Abstract
Voriconazole is frequently utilized for the prevention and treatment of invasive fungal infections (IFIs), and is extensively metabolized by the cytochrome P450 (CYP) system. The impact of activity of the genes encoding CYP3A4, CYP3A5, and CYP2C9 on the pharmacokinetics of voriconazole cannot be ignored because, second to CYP2C19, they are the most important enzymes involved in voriconazole metabolism. The influence of genetic polymorphisms in CYP3A4, CYP3A5, and CYP2C9 on the plasma concentrations of voriconazole was evaluated in the present study. The study cohort comprised 158 patients with IFIs in whom 22 single-nucleotide polymorphisms (SNPs) in CYP3A4, CYP3A5, and CYP2C9 were genotyped using the Sequenom MassARRAY RS1000 system, and voriconazole plasma concentrations were measured by high-performance liquid chromatography (HPLC). 40, 91, and 27 patients presented with low (<1 mg/L), normal (1–4 mg/L), and high (>4 mg/L) plasma voriconazole concentrations, respectively. Correlation analysis between polymorphisms and the plasma voriconazole concentration revealed an association between the presence of the rs4646437 T allele and a higher plasma voriconazole concentration [p = 0.033, odds ratio (OR) = 2.832, 95 % confidence interval (CI) = 1.086–7.384]. This study has identified a new SNP related to the metabolism of voriconazole, potentially providing novel insight into the influence of CYP3A4 on the pharmacokinetics of this antifungal agent.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Voriconazole is a broad-spectrum triazole antifungal agent that is available as both an oral and an intravenous formulation, with potent in vitro and in vivo activity against Aspergillus and fluconazole-resistant Candida and Fusarium species [1, 2], and it is used mainly to treat and prevent invasive fungal infections (IFIs) in clinical practice. Data obtained from a meta-analysis study suggested a target plasma concentration range of 1–4 mg/L [3]. Plasma voriconazole concentrations demonstrate a large variability, with concentrations ranging from 0.2 to 12 mg/L, having been observed in patients with IFIs who were treated according to the recommended dosage regimen [4–7]. Modulators of this variability include both comedication and genetic factors [8–13]. Low voriconazole trough concentrations have been associated with poor treatment outcome, while high concentrations are suggestive of an underlying predisposition toward toxicity [14, 15]; both of these situations should, therefore, be avoided.
Voriconazole is extensively metabolized by the cytochrome P450 (CYP) system. N-oxidation of voriconazole into its major circulating metabolite is performed mainly by the cytochrome CYP2C19 [8, 16]. It has been demonstrated that the variability of plasma voriconazole concentrations is related to the presence of CYP2C19 polymorphisms, and that the most common defective alleles are CYP2C19*2, CYP2C19*3, CYP2C19*4, and CYP2C19*17. The frequency of occurrence of the CYP2C19*4 allele is very low in Asians [8]. CYP2C19 genotype status affects the metabolic rate as follows: CYP2C19*1/*17: ultrarapid metabolizer; CYP2C19*1/*1: extensive metabolizer; CYP2C19*1/*2 and CYP2C19*1/*3: intermediate metabolizer; and CYP2C19*2/*2, CYP2C19*2/*3, and CYP2C19*3/*3: poor metabolizer [17].
A final conclusion has not yet been reached as to the effects of polymorphisms in CYP2C9 and CYP3A4 on the plasma voriconazole concentration, although the association between variants of CYP3A4 and CYP2C9 and voriconazole pharmacokinetics have been studied, especially for some missense mutations [13, 18, 19]. Since CYP2C9 and CYP3A4 are also involved in the metabolism of voriconazole, the impacts of CYP3A4 and CYP2C9 activity cannot be ignored. In addition, another major member of the CYP3A family, CYP3A5, like CYP3A4, also plays an important role in drug metabolism [20], and some studies have demonstrated an association between this CYP and the physiological disposition of voriconazole [13, 19, 21]. It is, therefore, rational to hypothesize that single-nucleotide polymorphisms (SNPs) in CYP2C9, CYP3A4, and CYP3A5 could influence their activities and, further, affect the plasma voriconazole concentration.
In the present study, SNPs were selected according to the Chinese Han population data from HapMap and a literature search, and their association with the plasma voriconazole concentration was evaluated, taking into account other factors that influence voriconazole pharmacokinetics, with a view to providing some insight into voriconazole metabolism and pharmacokinetics.
Materials and methods
Study population
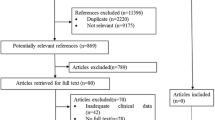
Chinese patients with a proven or probable IFI and who had received intravenous or oral voriconazole at loading or maintenance doses based on the package insert between January 2008 and August 2012 at the First Affiliated Hospital of Medical College, Xi’an Jiaotong University were studied. IFIs were classified as defined by the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group [22]. The exclusion criteria were as follows: (i) age ≤12 years old, (ii) pregnancy, and (iii) lack of compliance with the recommended dosage regimen.
Some of the characteristics potentially influencing the plasma voriconazole concentration were considered, including age, body weight, liver function, renal function, concomitant medications (inhibitors or inducers of CYP2C19, CYP2C9, or CYP3A4), and CYP2C19 genotype status (ultrarapid metabolizer, extensive metabolizer, intermediate metabolizer, or poor metabolizer).
The study protocol was designed in compliance with the principles of the Helsinki Accords, and was reviewed and approved by the local ethical committee. A statement of informed consent was obtained from all participants after the procedure had been explained to them in full.
Genotyping assay
The blood samples were collected into tubes containing ethylenediaminetetraacetic acid. After centrifugation, the samples were stored at −80 °C until analysis. The standard phenol–chloroform extraction method was used to extract genomic DNA from whole blood. DNA concentration was measured by spectrometry (DU-530 UV/Vis spectrophotometer, Beckman Instruments, Fullerton, CA, USA). Twenty-two SNPs (rs1799853, rs1057910, rs12772884, rs1934968, rs9332146, and rs1934967 in CYP2C9; rs2740574, rs4986910, rs4986908, rs4986909, rs12721634, rs4986907, rs4987161, rs4986913, rs4646440, rs4646437, and rs2246709 in CYP3A4; and rs776746, rs10264272, rs41303343, rs4646450, and rs3800959 in CYP3A5) that captured most of the known common CYP2C9, CYP3A4, and CYP3A5 variations were selected for the present study. We also tested the genotype status of CYP2C19 (genotype for *2,*3, and *17) to determine the metabolic types. Sequenom MassARRAY Assay Design 3.0 software was used to design a multiplexed SNP MassEXTEND assay [23–25]. SNPs genotyping was performed using the Sequenom MassARRAY RS1000 system according to the standard protocol recommended by the manufacturer [25]. The primers used for each SNP in the present study are listed in Table 1. Sequenom TYPER 4.0 software was used for data management and analyses [23–26].
Blood sampling and analytical assays
In total, 452 blood samples were collected from the 158 patients receiving voriconazole therapy. The plasma concentrations of voriconazole were determined using the high-performance liquid chromatography (HPLC) method. In brief, a 0.5-mL aliquot of each plasma sample was mixed with 20 μL of internal standard solution (50 mg/L ketoconazole). After vortexing, voriconazole and the internal standard solution were extracted from the plasma using 2 mL of ethyl acetate. The mixture was then centrifuged at 3,000 rpm/min for 10 min, and the supernatant was transferred to a clean culture tube and evaporated to dryness under a stream of nitrogen. The residue was then reconstituted in 100 μL of mobile phase [water (with the addition of 180 mg of ammonium acetate and 80 μL of triethylamine, and adjusted to pH 3.0 using phosphoric acid)/acetonitrile at 60:40, v/v], and a 50-μL aliquot was injected into the HPLC system. Voriconazole and the internal standard were eluted from a C18 column (25 cm × 4.6 mm; Phenomen, UK) using a mobile phase at a flow rate of 1.0 mL/min and with a run time of 20 min. The standard curve was linear from 0.06 to 8.0 mg/L (R 2 = 0.9998). The intra- and interday precisions were within 6.7 and 7.6 %, respectively [17]. Voriconazole was stable after being stored at room temperature for 24 h, after freezing for 20 or 40 days, and after three freeze–thaw cycles. The patients were divided into three groups according to their average plasma voriconazole concentration: low (<1 mg/L), normal (1–4 mg/L), and high (>4 mg/L).
Statistical analysis
The statistical analyses were performed using SPSS 18.0 for Windows (PASW Statistics, SPSS, Chicago, IL, USA). Differences in genotype and allele frequencies between the low and normal plasma concentration groups and between the high and normal plasma concentration groups were assessed using the χ2 test. Bonferroni correction for those multiple comparisons was applied to genotype and allele distribution analysis at the α = 0.025 level. In addition, the association between SNPs and plasma voriconazole concentration was assessed using logistic regression while considering the aforementioned potential confounding factors. The cutoff for statistical significance was set at p < 0.05 (two-sided).
Results
In total, 158 consecutive patients were included; their characteristics are listed in Table 2. Overall, 40, 91, and 27 patients exhibited low, normal, and high plasma voriconazole concentrations, respectively. Analysis of the genotyping assay results revealed that the following alleles were either not observed in the study population or the minor allele frequencies (MAF) were particularly low: rs1799853 (*2) and rs1057910 (*3) of CYP2C9; rs2740574 (*1B), rs4986910 (*3), rs4986908 (*10), rs4986909 (*13), rs12721634 (*14), rs4986907 (*15A) , rs4987161 (*17), and rs4986913 (*19) of CYP3A4; and rs10264272 (*6) and rs41303343 (*7) of CYP3A5 (MAF < 0.01; data not shown). These SNPs were, therefore, not studied further. The genotype distribution and allele frequencies of the other SNPs in the three plasma voriconazole concentration groups are given in Tables 3 and 4. Only rs4646437 was associated with the plasma voriconazole concentration, and the T allele was associated with a significant predisposition to a high plasma voriconazole concentration (p = 0.019). After adjustment for the confounding factors associated with the voriconazole concentration, the T allele was still associated with a higher plasma concentration [p = 0.033, odds ratio (OR) = 2.832, 95 % confidence interval (CI) = 1.086–7.384].
Discussion
The primary purpose of the present study was to determine the impact of CYP2C9, CYP3A5, and CYP3A4 polymorphisms on the plasma voriconazole concentration, with a view to further clarifying additional factors affecting voriconazole pharmacokinetics in a Chinese Han population. After adjustment for confounding factors, such as age, body weight, liver function, renal function, concomitant medications, and metabolic type, it was discovered that rs4646437 of the CYP3A4 gene significantly and independently affected the average plasma concentrations of voriconazole, and that the T-variant allele was related to a higher plasma concentration.
CYP2C9 also plays a role in voriconazole metabolism. While there are few published controlled clinical trials regarding the influence of different CYP2C9 genotypes on the pharmacokinetics and metabolism of voriconazole, there have been some indications that the CYP2C9 genotype might not be relevant to voriconazole pharmacokinetics. One study found no effect of the CYP2C9 genotype on voriconazole levels [27]. Furthermore, a case report found that the voriconazole pharmacokinetics of a patient who was homozygous for CYP2C9*2 were unchanged [18]. In addition, a multiple regression analysis of voriconazole pharmacokinetics found no influence of CYP2C9 among the 35 healthy study participants [13]. In the present study, we found that the minor allele frequency of CYP2C9*2 and CYP2C9*3 SNPs in our population was very low, in accordance with that seen in the Chinese Han population through the HapMap database. This can also be interpreted as the *2 and *3 SNPs not affecting the plasma voriconazole concentration in the Chinese population, in accordance with the findings of the three aforementioned studies [13, 18, 27]. No effect was found for the other SNPs of CYP2C9.
The rs776746 (*3) SNP was the most important for the CYP3A5 gene, which was found in all ethnic groups [28, 29]. Studies of the influence of CYP3A5 on voriconazole pharmacokinetics have yielded contradictory findings. Weiss et al. found no correlation between the CYP3A5*3 variant and voriconazole pharmacokinetics [13], and Levin et al. regarded the serum concentrations of a hepatic drug metabolizing enzyme to be a marker of liver toxicity induced by high voriconazole plasma concentrations and found that liver toxicity was not associated with the CYP3A5*3 allele [19]. However, in vitro studies have shown that voriconazole causes an approximately three-fold higher increase in area under the plasma concentration time curve in individuals with CYP3A5*3/*3 compared to those expressing at least one functional allele [21]. In the present study, the frequency of the rs776746 GA genotype was higher in the low concentration group than in the normal concentration group, and the probability value was close to reaching marginal significance (p = 0.028). However, it is important to note that the sample was small, and, so, the effect of this SNP needs to be verified in large samples.
Voriconazole N-oxidation and 4-hydroxylation are catalyzed more efficiently by CYP3A4 than by CYP3A5 [30]. Numerous variants of CYP3A4 have been described, but none of them can be held accountable for the high phenotypic variation observed [8]. In the present study, we found that the mutation allele frequencies of the known functional SNPs of CYP3A4 were very low, in accordance with that seen in the Chinese Han population through the HapMap database. Therefore, these known functional SNPs do not appear to influence voriconazole metabolism in Chinese people. We also studied another three SNPs located in the intron area. Their mutant allele frequencies in our population were high. We found that the rs4646437 mutant allele was associated with a high plasma voriconazole concentration, which represents a novel finding. The influence of this SNP on the plasma concentrations of risperidone and metabolite 9-hydroxy-risperidone, and adverse effects have been studied, but no significant difference was found [31]. This conclusion was not surprising, since risperidone is metabolized mainly by CYP2D6 [32]. Another study found that the rs4646437 C>T polymorphism could potentially affect the interindividual variability in tacrolimus metabolism among Chinese renal transplant recipients [33]. Tacrolimus is metabolized mainly by CYP3A4 and CYP3A5 [34]. This finding is consistent with the present results. Therefore, although this SNP occurs in an intron, it may affect drugs that are metabolized via CYP3A.
The rs4646437 allele frequency differed significantly between the normal and high plasma voriconazole concentration groups, and the genotype distributions were close to differing significantly. Since the number of mutant homozygotes was small in our studied population, it was difficult to judge whether there was a linear relationship between the number of mutant alleles being carried and the voriconazole concentration. Research involving a large number of samples is needed in order to confirm this kind of contact. We used logistic regression while correcting for some potentially confounding factors to further confirm the association between alleles and the plasma voriconazole concentration. The association between the T allele and a high plasma voriconazole concentration persisted after applying logistic regression.
Voriconazole has been widely used clinically. Since it is metabolized mainly via CYP450, many factors could affect its metabolic processes in the body, among which the CYP450 gene mutation is an important factor. There have been many investigations of how CYP450 gene SNPs influence voriconazole efficacy, both before and after this drug was introduced into the clinical setting, although most of these studies have focused on SNPs in exons and promoters. In the present study, the significant SNPs were located in the intron area, and, therefore, did not (in theory) induce a change in the encoding protein. However, Choi et al. indicated that it is possible for an intronic SNP to alter the splicing of primary transcripts or gene expression [35]. Many other studies have also revealed that specific subsets of exonic splicing silencers exert distinct effects on a multifunctional intron retention reporter, and that one of these subsets is the most likely to be involved in the regulation of endogenous intron retention events. Intron retention could, therefore, affect alternative splicing of pre-mRNA by influencing the exonic splicing silencers, thus producing different mature mRNAs and then translating proteins with different functions [36–38]. Therefore, in the present study, we selected SNPs associated with the voriconazole concentration from the statistical perspective; these SNPs may also have an impact on protein function. Further studies are needed in order to ascertain the biological function of the proteins encoded by these SNPs.
The smallness of the sample was the main limitation of this study. Some homozygous mutations did not appear, and, thus, it was not possible to determine the real genotype distribution. The limitation of the small sample also made it difficult to determine whether there was a linear relationship between gene mutation and the plasma voriconazole concentration. In addition, due to technical reasons, some SNPs were not genotyped successfully in several patients.
In summary, this study provides preliminary evidence that rs14646437 could be a factor underlying a higher plasma voriconazole concentration, suggesting that more data are needed to verify the correct voriconazole dosing in patients. But it needs to be confirmed using large-scale data validation, and more research is needed into the mechanism underlying the function of this SNP.
References
Chiou CC, Groll AH, Walsh TJ (2000) New drugs and novel targets for treatment of invasive fungal infections in patients with cancer. Oncologist 5(2):120–135
Cocchi S, Codeluppi M, Venturelli C, Bedini A, Grottola A, Gennari W, Cavrini F, Di Benedetto F, De Ruvo N, Rumpianesi F, Gerunda GE, Guaraldi G (2011) Fusarium verticillioides fungemia in a liver transplantation patient: successful treatment with voriconazole. Diagn Microbiol Infect Dis 71(4):438–441
Hamada Y, Seto Y, Yago K, Kuroyama M (2012) Investigation and threshold of optimum blood concentration of voriconazole: a descriptive statistical meta-analysis. J Infect Chemother 18(4):501–507
Walsh TJ, Karlsson MO, Driscoll T, Arguedas AG, Adamson P, Saez-Llorens X, Vora AJ, Arrieta AC, Blumer J, Lutsar I, Milligan P, Wood N (2004) Pharmacokinetics and safety of intravenous voriconazole in children after single- or multiple-dose administration. Antimicrob Agents Chemother 48(6):2166–2172
Lazarus HM, Blumer JL, Yanovich S, Schlamm H, Romero A (2002) Safety and pharmacokinetics of oral voriconazole in patients at risk of fungal infection: a dose escalation study. J Clin Pharmacol 42(4):395–402
Eiden C, Cociglio M, Hillaire-Buys D, Eymard-Duvernay S, Ceballos P, Fegueux N, Peyrière H (2010) Pharmacokinetic variability of voriconazole and N-oxide voriconazole measured as therapeutic drug monitoring. Xenobiotica 40(10):701–706
Boyd AE, Modi S, Howard SJ, Moore CB, Keevil BG, Denning DW (2004) Adverse reactions to voriconazole. Clin Infect Dis 39(8):1241–1244
Mikus G, Scholz IM, Weiss J (2011) Pharmacogenomics of the triazole antifungal agent voriconazole. Pharmacogenomics 12(6):861–872
Geist MJ, Egerer G, Burhenne J, Riedel KD, Mikus G (2007) Induction of voriconazole metabolism by rifampin in a patient with acute myeloid leukemia: importance of interdisciplinary communication to prevent treatment errors with complex medications. Antimicrob Agents Chemother 51(9):3455–3456
Mikus G, Schöwel V, Drzewinska M, Rengelshausen J, Ding R, Riedel KD, Burhenne J, Weiss J, Thomsen T, Haefeli WE (2006) Potent cytochrome P450 2C19 genotype-related interaction between voriconazole and the cytochrome P450 3A4 inhibitor ritonavir. Clin Pharmacol Ther 80(2):126–135
Rengelshausen J, Banfield M, Riedel KD, Burhenne J, Weiss J, Thomsen T, Walter-Sack I, Haefeli WE, Mikus G (2005) Opposite effects of short-term and long-term St John’s wort intake on voriconazole pharmacokinetics. Clin Pharmacol Ther 78(1):25–33
Hafner V, Albermann N, Haefeli WE, Ebinger F (2008) Inhibition of voriconazole metabolism by chloramphenicol in an adolescent with central nervous system aspergillosis. Antimicrob Agents Chemother 52(11):4172–4174
Weiss J, Ten Hoevel MM, Burhenne J, Walter-Sack I, Hoffmann MM, Rengelshausen J, Haefeli WE, Mikus G (2009) CYP2C19 genotype is a major factor contributing to the highly variable pharmacokinetics of voriconazole. J Clin Pharmacol 49(2):196–204
Prakash G, Sharma N, Goel M, Titiyal JS, Vajpayee RB (2008) Evaluation of intrastromal injection of voriconazole as a therapeutic adjunctive for the management of deep recalcitrant fungal keratitis. Am J Ophthalmol 146(1):56–59
Andes D, Pascual A, Marchetti O (2009) Antifungal therapeutic drug monitoring: established and emerging indications. Antimicrob Agents Chemother 53(1):24–34
Geist MJ, Egerer G, Burhenne J, Riedel KD, Weiss J, Mikus G (2013) Steady-state pharmacokinetics and metabolism of voriconazole in patients. J Antimicrob Chemother 68(11):2592–2599
Wang T, Chen S, Sun J, Cai J, Cheng X, Dong H, Wang X, Xing J, Dong W, Yao H, Dong Y (2014) Identification of factors influencing the pharmacokinetics of voriconazole and the optimization of dosage regimens based on Monte Carlo simulation in patients with invasive fungal infections. J Antimicrob Chemother 69(2):463–470
Geist MJ, Egerer G, Burhenne J, Mikus G (2006) Safety of voriconazole in a patient with CYP2C9*2/CYP2C9*2 genotype. Antimicrob Agents Chemother 50(9):3227–3228
Levin MD, den Hollander JG, van der Holt B, Rijnders BJ, van Vliet M, Sonneveld P, van Schaik RH (2007) Hepatotoxicity of oral and intravenous voriconazole in relation to cytochrome P450 polymorphisms. J Antimicrob Chemother 60(5):1104–1107
Wojnowski L (2004) Genetics of the variable expression of CYP3A in humans. Ther Drug Monit 26(2):192–199
Yamazaki H, Nakamoto M, Shimizu M, Murayama N, Niwa T (2010) Potential impact of cytochrome P450 3A5 in human liver on drug interactions with triazoles. Br J Clin Pharmacol 69(6):593–597
De Pauw B, Walsh TJ, Donnelly JP, Stevens DA, Edwards JE, Calandra T, Pappas PG, Maertens J, Lortholary O, Kauffman CA, Denning DW, Patterson TF, Maschmeyer G, Bille J, Dismukes WE, Herbrecht R, Hope WW, Kibbler CC, Kullberg BJ, Marr KA, Muñoz P, Odds FC, Perfect JR, Restrepo A, Ruhnke M, Segal BH, Sobel JD, Sorrell TC, Viscoli C, Wingard JR, Zaoutis T, Bennett JE; European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group; National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group (2008) Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis 46(12):1813–1821
He GH, Lu J, Shi PP, Xia W, Yin SJ, Jin TB, Chen DD, Xu GL (2013) Polymorphisms of human histamine receptor H4 gene are associated with breast cancer in Chinese Han population. Gene 519(2):260–265
Li S, Jin T, Zhang J, Lou H, Yang B, Li Y, Chen C, Zhang Y (2012) Polymorphisms of TREH, IL4R and CCDC26 genes associated with risk of glioma. Cancer Epidemiol 36(3):283–287
Gabriel S, Ziaugra L, Tabbaa D (2009) SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet Chapter 2:Unit 2.12
Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, Wang M, Feng W, Zander T, MacConaill L, Lee JC, Nicoletti R, Hatton C, Goyette M, Girard L, Majmudar K, Ziaugra L, Wong KK, Gabriel S, Beroukhim R, Peyton M, Barretina J, Dutt A, Emery C, Greulich H, Shah K, Sasaki H, Gazdar A, Minna J, Armstrong SA, Mellinghoff IK, Hodi FS, Dranoff G, Mischel PS, Cloughesy TF, Nelson SF, Liau LM, Mertz K, Rubin MA, Moch H, Loda M, Catalona W, Fletcher J, Signoretti S, Kaye F, Anderson KC, Demetri GD, Dummer R, Wagner S, Herlyn M, Sellers WR, Meyerson M, Garraway LA (2007) High-throughput oncogene mutation profiling in human cancer. Nat Genet 39(3):347–351
Zonios D, Yamazaki H, Murayama N, Natarajan V, Palmore T, Childs R, Skinner J, Bennett JE (2014) Voriconazole metabolism, toxicity, and the effect of cytochrome P450 2C19 genotype. J Infect Dis 209(12):1941–1948
Schuetz EG, Relling MV, Kishi S, Yang W, Das S, Chen P, Cook EH, Rosner GL, Pui CH, Blanco JG, Edick MJ, Hancock ML, Winick NJ, Dervieux T, Amylon MD, Bash RO, Behm FG, Camitta BM, Raimondi SC, Goh BC, Lee SC, Wang LZ, Fan L, Guo JY, Lamba J, Lim R, Lim HL, Ong AB, Lee HS, Kuehl P, Zhang J, Lin Y, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, Maurel P, Brimer C, Yasuda K, Venkataramanan R, Strom S, Thummel K, Boguski MS (2004) PharmGKB update: II. CYP3A5, cytochrome P450, family 3, subfamily A, polypeptide 5. Pharmacol Rev 56(2):159
Daly AK (2006) Significance of the minor cytochrome P450 3A isoforms. Clin Pharmacokinet 45(1):13–31
Murayama N, Imai N, Nakane T, Shimizu M, Yamazaki H (2007) Roles of CYP3A4 and CYP2C19 in methyl hydroxylated and N-oxidized metabolite formation from voriconazole, a new anti-fungal agent, in human liver microsomes. Biochem Pharmacol 73(12):2020–2026
Choong E, Polari A, Kamdem RH, Gervasoni N, Spisla C, Jaquenoud Sirot E, Bickel GG, Bondolfi G, Conus P, Eap CB (2013) Pharmacogenetic study on risperidone long-acting injection: influence of cytochrome P450 2D6 and pregnane X receptor on risperidone exposure and drug-induced side-effects. J Clin Psychopharmacol 33(3):289–298
Mannheimer B, Holm J, Koukel L, Bertilsson L, Osby U, Eliasson E (2014) Risperidone metabolic ratio as a biomarker of individual CYP2D6 genotype in schizophrenic patients. Eur J Clin Pharmacol 70(6):695–699
Li CJ, Li L, Lin L, Jiang HX, Zhong ZY, Li WM, Zhang YJ, Zheng P, Tan XH, Zhou L (2014) Impact of the CYP3A5, CYP3A4, COMT, IL-10 and POR genetic polymorphisms on tacrolimus metabolism in Chinese renal transplant recipients. PLoS One 9(1):e86206
Gijsen VM, van Schaik RH, Elens L, Soldin OP, Soldin SJ, Koren G, de Wildt SN (2013) CYP3A4*22 and CYP3A combined genotypes both correlate with tacrolimus disposition in pediatric heart transplant recipients. Pharmacogenomics 14(9):1027–1036
Choi JW, Park CS, Hwang M, Nam HY, Chang HS, Park SG, Han BG, Kimm K, Kim HL, Oh B, Kim Y (2008) A common intronic variant of CXCR3 is functionally associated with gene expression levels and the polymorphic immune cell responses to stimuli. J Allergy Clin Immunol 122(6):1119–1126.e7
Howe KJ, Ares M Jr (1997) Intron self-complementarity enforces exon inclusion in a yeast pre-mRNA. Proc Natl Acad Sci U S A 94(23):12467–12472
Pohl M, Bortfeldt RH, Grützmann K, Schuster S (2013) Alternative splicing of mutually exclusive exons—a review. Biosystems 114(1):31–38
Wang Z, Xiao X, Van Nostrand E, Burge CB (2006) General and specific functions of exonic splicing silencers in splicing control. Mol Cell 23(1):61–70
Acknowledgments
This work was supported by the Fundamental Research Funds for the Central Universities of China (grant no. 08143047).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Hai-Rong He and Jin-Yue Sun contributed equally to the study.
Rights and permissions
About this article
Cite this article
He, HR., Sun, JY., Ren, XD. et al. Effects of CYP3A4 polymorphisms on the plasma concentration of voriconazole. Eur J Clin Microbiol Infect Dis 34, 811–819 (2015). https://doi.org/10.1007/s10096-014-2294-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-014-2294-5