Abstract
Background
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease associated with both genetic and environmental risk factors. Previous studies trying to find an association between ALS and unc-13 homolog A (UNC13A) gene variants have shown inconsistent results. This study aimed to conduct a meta-analysis of the association between the C allele of rs12608932, a single-nucleotide polymorphism located in an intron of UNC13A, and risk of ALS and patient survival.
Methods
PubMed, Web of Science, Embase, Chinese National Knowledge Infrastructure, Wanfang, and SinoMed databases were systematically searched for genome-wide association studies or case-control studies published up to January 2019 on the association between this variant in UNC13A and risk and/or prognosis of ALS. Data from eligible studies were extracted and analyzed.
Results
The pooled data (28,072 patients with sporadic ALS and 56,545 controls) showed that rs12608932(C) was associated with an increased risk of ALS (OR = 1.13, 95%CI 1.07–1.20). Subgroup analysis revealed that rs12608932(C) increased the risk of sporadic ALS in non-Asian individuals, including those from the USA and Europe (OR 1.17, 95%CI 1.10–1.25, P < 0.000), but not in Japanese or Chinese subjects (OR 1.01, 95%CI 0.92–1.10, P = 0.85). The available data demonstrated that the CC genotype decreased the survival time of patients with ALS (OR 1.33, 95%CI 1.19–1.49, P < 0.001).
Conclusion
The present meta-analysis suggests that rs12608932(C) is associated with increased ALS susceptibility, especially in Caucasian and European subjects, and that the CC genotype of rs12608932 is associated with reduced ALS patient survival.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disease characterized by muscle weakness and loss of control voluntary movement caused by degeneration of both the upper and lower motor neurons [1]. The progression of ALS is rapid and usually results in paralysis and death from respiratory failure [2]. The average survival time of patients with ALS ranges from 3 to 5 years [3]. The pathophysiology of the disease remains partially elusive, although it has been reported that genetic and environmental risk factors contribute to ALS development [3].
In some patients, the disease is considered an inherited disorder following a Mendelian pattern. Within populations of European descent, up to 20% of individuals with ALS have a family history of either ALS or frontotemporal dementia. Seventy percent of familial ALS cases are linked to mutations in four genes: superoxide dismutase 1 (SOD1), C9orf72, TAR DNA-binding protein 43 (TARDBP-43), and FUS. Based on population-based registers, genome-wide association studies (GWAS) of sporadic ALS suggested that several genetic variants are associated with an increased risk of ALS. These studies have shed light on the association of DPP6, ITPR2, SUNC1, 9p21.2, and UNC13A genes with ALS [4]. Among these genetic risk factors, one locus in chromosome 9p21 is the only one successfully identified across all the studies. The rs12608932 variant in UNC13A gene was first identified as a risk single-nucleotide polymorphism (SNP) associated with ALS [4], and similar results were obtained in ALS patients from the Netherlands [5], Italy [6], USA, and Europe [7], but these results were not replicated in studies in France [8], UK [9], Japan [10], or China [10, 11]. Interestingly, another publication using an independent cohort and data from the original study found that a minor allele in rs12608932 was associated with a shorter survival of ALS patients [5]. Similar observations were made in ALS patients from Italy [6] but not Spain [12]. In view of the inconsistence of these studies, we conducted a meta-analysis to further verify whether rs12608932 was a risk factor for ALS and/or modify the survival of ALS patients.
Materials and methods
Literature search strategy
Pubmed, Embase, Web of Science, Wanfang, Chinese National Knowledge Infrastructure, and SinoMed databases were searched for eligible studies published up to January 2019. The following search terms were applied: “UNC13A,” “unc-13 homologue A gene,” “rs12608932,” “Genome-wide association studies,” and “amyotrophic lateral sclerosis.”. No language restrictions were applied.
Study selection criteria
To be included in the meta-analysis, studies had to fulfill all the following criteria: (i) use a GWAS design or case-control study design to analyze patients with ALS (diagnosed according to the El Escorial criteria) [13] and healthy controls; (ii) evaluate the association between the rs12608932 variant and ALS risk and/or the association between rs12608932 and prognosis of ALS; and (iii) report the minor allele frequency (MAF) distributions for both cases and controls. If there were studies on overlapping cohorts, only the largest study was included. Studies were excluded if they did not report the MAF of rs12608932 or if it was impossible to calculate the MAF based on the data provided by the authors.
Data extraction
Two investigators (BY Yang and B Liu) independently searched the literature and extracted data. Disagreements were resolved by the corresponding author (XL Yang). The following data were extracted from the original article: surname of the first author, year of publication, ethnicity of the study cohort, sample size of ALS and control groups, allele frequencies in cases and controls, the odds ratio (OR), and 95% confidence intervals (CI) for survival.
Statistical analysis
We evaluated the association between ALS risk with the C allele of rs12608932 expressed as OR and 95% CI. Cochran’s Q test and I2 statistics were used to assess the heterogeneity among studies. If I2 < 25%, no heterogeneity among studies was considered; 25% ≤ I2 < 50% indicated low heterogeneity; 50% ≤ I2 < 75% indicated moderated heterogeneity; while I2 ≥ 75% indicated substantial heterogeneity [14]. A fixed-effect model was used to analyze pooled data classified as homogeneous or with low heterogeneity, and a random-effect model was applied to assess data classified as of moderate or substantial heterogeneity [15]. Egger’s and Begg’s tests were applied to evaluate the publication bias [15, 16]. Stata version 12.0 (StataCorp, College Station, TX, USA) was used to perform the analyses. A P value of less than 0.05 was considered the threshold for statistical significance in all the analyses.
Results
Literature search and study selection
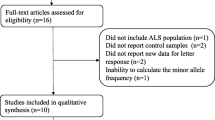
A total of 49 potentially eligible articles were identified after database search and removal of duplicates. After eliminating 28 articles based on the title and abstract, the remaining 21 were read and the following publications were excluded: 2 review articles, 3 GWAS studies not providing detailed information on rs12608932 [17,18,19], 1 GWAS study conducted in China that did not detect rs12608932 and that deviated significantly from the Hardy-Weinberg equilibrium [20], 1 study not following a case-control design, and 4 publications not assessing the relationship between the rs12608932 variant and ALS risk or survival.
The remaining 10 publications included a total of 28,072 patients with sporadic ALS and 56,545 controls. These 10 studies included 12 cohorts and evaluated the possible association between the rs12608932 C variant and ALS risk in Europe [4], France [8], UK and other European countries [9], The Netherlands [5], Italy [6], USA and Europe [7, 21], Japan [10], and China [10, 11] (Fig. 1). Four studies including six cohorts examined the association between the rs12608932 variant and survival of patients with ALS [4,5,6, 12].
The rs12608932(C) variant and risk of sporadic ALS
Data from the nine GWAS and case-control studies were pooled to analyze the potential association between the rs12608932 variant and risk of sporadic ALS (Table 1). Frequencies of the C allele showed heterogeneity based on a chi-squared test (χ2 = 42.79; degrees of freedom, DF = 11; P < 0.001) and the I2 value (74.3.1%). Therefore, allele data were analyzed using a random-effects model.
The pooled OR for ALS risk in the presence of the C allele relative to the A allele was 1.13 (95%CI 1.07–1.20; Fig. 2), and the Z test for overall effect was 4.26 (P < 0.000). Sensitivity analysis indicated that none of the studies significantly affected the results in an independent fashion (Figs. 3 and 4). The funnel plot was visually symmetrical indicating that there was no significant publication bias (Fig. 3). Neither Egger’s test (P = 0.945) nor Begg’s test (P = 0.598) showed significant risk of publication bias.
Geographical differences were also evaluated between rs12608932(C) and risk of sporadic ALS. Our analyses revealed that the C allele of rs12608932 increased the risk of sporadic ALS in non-Asian subjects, including those from the USA and Europe (OR 1.17, 95%CI 1.10–1.25, P < 0.000), while a significant association was not found in Japanese or Chinese (OR 1.01, 95%CI 0.92–1.10, P = 0.85) (Fig. 2).
The rs12608932(C) variant and survival of ALS patients
Data were pooled from six cohorts in four studies (Table 2), and a fixed-effect model was applied because the data were homogeneous (I2 = 0, DF = 4, P = 0.587). Pooled hazard ratio (HR) for survival of ALS patients with the CC genotype relative to those without this genotype was 1.33 (95%CI 1.19–1.49, P < 0.001; Fig. 5). The funnel plot was visually symmetrical, indicating that there was no significant publication bias (Fig. 6). Neither Egger’s test (P = 0.110) nor Begg’s test (P = 0.452) showed significant risk of publication bias.
Discussion
Results from the present meta-analysis revealed that rs12608932(C) variant in UNC13A gene was significantly associated with a higher risk of developing ALS in Caucasian and European subjects, but not in Japanese or Chinese cohorts. Besides, the present data support UNC13A as a modifier of prognosis among sporadic ALS.
These results are in line with previous evidence suggesting that UNC13A may play a role in the pathophysiology of ALS. UNC13A is a member of a large family of presynaptic brain proteins. UNC13A protein regulates not only presynaptic vesicle priming but also glutamate release at neuromuscular synapses. In vitro experiments using glutamatergic hippocampal neurons from mice lacking the UNC13A gene showed arrested synaptic vesicle maturation and disrupted glutamatergic transmission [22]. A dysfunction in glutamate metabolism has been hypothesized to play a key role in the pathophysiology of ALS and, in fact, the only drug currently approved for ALS treatment, riluzole, is a glutamate release inhibitor [23]. Experiments with Caenorhabditis elegans suggest that UNC13A contributes to motor neuron degeneration through interaction with TARDBP-43 [24], which has been proposed as a pathological biomarker of ALS [1].
Our subgroup analysis indicated that rs12608932(C) influences the risk of sporadic ALS in Caucasian and European populations (OR 1.19, 95%CI 1.13–1.26, P < 0.001), but not in Japanese or Chinese subjects. These results are consistent with numerous individual studies in the literature. One reason for this ethnic bias may be the fact that the Japanese and Chinese subjects had relatively small samples, increasing risk of non-generalizable results. Another reason may be ethnic differences: as evidence from the public database among Caucasian and European healthy study subjects in our meta-analysis, even among the seven studies based on Caucasian and European subjects, studies from France and UK did not detect an association between the (C) variant and risk of ALS. We therefore hypothesize that the rs12608932(C) variant may be a susceptibility SNP for ALS in some specific populations. In fact, a role for ethnic-specific gene variants has been described in other neurodegenerative diseases. For example, several SNPs in the microtubule-associated protein tau (MAPT) locus have been strongly associated with increased risk of Parkinson’s disease in the European population, while a recent GWAS study conducted in multiple Asian nations failed to identify any association [25]. The SNP rs10139154 in the gene encoding Sec1 family domain containing 1 (SCFD1) protein was identified as a risk SNP for ALS in European populations [17] but not in a large cohort of Chinese patients [26]. ALS is a complex disease involving genetic and environmental risk factors and, therefore, it is extremely difficult to find risk SNPs common to different regions or ethnicities.
Although the mechanisms by which UNC13A influences survival remain unclear, the present study found that the CC genotype of rs12608932 significantly decreased the survival time of ALS patients. In the past 20 years, significant efforts have been directed at exploring the factors determining ALS patient survival. Survival is longer in patients who are younger or in whom symptom onset begins in the limbs, as well as in individuals who are diagnosed longer after symptom onset [27]. In addition, missense mutations in the FUS gene and pathogenic repeat expansions of C9ORF72 gene have been shown to adversely influence survival. Previous research conducted by our group indicated that Chinese ALS patients carrying the AA genotype of rs9268856 have shorter survival [28]. However, larger independent studies are still necessary to establish a definitive role for UNC13A in ALS survival.
While our meta-analysis offers the largest pooled comprehensive evaluation of the rs12608932(C) variant and ALS risk and survival, some limitations should be taken into consideration. Firstly, although we searched six international and Chinese databases, and Egger’s and Begg’s tests suggested no significant risk of such publication bias; this risk is always present. Although this meta-analysis included 28,072 patients with sporadic ALS and 56,545 controls from multiple ethnicities, potential heterogeneity may still be present, even if a random-effect model was applied. Therefore, our results should be interpreted with caution. In addition, there were only four studies involving six cohorts that assessed the relationship between rs12608932 and prognosis of ALS. These studies were performed only in Caucasian and European populations; whether rs12608932 confers survival in other ethnicities requires further verification.
In conclusion, our results suggest that the C allele of rs12608932 is associated with ALS susceptibility, especially in Caucasian and European populations. Furthermore, the CC genotype was confirmed as a modifier of ALS patient survival. However, further well-designed, larger studies involving more ethnicities are needed to validate these results. Similarly, functional studies addressing the pathogenic mechanisms linking UNC13A and ALS are required. Ultimately, the UNC13A pathophysiological pathway may be identified as a potential therapeutic target that could help prolong the survival of ALS patients.
References
Hardiman O, Al-Chalabi A, Chio A et al (2017) Amyotrophic lateral sclerosis. Nat Rev Dis Primers 3:17071
Chio A, Battistini S, Calvo A, Caponnetto C, Conforti FL, Corbo M, Giannini F, Mandrioli J, Mora G, Sabatelli M, the ITALSGEN Consortium, Monsurro MR, Tedeschi G, Conte A, Luigetti M, Lattante S, Marangi G, Volanti P, Marinou K, Papetti L, Lunetta C, Pintor GL, Salvi F, Bartolomei I, Quattrone A, Gambardella A, Logroscino G, Simone I, Pisano F, Spataro R, la Bella V, Colletti T, Mancardi G, Origone P, Sola P, Borghero G, Marrosu F, Marrosu MG, Murru MR, Floris G, Cannas A, Piras V, Costantino E, Pani C, Sotgiu MA, Pugliatti M, Parish LD, Cossu P, Ticca A, Rodolico C, Portaro S, Ricci C, Moglia C, Ossola I, Brunetti M, Barberis M, Canosa A, Cammarosano S, Bertuzzo D, Fuda G, Ilardi A, Manera U, Pastore I, Sproviero W, Logullo F, Tanel R, Ajmone C, Mastro E, Pain D, Mandich P, Penco S, Restagno G, Zollino M, Surbone A (2014) Genetic counselling in ALS: facts, uncertainties and clinical suggestions. J Neurol Neurosurg Psychiatry 85(5):478–485
Oskarsson B, Gendron TF, Staff NP (2018) Amyotrophic lateral sclerosis: an update for 2018. Mayo Clin Proc 93(11):1617–1628
van Es MA, Veldink JH, Saris CG, Blauw HM, van Vught P, Birve A, Lemmens R, Schelhaas HJ, Groen EJ, Huisman MH, van der Kooi A, de Visser M, Dahlberg C, Estrada K, Rivadeneira F, Hofman A, Zwarts MJ, van Doormaal P, Rujescu D, Strengman E, Giegling I, Muglia P, Tomik B, Slowik A, Uitterlinden AG, Hendrich C, Waibel S, Meyer T, Ludolph AC, Glass JD, Purcell S, Cichon S, Nöthen MM, Wichmann HE, Schreiber S, Vermeulen SH, Kiemeney LA, Wokke JH, Cronin S, McLaughlin R, Hardiman O, Fumoto K, Pasterkamp RJ, Meininger V, Melki J, Leigh PN, Shaw CE, Landers JE, al-Chalabi A, Brown RH Jr, Robberecht W, Andersen PM, Ophoff RA, van den Berg L (2009) Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat Genet 41(10):1083–1087
Diekstra FP, van Vught PW, van Rheenen W et al (2012) UNC13A is a modifier of survival in amyotrophic lateral sclerosis. Neurobiol Aging 33(3):630 e3–630 e8
Chio A, Mora G, Restagno G et al (2013) UNC13A influences survival in Italian amyotrophic lateral sclerosis patients: a population-based study. Neurobiol Aging 34(1):357 e1–357 e5
Ahmeti KB, Ajroud-Driss S, Al-Chalabi A et al (2013) Age of onset of amyotrophic lateral sclerosis is modulated by a locus on 1p34.1. Neurobiol Aging 34(1):357 e7–357 19
Daoud H, Belzil V, Desjarlais A, Camu W, Dion PA, Rouleau GA (2010) Analysis of the UNC13A gene as a risk factor for sporadic amyotrophic lateral sclerosis. Arch Neurol 67(4):516–517
Shatunov A, Mok K, Newhouse S, Weale ME, Smith B, Vance C, Johnson L, Veldink JH, van Es MA, van den Berg LH, Robberecht W, van Damme P, Hardiman O, Farmer AE, Lewis CM, Butler AW, Abel O, Andersen PM, Fogh I, Silani V, Chiò A, Traynor BJ, Melki J, Meininger V, Landers JE, McGuffin P, Glass JD, Pall H, Leigh PN, Hardy J, Brown RH Jr, Powell JF, Orrell RW, Morrison KE, Shaw PJ, Shaw CE, al-Chalabi A (2010) Chromosome 9p21 in sporadic amyotrophic lateral sclerosis in the UK and seven other countries: a genome-wide association study. Lancet Neurol 9(10):986–994
Iida A, Takahashi A, Deng M et al (2011) Replication analysis of SNPs on 9p21.2 and 19p13.3 with amyotrophic lateral sclerosis in East Asians. Neurobiol Aging 32(4):757 e13–757 e14
Chen X, Huang R, Chen Y, Zheng Z, Chen K, Song W, Zhao B, Yang Y, Yuan L, Shang H (2014) Association analysis of four candidate genetic variants with sporadic amyotrophic lateral sclerosis in a Chinese population. Neurol Sci 35(7):1089–1095
Vidal-Taboada JM, Lopez-Lopez A, Salvado M, Lorenzo L, Garcia C, Mahy N, Rodríguez MJ, Gamez J (2015) UNC13A confers risk for sporadic ALS and influences survival in a Spanish cohort. J Neurol 262(10):2285–2292
Brooks BR, Miller RG, Swash M, Munsat TL (2000) El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord 1(5):293–299
Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ. 327(7414):557–560
Zhang TS, Zhong WZ (2012) Applied methodology for evidence-based medicine, pp 166–175
Egger M, Smith GD, Phillips AN (1997) Meta-analysis: principles and procedures. BMJ. 315(7121):1533–1537
Laaksovirta H, Peuralinna T, Schymick JC, Scholz SW, Lai SL, Myllykangas L, Sulkava R, Jansson L, Hernandez DG, Gibbs JR, Nalls MA, Heckerman D, Tienari PJ, Traynor BJ (2010) Chromosome 9p21 in amyotrophic lateral sclerosis in Finland: a genome-wide association study. Lancet Neurol 9(10):978–985
Nicolas A, Kenna KP, Renton AE, Ticozzi N, Faghri F, Chia R, Dominov JA, Kenna BJ, Nalls MA, Keagle P, Rivera AM, van Rheenen W, Murphy NA, van Vugt JJFA, Geiger JT, van der Spek RA, Pliner HA, Shankaracharya, Smith BN, Marangi G, Topp SD, Abramzon Y, Gkazi AS, Eicher JD, Kenna A, Mora G, Calvo A, Mazzini L, Riva N, Mandrioli J, Caponnetto C, Battistini S, Volanti P, la Bella V, Conforti FL, Borghero G, Messina S, Simone IL, Trojsi F, Salvi F, Logullo FO, D’Alfonso S, Corrado L, Capasso M, Ferrucci L, Moreno CAM, Kamalakaran S, Goldstein DB, Gitler AD, Harris T, Myers RM, Phatnani H, Musunuri RL, Evani US, Abhyankar A, Zody MC, Kaye J, Finkbeiner S, Wyman SK, LeNail A, Lima L, Fraenkel E, Svendsen CN, Thompson LM, van Eyk JE, Berry JD, Miller TM, Kolb SJ, Cudkowicz M, Baxi E, Benatar M, Taylor JP, Rampersaud E, Wu G, Wuu J, Lauria G, Verde F, Fogh I, Tiloca C, Comi GP, Sorarù G, Cereda C, Corcia P, Laaksovirta H, Myllykangas L, Jansson L, Valori M, Ealing J, Hamdalla H, Rollinson S, Pickering-Brown S, Orrell RW, Sidle KC, Malaspina A, Hardy J, Singleton AB, Johnson JO, Arepalli S, Sapp PC, McKenna-Yasek D, Polak M, Asress S, al-Sarraj S, King A, Troakes C, Vance C, de Belleroche J, Baas F, ten Asbroek ALMA, Muñoz-Blanco JL, Hernandez DG, Ding J, Gibbs JR, Scholz SW, Floeter MK, Campbell RH, Landi F, Bowser R, Pulst SM, Ravits JM, MacGowan DJL, Kirby J, Pioro EP, Pamphlett R, Broach J, Gerhard G, Dunckley TL, Brady CB, Kowall NW, Troncoso JC, le Ber I, Mouzat K, Lumbroso S, Heiman-Patterson TD, Kamel F, van den Bosch L, Baloh RH, Strom TM, Meitinger T, Shatunov A, van Eijk KR, de Carvalho M, Kooyman M, Middelkoop B, Moisse M, McLaughlin RL, van Es MA, Weber M, Boylan KB, van Blitterswijk M, Rademakers R, Morrison KE, Basak AN, Mora JS, Drory VE, Shaw PJ, Turner MR, Talbot K, Hardiman O, Williams KL, Fifita JA, Nicholson GA, Blair IP, Rouleau GA, Esteban-Pérez J, García-Redondo A, al-Chalabi A, Rogaeva E, Zinman L, Ostrow LW, Maragakis NJ, Rothstein JD, Simmons Z, Cooper-Knock J, Brice A, Goutman SA, Feldman EL, Gibson SB, Taroni F, Ratti A, Gellera C, van Damme P, Robberecht W, Fratta P, Sabatelli M, Lunetta C, Ludolph AC, Andersen PM, Weishaupt JH, Camu W, Trojanowski JQ, van Deerlin VM, Brown RH Jr, van den Berg LH, Veldink JH, Harms MB, Glass JD, Stone DJ, Tienari P, Silani V, Chiò A, Shaw CE, Traynor BJ, Landers JE, Logullo FO, Simone I, Logroscino G, Salvi F, Bartolomei I, Borghero G, Murru MR, Costantino E, Pani C, Puddu R, Caredda C, Piras V, Tranquilli S, Cuccu S, Corongiu D, Melis M, Milia A, Marrosu F, Marrosu MG, Floris G, Cannas A, Tranquilli S, Capasso M, Caponnetto C, Mancardi G, Origone P, Mandich P, Conforti FL, Cavallaro S, Mora G, Marinou K, Sideri R, Penco S, Mosca L, Lunetta C, Pinter GL, Corbo M, Riva N, Carrera P, Volanti P, Mandrioli J, Fini N, Fasano A, Tremolizzo L, Arosio A, Ferrarese C, Trojsi F, Tedeschi G, Monsurrò MR, Piccirillo G, Femiano C, Ticca A, Ortu E, la Bella V, Spataro R, Colletti T, Sabatelli M, Zollino M, Conte A, Luigetti M, Lattante S, Marangi G, Santarelli M, Petrucci A, Pugliatti M, Pirisi A, Parish LD, Occhineri P, Giannini F, Battistini S, Ricci C, Benigni M, Cau TB, Loi D, Calvo A, Moglia C, Brunetti M, Barberis M, Restagno G, Casale F, Marrali G, Fuda G, Ossola I, Cammarosano S, Canosa A, Ilardi A, Manera U, Grassano M, Tanel R, Pisano F, Harms MB, Goldstein DB, Shneider NA, Goutman S, Simmons Z, Miller TM, Chandran S, Pal S, Manousakis G, Appel SH, Simpson E, Wang L, Baloh RH, Gibson S, Bedlack R, Lacomis D, Sareen D, Sherman A, Bruijn L, Penny M, Allen AS, Appel S, Baloh RH, Bedlack RS, Boone BE, Brown R, Carulli JP, Chesi A, Chung WK, Cirulli ET, Cooper GM, Couthouis J, Day-Williams AG, Dion PA, Gibson S, Gitler AD, Glass JD, Goldstein DB, Han Y, Harms MB, Harris T, Hayes SD, Jones AL, Keebler J, Krueger BJ, Lasseigne BN, Levy SE, Lu YF, Maniatis T, McKenna-Yasek D, Miller TM, Myers RM, Petrovski S, Pulst SM, Raphael AR, Ravits JM, Ren Z, Rouleau GA, Sapp PC, Shneider NA, Simpson E, Sims KB, Staropoli JF, Waite LL, Wang Q, Wimbish JR, Xin WW, Phatnani H, Kwan J, Sareen D, Broach JR, Simmons Z, Arcila-Londono X, Lee EB, van Deerlin VM, Shneider NA, Fraenkel E, Ostrow LW, Baas F, Zaitlen N, Berry JD, Malaspina A, Fratta P, Cox GA, Thompson LM, Finkbeiner S, Dardiotis E, Miller TM, Chandran S, Pal S, Hornstein E, MacGowan DJ, Heiman-Patterson T, Hammell MG, Patsopoulos NA, Dubnau J, Nath A, Kaye J, Finkbeiner S, Wyman S, LeNail A, Lima L, Fraenkel E, Rothstein JD, Svendsen CN, Thompson LM, van Eyk J, Maragakis NJ, Berry JD, Glass JD, Miller TM, Kolb SJ, Baloh RH, Cudkowicz M, Baxi E, Benatar M, Taylor JP, Wu G, Rampersaud E, Wuu J, Rademakers R, Züchner S, Schule R, McCauley J, Hussain S, Cooley A, Wallace M, Clayman C, Barohn R, Statland J, Ravits J, Swenson A, Jackson C, Trivedi J, Khan S, Katz J, Jenkins L, Burns T, Gwathmey K, Caress J, McMillan C, Elman L, Pioro E, Heckmann J, So Y, Walk D, Maiser S, Zhang J, Silani V, Ticozzi N, Gellera C, Ratti A, Taroni F, Lauria G, Verde F, Fogh I, Tiloca C, Comi GP, Sorarù G, Cereda C, D’Alfonso S, Corrado L, de Marchi F, Corti S, Ceroni M, Mazzini L, Siciliano G, Filosto M, Inghilleri M, Peverelli S, Colombrita C, Poletti B, Maderna L, del Bo R, Gagliardi S, Querin G, Bertolin C, Pensato V, Castellotti B, Camu W, Mouzat K, Lumbroso S, Corcia P, Meininger V, Besson G, Lagrange E, Clavelou P, Guy N, Couratier P, Vourch P, Danel V, Bernard E, Lemasson G, al Kheifat A, al-Chalabi A, Andersen P, Basak AN, Blair IP, Chio A, Cooper-Knock J, Corcia P, Couratier P, de Carvalho M, Dekker A, Drory V, Redondo AG, Gotkine M, Hardiman O, Hide W, Iacoangeli A, Glass J, Kenna K, Kiernan M, Kooyman M, Landers J, McLaughlin R, Middelkoop B, Mill J, Neto MM, Moisse M, Pardina JM, Morrison K, Newhouse S, Pinto S, Pulit S, Robberecht W, Shatunov A, Shaw P, Shaw C, Silani V, Sproviero W, Tazelaar G, Ticozzi N, van Damme P, van den Berg L, van der Spek R, van Eijk K, van Es M, van Rheenen W, van Vugt J, Veldink J, Weber M, Williams KL, Zatz M, Bauer DC, Twine NA (2018) Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron. 97(6):1268–83 e6
Chen CJ, Chen CM, Pai TW, Chang HT, Hwang CS (2016) A genome-wide association study on amyotrophic lateral sclerosis in the Taiwanese Han population. Biomark Med 10(6):597–611
Deng M, Wei L, Zuo X, Tian Y, Xie F, Hu P, Zhu C, Yu F, Meng Y, Wang H, Zhang F, Ma H, Ye R, Cheng H, du J, Dong W, Zhou S, Wang C, Wang Y, Wang J, Chen X, Sun Z, Zhou N, Jiang Y, Liu X, Li X, Zhang N, Liu N, Guan Y, Han Y, Han Y, Lv X, Fu Y, Yu H, Xi C, Xie D, Zhao Q, Xie P, Wang X, Zhang Z, Shen L, Cui Y, Yin X, Cheng H, Liang B, Zheng X, Lee TMC, Chen G, Zhou F, Veldink JH, Robberecht W, Landers JE, Andersen PM, al-Chalabi A, Shaw C, Liu C, Tang B, Xiao S, Robertson J, Zhang F, van den Berg LH, Sun L, Liu J, Yang S, Ju X, Wang K, Zhang X (2013) Genome-wide association analyses in Han Chinese identify two new susceptibility loci for amyotrophic lateral sclerosis. Nat Genet 45(6):697–700
van Rheenen W, Shatunov A, Dekker AM et al (2016) Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat Genet 48(9):1043–1048
Augustin I, Rosenmund C, Sudhof TC, Brose N (1999) Munc13-1 is essential for fusion competence of glutamatergic synaptic vesicles. Nature. 400(6743):457–461
Liu J, Wang LN (2018) The efficacy and safety of riluzole for neurodegenerative movement disorders: a systematic review with meta-analysis. Drug Deliv 25(1):43–48
Veriepe J, Fossouo L, Parker JA (2015) Neurodegeneration in C. elegans models of ALS requires TIR-1/Sarm1 immune pathway activation in neurons. Nat Commun 6:7319
Foo JN, Tan LC, Irwan ID, Au WL, Low HQ, Prakash KM, Ahmad-Annuar A, Bei J, Chan AY, Chen CM, Chen YC, Chung SJ, Deng H, Lim SY, Mok V, Pang H, Pei Z, Peng R, Shang HF, Song K, Tan AH, Wu YR, Aung T, Cheng CY, Chew FT, Chew SH, Chong SA, Ebstein RP, Lee J, Saw SM, Seow A, Subramaniam M, Tai ES, Vithana EN, Wong TY, Heng KK, Meah WY, Khor CC, Liu H, Zhang F, Liu J, Tan EK (2017) Genome-wide association study of Parkinson’s disease in East Asians. Hum Mol Genet 26(1):226–232
Chen Y, Zhou Q, Gu X, Wei Q, Cao B, Liu H, Hou Y, Shang H (2018) An association study between SCFD1 rs10139154 variant and amyotrophic lateral sclerosis in a Chinese cohort. Amyotroph Lateral Scler Frontotemporal Degener 19(5–6):413–418
Salameh JS, Brown RH Jr, Berry JD (2015) Amyotrophic lateral sclerosis: review. Semin Neurol 35(4):469–476
Yang X, Zheng J, Tian S, Chen Y, An R, Zhao Q, Xu Y (2017) HLA-DRA/HLA-DRB5 polymorphism affects risk of sporadic ALS and survival in a southwest Chinese cohort. J Neurol Sci 373:124–128
Funding
This research was supported by the Yunnan Applied Basic Research Project-Union Foundation of China (201801CH00572), National Natural Science Foundation of China (81860247), Yunnan Province Medical Health Research Institute Project (2018NS0102), and The First Affiliated Hospital of Kunming Medical University Doctoral Research Fund Project (2017BS005).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yang, B., Jiang, H., Wang, F. et al. UNC13A variant rs12608932 is associated with increased risk of amyotrophic lateral sclerosis and reduced patient survival: a meta-analysis. Neurol Sci 40, 2293–2302 (2019). https://doi.org/10.1007/s10072-019-03951-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-019-03951-y