on the pivotal problem of Named Entity Recognition (NER), a fundamental task in Natural Language Processing (NLP), and employs the Distil-BERT model in its approach. The cruciality of NER in NLP is not to be understated, given its applicability across a wide spectrum of advanced applications, encompassing areas like Knowledge Graphs and Ontology. Hence, our research emphasis is clearly set on tackling the NER problem, thereby advancing the state-of-the-art in this essential NLP task.
In this study, we employ CORD-NER [6], an earlier CORD-19 dataset of named entities, to fine-tune BERT. Subsequently, this refined model is applied to extract named entities from the corpus of CORD-19 literature. Nevertheless, the significant computational resources needed to fine-tune BERT on vast datasets can be prohibitive, particularly for researchers with limited computational capabilities. To mitigate this challenge, we implement two distinct sampling techniques on each dataset. We commence with the greedy selection of data subsets from 25 entities of the CORD-NER dataset, followed by the fine-tuning of this prepared NER dataset with Distil-BERT. The second part of the study effectively samples the CORD-19 dataset using a topic modeling method, namely, Latent Dirichlet Allocation (LDA) [8]. We then generate a comprehensive NER dataset by applying the fine-tuned model to the sampled CORD-19 dataset. The resultant named entity dataset has the potential to be extended for other advanced applications.
In sum, this study aims to tackle the Named Entity Recognition problem for the COVID-19 open research dataset. We achieve this goal by fine-tuning BERT on the CORD-NER dataset to better comprehend the context and enhance with the biomedical semantics of named entities. We investigate the effectiveness of different sampling techniques for fine-tuning BERT. Our research results provide a valuable resource for NER on the CORD-19 document-level corpus.
2 Related Work
2.1 Bidirectional Encoder Representations from Transformers
Large language models (LLMs) have revolutionized natural language processing tasks. Bidirectional Encoder Representations from Transformers (BERT) [9] is one such LLM that has achieved state-of-the-art performance on several natural language processing tasks including NER [1, 2], and text summarization [4]. BERT is a transformer-based model that uses a pre-trained language representation to generate context-aware representations of input text. Fine-tuning BERT on specific NER datasets has shown promising results [1, 2]. The study applies BERT on the CORD-19 literature corpus without supercomputers, which could have significant implications for natural language processing research.
2.2 Document-Level Entity Extraction
Document-level entity extraction has been a topic of interest in the natural language processing community for several years [6, 10, 11]. Named entity recognition (NER) is the task of extracting entities such as people, organizations, and locations from a given text. Several approaches have been proposed for NER, including rule-based, statistical, and machine-learning methods [12, 13]. With the emergence of large-scale language models like BERT, there has been renewed interest in using these models for NER tasks [1, 2].
Named Entity Recognition is a sub-task in the construction of a knowledge graph. Giles et al. [10] applied a TERMite commercial NER engine to recognize the respective entity types of the entities obtained from the CORD-19 dataset. X. Wang et al. [6] created a NER dataset, called CORD-NER, with 75 entity types based on the earliest version of the CORD-19 dataset. This CORD-NER dataset is then used as training data for entity recognition in this study. Wu et al., 2020 [11] tried to extract the entities of the ‘PERSON’ and ‘ORG’ entity types in the CORD-19 dataset using Python built-in library named ‘Stanza’.
2.3 Dataset Sampling
Dataset sampling is an important aspect of NER tasks, especially when dealing with large corpora such as CORD-19 [14]. Various approaches have been utilized for dataset sampling [15], including random sampling, stratified sampling, and cluster-based sampling. In this research, two different sampling approaches are used for CORD-19 and CORD-NER. For CORD-19, a Latent Dirichlet allocation algorithm is used to focus on closely related contents, whereas, a simple greedy approach is used in case of CORD-NER to collect the most informative data of 25 entity types. These sampling techniques are designed to make the NER process more efficient and effective, especially when dealing with large datasets.
Researchers also tried to cluster the literatures based on their content similarity. A sort of statistical modeling called topic modeling is used to identify the abstract topics that appear in a corpus of documents. Latent Dirichlet Allocation (LDA) is commonly used to build a topic model from titles and abstracts of the research corpus [16]. Different versions of the pre-trained model BERT can also be applied in paper clustering [4].
3 Methodology
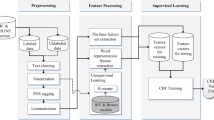
Our methodology consists of two main stages. fine-tuning the pre-trained BERT model on the NER dataset (in Sect. 3.1) and extracting named entities from the Covid-19-related literatures using the fine-tuned model (in Sect. 3.2). Each section involves the dataset preparation step (sampling). The detailed methodology is discussed in the next sections. The overview workflow of the proposed system is illustrated in Fig. 1.
3.1 Named Entity Recognition
This section discusses the details of the NER dataset, training model, and sampling method. The NER process is performed by fine-tuning the BERT model on the selective types of the most informative entities of the CORD-NER dataset.
NER Dataset. The CORD-NER is defined based on the earliest version of the CORD-19 dataset published in March 2020. The CORD-19 dataset which is used as the main corpus in this research was published in March 2022. CORD-NER has 75 fine-grained entity types. Examples of some annotated entities in CORD-NER are illustrated in Table 1.
Deep Learning Model for NER. There are different methodologies that can provide a well-trained NER model, such as CNN, LSTM, Bi-LSTM, GPT-n, and BERT. Based on our preliminary study, we have decided to employ BERT in this study. The fine-tuning process requires huge processing power and takes time since the NER dataset is big and the BERT model has millions of parameters. We discuss our strategy to handle this problem in the next section.
Dataset Sampling. Since the CORD-NER dataset is big, it will require a huge processing power to fine-tune all data. Therefore, a subset of the dataset is used to fine-tune the BERT transformer. We propose to experiment with two approaches to dataset sampling for CORD-NER in this study.
Sampling Method - 1. The first and simple attempt is to segment the dataset into specific parts (2, 4, 8, 16, ..., 256) and take the part with the most information (entity counts for each type). An example segmentation work is illustrated in Fig. 2. The idea behind this sampling method can be described as follows:
-
1.
Segmentation: the CORD-NER dataset is segmented into two equal parts.
-
2.
Selection: the statistical information (number of documents, number of sentences, number of entity types, overall number of entities) on segmented parts are compared. And the segment that contains more information is selected.
-
3.
Segmentation: then, the selected segment is divided into half again. And followed by Selection as in step 2.
-
4.
This process is carried out until a specified segment threshold is reached.
Sampling Method - 2. The CORD-NER has 75 entity types: 10 covid-19 related types (e.g. Coronavirus, Material, Immune response), 18 Scispacy types (e.g. Loc, Cardinal, Quantity) [18], and 47 UMLS types (e.g. Cell, Bacterium, Social Behavior) [17]. To handle the unbalance distribution of entities for each type and sample a compact NER dataset, 15 biomedical-related types and 10 general types are selected manually. The selected types along with respective categories are shown in Fig. 3. Out of 25 types, the 5 Spacy types are written in abbreviations. FAC type represents for buildings, airports, highways, bridges, etc. GPE represents countries, cities, and states. LOC includes Non-GPE locations, mountain ranges, and bodies of water. Nationalities, religious, or political groups are in NORP. ORG includes companies, agencies, institutions, etc.
The balanced dataset for the selected 25 types is performed in the following procedure: (The process is illustrated with 15 entity types in Table 2.)
-
1.
Given the CORD-NER dataset, we first filter sentences that only include selected 25 types. If we utilize the sentences that also have ignored types, we have to delete those types and this might affect the meaning and structure of sentences (cf. the ‘Selected Types’ column in Table 2).
-
2.
Sort entities count of the filtered corpus in descending order.
-
3.
Cut off the smallest N types to take all and get all sentences involving selected types. The smallest five types are marked as base types and all the entities of these types will be taken (cf. the ‘Smallest types’ column in Table 2).
-
4.
Count entities in the cut-off dataset. Sort again as in step 2.
-
5.
Fill entities with lower counts by adding more sentences that contain those types. [’added_1’ column].
-
6.
Continue steps 4 and 5 until the desired entity count is satisfied. In the current case, the desired entity range is [2000, 20000] and it is satisfied at the added_1 stage.
3.2 Entity Extraction
The fine-tuned model is applied to the corpus of biomedical literatures, CORD-19. The CORD-19 dataset is sampled by using a topic modeling method, LDA, to reduce the size of a big corpus while maintaining sentence structure in a co-related topic. Then, entities and respective types are extracted. The NER process is finalized by a simple post-processing step.
Sampling on the CORD-19 Dataset. The CORD-19 dataset [14] used in the proposed system was published on 5 March 2022. This dataset is composed of 902,589 articles and encompasses various information like ‘source’, ‘title’, ‘doi’, ‘abstract’, ‘publish_time’, ‘authors’, ‘journal’, ‘pdf_json_files’. For the purposes of our system, only the ‘title’, ‘abstract’, and ‘publish time’ will be utilized. To determine dominant topics within the extracted literature, the standard Latent Dirichlet Allocation (LDA) approach [8] is employed. LDA is a generative probabilistic model typically used for collections of discrete data, such as text corpora, wherein each word is considered a blend of an underlying set of topics, and each topic is a blend of a set of topic probabilities. We then select one of the most intriguing topics, and the papers related to this topic form the dataset for the proposed system.
NER Post-Processing. Once the fine-tuning process is completed, the model is employed to classify abstracts from the refined corpus. The model’s output will be an entity and its associated type. Two prefixes (B-, I-) are used for each recognized type, where (B-) denotes the start of an entity and (I-) indicates a subsequent entity. The use of such prefixes allows the identification of entities composed of multiple words. Subsequently, post-processing is performed on these recognized entities and types to yield finalized and clearly named entities. The principles behind the NER post-processing are illustrated in Fig. 4. The (I-) prefix cannot exist independently; it is only accepted if it matches the type of its preceding entity, otherwise, the identified (I-) entity is omitted. All entities with (B-) prefixes are considered. If a (B-) prefix is already matched with an (I-), it is not considered as a standalone word. If not, the (B-) entity is recognized as a single word of its corresponding type.
4 Result and Discussion
4.1 Named Entity Recognition
Named Entity Recognition (NER) is the process of extracting entities along with their respective types from the input sentence. For example, for the sentence “Pfizer effective reducing incidence severe SARS-COV 2 hypoxia critical illness death”, NER will provide some pairs of entities and types such as (Pfizer, vaccine), (SARS-COV 2, covid-19) and (illness, disease). To perform this process, the BERT model is fine-tuned on the existing CORD-NER dataset.
NER Dataset Preparation. The BERT model will recognize word by word. In order to keep two or more words that can be merged to be an entity, we have to add some labels (B- as start of an entity, I- as a subsequent entity). Therefore, the CORD-NER Dataset is reformatted to a suitable format for the fine-tuning of the BERT model, as depicted in Table 3. The Sampling Method-1, detailed in Sect. 3.1, is applied to the CORD-NER Dataset. DistilBERT is fine-tuned using two segmentation sizes: 512 and 32, and with a single epoch. The associated performance metrics and runtime are documented in Table 4. However, this approach possesses two key limitations:
-
1.
Given that segmentation is conducted on sequential data, potential bias towards similar domain topics might arise.
-
2.
The selection procedure employs a greedy algorithm, implying that the predominant types are determined solely based on a segmented portion, not on the entire dataset.
To address these shortcomings, we propose to employ our Sampling Method-2. This approach places emphasis on the entity types, in contrast to the count-centric focus of the first method. Within the CORD-NER Dataset, a total of 75 entity types are present. The entity count for each type exhibits a significant imbalance. Hence, the dataset is balanced by selecting 25 crucial entity types and adjusting the count of unique entities within each type to fall between 2,000 and 20,000, as outlined in Sect. 3.1, using sampling method-2 under the dataset sampling subsection. The entity distribution for the selected types is depicted in Fig. 5. The balanced dataset comprises a total of 43,432 sentences.
Fine-Tuning BERT Model for NER. The balanced CORD-NER dataset (resulting from Sect. 4.1) is fine-tuned using a smaller version of the BERT model, i.e., the distilbert-base-uncased. Three different BERT transformers are trained and compared (see Table 5). The experiment made use of the NER model provided by the Simple Transformers package. An allocation of 80% of the dataset was made for training, with the remaining 20% set aside for testing purposes. The model is trained for 30 epochs at a learning rate of 1e-4, resulting in a training duration of approximately 22 h and 14 minutes. The trend of loss during the fine-tuning task across each epoch is graphically represented in Fig. 6. Performance metrics for the 20% randomly selected test data demonstrated a precision of 0.7494, recall of 0.7366, and an F1 score of 0.7429.
The fine-tuned model performed well during the training (i.e., loss \(\approx \) 0.00) but the accuracy of testing on unseen random test data was around 75%. This suggests that the fine-tuned model is overfitted. Eight simple techniques to prevent overfitting are discussed in [19]. They are Hold-out, Cross-validation, Data augmentation, Feature selection, L1 / L2 regularization, Removing layers/number of units per layer, Dropout, and Early stopping. In our proposed system, two techniques had already been done in the dataset preparation stage: (1) Hold-out which is an approach for separation of training and testing parts, and (2) Feature selection. We will consider other techniques to obtain a better model in future.
4.2 Entity Extraction
The latest CORD-19 dataset [14], published on March 2022, consists of 902,589 papers in terms of 19 attributes: ‘cord_uid’, ‘sha’, ‘source_x’, ‘title’, ‘doi’, ‘pmcid’, ‘pubmed_id’, ‘license’, ‘abstract’, ‘publish_time’, ‘authors’, ‘journal’, ‘mag_id’, ‘who_covidence_id’, ‘arxiv_id’, ‘pdf_json_files’, ‘pmc_json_files’, ‘url’, and ‘s2_id’. Three of 19 attributes are extracted for this study, i.e., ‘title’, ‘abstract’, and ‘publish_time’. Papers published before 2020 are filtered out in order to focus on recent research. Also, 396,374 papers are left after removing null and duplicate files. Then, the vaccine-related dataset is extracted following the procedure presented in the next section.
Dataset Preprocessing. The abstracts from the filtered dataset are taken and preprocessed. The stop words are removed from the abstract sentences. The pre-defined stop words list is acquired from the Natural Language Toolkit (NLTK), a built-in library provided in Python. There are 179 words in the pre-defined stop words list. Some of them are ‘about’, ‘an’, ‘didn’t’, ‘each’, ‘hers’, and ‘most’. Then, the data are tokenized. Tokenization is a way of separating a piece of text into smaller units called ‘tokens’. Then, non-English words are removed by checking whether each word is an alphabet or not. Finally, words with single characters are also removed. The list of vaccine-related words including ‘vaccinated’, and ‘vaccination’ is searched through the cleaned abstracts. A total number of 30,208 vaccine-related papers are found after this process. Table 6 shows the statistical information of filtering papers for the dataset.
Literature Clustering. The obtained vaccine-related dataset is clustered into specific topics. The Latent Dirichlet Allocation (LDA) method [8] is used to do topic modeling. The abstract data are lemmatized and chunked into uni-gram and bi-gram word phrases of ‘NOUN’, ‘ADJ’, ‘VERB’, and ‘ADV’. Examples of extracted phrases include ‘urgently_need’, ‘immune_response’, and ‘infection’.
LDA considers each document as a collection of topics, and each topic is a collection of keywords. Once the number of topics is provided, the algorithm rearranges the topics distribution within the documents and keywords distribution within the topics to obtain a good composition of the topic-keywords distribution. Note that each topic’s name is not defined by the algorithm. We have to define each topic by looking at the dominant terms under the specific topic.
The coherence value defines the accuracy of the LDA model. We can adjust the number of topics and compare the result coherence value to select the optimal topic number. The higher the coherence value, the better the clustering. The data is trained for the topic numbers from three to ten. The result coherence scores are plotted via a line chart in Fig. 7. Although eight is the highest score, there is a gradual improvement from topic numbers four to six. Therefore, we analyzed the topics under six clusters.
We train LDA based on the configuations: \( random\_state =98, \textit{chunksize}=200, \textit{passes}=30, \textit{iterations}=150, \textit{decay}=0.7\), and \( num\_topics =6\). Using LDA, the probability of each cluster for each paper is obtained. The sample data on clustering on six topics of five papers are shown in Table 7.
According to the results shown in Table 8, the papers under Topic 4 are selected to be used as the core dataset.
Post-processing Recognized Named Entities. The CORD-NER dataset also provides information for two-word entities by identifying them using two prefixes (B-) and (I-). The procedure to clean the types has been discussed in NER Post-Processing under Sect. 3.2. Results are illustrated in Fig. 8. In this example, only the words ‘submacular hemorrhage’ will be combined as the ‘DISEASE_OR_SYNDROME’ type. Other entities with the prefix (I-) are discarded. Other entities with the prefix (B-) are taken together with respective entity types. The finalized NER dataset is accessible through IEEE Dataportal [20]. The overall implementation of this research is available online [21].
5 Conclusion
In conclusion, this research presents a method for document-level named entity recognition (NER) using BERT, a large language model (LLM), on the CORD-19 dataset. The study utilizes different sampling approaches for the two datasets, with the Latent Dirichlet allocation algorithm applied to CORD-19 and a simple greedy approach used to collect the most informative data of 25 entity types for CORD-NER. By extracting named entities from the CORD-19 dataset, a baseline dataset for further advanced applications such as knowledge base and ontology is established. Furthermore, the study demonstrates the possibility of utilizing the content understanding ability of LLMs on low-performance machines without using supercomputers, which is an important aspect of making NER accessible to a wider range of researchers. Overall, this research provides a valuable contribution to the field of NER and LLMs, and it has implications for a wide range of applications, from information retrieval and knowledge management to natural language processing and artificial intelligence.
References
Scherbakov, V., Mayorov, V.: Finetuning BERT on partially annotated NER corpora. arXiv. (2022). https://doi.org/10.48550/arXiv.2211.14360
Park, Y.I., Lee, M., Yang, G., Park, S.J., Sohn, C.: Biomedical text NER tagging tool with web interface for generating BERT-based fine-tuning dataset. Appl. Sci. 12, 12012 (2022)
Balkus, S.V., Yan, D.: Improving short text classification with augmented data using GPT-3. ArXiv, abs/2205.10981 (2022)
Kieuvongngam, V., Tan, B., Niu, Y.: Automatic text summarization of COVID-19 medical research articles using BERT and GPT-2. ArXiv, abs/2006.01997 (2020)
Maltoudoglou, L., Paisios, A., Papadopoulos, H.: BERT-based conformal predictor for sentiment analysis. In Conformal and Probabilistic Prediction and Applications, pp. 269–284. PMLR (2020)
Wang, X., Song, X., Guan, Y., Li, B., Han, J.: Comprehensive named entity recognition on CORD-19 with distant or weak supervision. ArXiv, abs/2003.12218 (2020)
Pestryakova, S., et al.: CovidPubGraph: a FAIR knowledge graph of COVID-19 publications. Sci. Data 9, 389 (2022)
Blei, D.M., Ng, A., Jordan, M.I.: Latent dirichlet allocation. J. Mach. Learn. Res. 3, 993–1022 (2001). https://doi.org/10.1016/B978-0-12-411519-4.00006-9
Devlin, J., Chang, M., Lee, K., Toutanova, K.: BERT: pre-training of deep bidirectional transformers for language understanding. ArXiv (2019). https://doi.org/10.48550/arXiv.1810.04805
Giles, O., Huntley, R.P., Karlsson, A., Lomax, J., Malone, J.: Reference ontology and database annotation of the COVID-19 Open Research Dataset (CORD-19). bioRxiv (2020). https://doi.org/10.1101/2020.10.04.325266
Wu, J., Wang, P., Wei, X., Rajtmajer, S.M., Giles, C.L., Griffin, C.: Acknowledgement entity recognition in CORD-19 papers. In: SDP, pp. 10-19 (2020). https://doi.org/10.18653/v1/2020.sdp-1.3
Popovski, G., Kochev, S., Korousic-Seljak, B., Eftimov, T.: FoodIE: a rule-based named-entity recognition method for food information extraction. Int. Conf. Pattern Recogn. Appl. Meth. 12, 915 (2019)
Dekhili, G., Sadat, F.: Hybrid statistical and attentive deep neural approach for named entity recognition in historical newspapers. In: Conference and Labs of the Evaluation Forum (2020)
Wang, L.L., et al.: CORD-19: the COVID-19 open research dataset. ArXiv (2020)
5 Probabilistic Training Data Sampling Methods in Machine Learning. https://towardsdatascience.com/5-probabilistic-training-data-sampling-methods-in-machine-learning-460f2d6ffd9. Accessed 1 July 2023
Liu, J., et al.: Tracing the pace of COVID-19 research: topic modeling and evolution. Big Data Res. 25, 100236–100236 (2021). https://doi.org/10.1016/j.bdr.2021.100236
Unified Medical Language System(UMLS). https://www.nlm.nih.gov/research/umls/ knowledge_sources/metathesaurus/index.html. Accessed 8 July 2023
SpaCy models for biomedical text processing. https://allenai.github.io/scispacy/. Accessed 8 July 2023
David Chuan-En Lin, 8 Simple Techniques to Prevent Overfitting. https://towardsdatascience.com/8-simple-techniques-to-prevent-overfitting-4d443da2ef7d. Accessed 1 July 2023
Thant, S., Anutariya, C., Andres, F., Racharak, T.: BERT fine-tuned CORD-19 NER dataset, IEEE Dataport (2023). https://doi.org/10.21227/m7gj-ks21
ShinThant3010, ‘ShinThant3010/Deep-Learning-based-KG-for-Covid19-Vaccination: Deep Learning based KG for Covid19 Vaccination’. Zenodo, 02 November 2023. https://doi.org/10.5281/zenodo.10066965, https://github.com/ShinThant3010/Deep-Learning-based-KG-for-Covid19-Vaccination
Acknowledgment
The authors would like to thank Dr. Chutiporn Anutariya for the fruitful discussion, and her inspirational comments. We would also like to thank the National Institute of Informatics (Tokyo, Japan) for the support of this research. We would like to express all our gratitude to Nicolas Greneche and Prof. Christophe Cerin (the Institut Galilée - LIPN UMR CNRS 7030, UNIVERSITÉ SORBONNE PARIS NORD) for their helpful advice and cooperation in the use of MAGI cloud. This work was also partially supported by JSPS KAKENHI Grant Number JP22K18004.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Thant, S., Racharak, T., Andres, F. (2023). BERT Fine-Tuning the Covid-19 Open Research Dataset for Named Entity Recognition. In: Anutariya, C., Bonsangue, M.M. (eds) Data Science and Artificial Intelligence. DSAI 2023. Communications in Computer and Information Science, vol 1942. Springer, Singapore. https://doi.org/10.1007/978-981-99-7969-1_19
Download citation
DOI: https://doi.org/10.1007/978-981-99-7969-1_19
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-7968-4
Online ISBN: 978-981-99-7969-1
eBook Packages: Computer ScienceComputer Science (R0)