Abstract
Background
Amyotrophic lateral sclerosis (ALS), one of the motor neuron diseases, appears to be caused by genetic and environmental risk factors. However, the influence of Pro34Ser variant of CHCHD10 gene in increasing risk of ALS remains indeterminate. This study conducted a meta-analysis to establish the association between Pro34Ser variant of CHCHD10 gene and risk of ALS.
Methods
PubMed, Web of Science, and Embase databases were systematically searched for genome-wide association studies or case–control studies published up to March 28, 2020, on the association between Pro34Ser variant and risk of ALS. Data from eligible studies were extracted and analyzed.
Results
Twelve case–control studies involving 7442 patients with sporadic ALS and 75,371 controls were analyzed. The Pro34Ser variant was not associated with increased risk of ALS disease based on fixed-effects meta-analysis (Pro34Ser-positive vs Pro34Ser-negative: OR 1.23, 95% CI 0.90 to 1.69, P = 0.201).
Conclusion
Existing evidence suggests that Pro34Ser variant in CHCHD10 is not associated with risk of ALS, particularly in Caucasian participants. However, our results ought to be validated using large, well-designed studies, especially in Asian and African populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Amyotrophic lateral sclerosis (ALS) is an idiopathic, fatal neurodegenerative disease of the human motor system [1]. Clinically, ALS is characterized by the presence of upper and lower motor neuron features involving brainstem and multiple spinal cord regions of innervation [1]. Progression of ALS is rapid, and most patients die within 3–5 years after onset of symptoms due to respiratory failure [2]. Pathogenesis of ALS has not been fully understood, although it has been reported that genetics and environmental risk factors contribute to its development [3].
Approximately 5–10% of ALS cases are familial, with a Mendelian pattern of inheritance, whereas the remaining 90% of ALS cases are sporadic [1]. Familial ALS has been associated with several gene mutations including superoxide dismutase 1 (SOD1), C9orf72, TARDNA-binding protein 43 (TARDBP-43), and FUS [4]. Seven novel genes, MATR3, CHCHD10, TBK1, TUBA4A, NEK1, C21orf2, and CCNF, which are associated with sporadic and familial ALS, have been identified based on population-based registers, genome-wide association studies (GWAS), whole genome studies, and exome sequencing technologies since 2014 [5]. CHCHD10 is a 14-kDa nuclear-encoded, mitochondrial protein localized to the mitochondrial intermembrane space [6]. This protein is important for maintenance of mitochondrial dynamics and cellular bioenergetics [6]. CHCHD10 was first linked to ALS after a study of a large French family, which identified a single heterozygous missense mutation p.S59L in exon 2 of CHCHD10 [7]. CHCHD10 was sequenced in 94 patients, and results revealed the presence of a novel heterozygous missense variant Pro34Ser in 2 unrelated frontotemporal dementia (FTD)-ALS individuals [8]. Consequently, there have been increasing studies to investigate Pro34Ser in other populations with ALS such as Italy [9], UK [10], and the Netherlands [11]. However, these results have not been consistent. We therefore conducted a meta-analysis to further evaluate whether CHCHD10 Pro34Ser variant is a risk factor of ALS because of the inconsistencies in previous studies.
Materials and methods
Literature search and strategy
We searched PubMed, Web of Science, and Embase databases for studies on potential association between CHCHD10 Pro34Ser variant and risk of ALS. The following search terms were used: “Amyotrophic lateral sclerosis” or “ALS” and “Pro34Ser” or “CHCHD10.” No language restrictions were imposed. Our final search date was March 28, 2020.
Selection and exclusion criteria
Eligible studies had to meet the following criteria: (a) use of a case–control study design or a GWAS design to analyze patients with ALS and healthy controls, (b) evaluation of the association between Pro34Ser variant and ALS risk, and (c) report on frequency of a minor allele distribution for both cases and controls or other data necessary for estimating odds radio (OR) at 95% confidence interval (CI). Only large studies were included for studies with overlapping cohorts. Studies that did not report on the frequency of minor allele in Pro34Ser and studies in which it was impossible to calculate minor allele frequency based on data provided by the authors were excluded.
Data extraction
Two authors (BY Yang, CH Yang) independently assessed studies for inclusion or exclusion, and discrepancies were resolved by holding discussions with a third reviewer (ZF Lin). The following data were extracted from each study: name of the first author, publication year, ethnicity of the study cohort, sample size of ALS and control groups, allele distribution in case and controls, OR, and 95% CI.
Statistical analysis
Potential association between CHCHD10 Pro34Ser variant and ALS risk was assessed using Stata 14.0 (StataCorp, College Station, TX, USA). Strength of the association was estimated using OR and 95% CI. OR was assessed using the allele model (Pro34Ser-positive vs Pro34Ser-negative). A P value equal to or less than 0.05 was considered to be statistically significant for all analyses. Cochran’s Q test and I2 statistic were used to assess heterogeneity among studies. Heterogeneity of studies was not considered if I2 was < 25%, low heterogeneity was indicated by 25% ≤ I2 < 50%, moderate heterogeneity was indicated by 50% ≤ I2 < 75%, and I2 ≥ 75% indicated substantial heterogeneity [12]. We applied fixed-effects model to perform meta-analysis of pooled data considered to be homogeneous or of low heterogeneity, and random-effects model to perform meta-analysis of data considered to be of moderate or substantial heterogeneity [13]. Begg’s test was applied to evaluate publication bias [14].
Results
Literature search and included studies
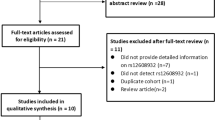
A total of 65 potentially eligible articles were identified after searching PubMed, Web of Science, and Embase databases, and removing duplicates. We eliminated 49 articles based on titles and abstracts and read the remaining 16 articles excluding the following publications: 1 study which did not include ALS population [15], 2 studies which had no control population to compare with ALS patients [16, 17], 2 studies which did not have new data for letter response [18, 19], and 1 study which did not report minor allele frequency within a population of ALS, and it could not be derived from provided data [20].
The remaining 10 publications had a total of 7442 patients with sporadic ALS and 75,371 as controls. The 10 studies included 12 cohorts, which evaluated possible association between Pro34Ser variant and ALS risk (Fig. 1).
Characteristics of included studies
Tables 1 and 2 summarize key characteristics of studies included in the meta-analysis. Studies were conducted in populations from France [8], Italy [9, 21], UK [10, 22], Germany [23], Spain [24], Belgium [25], Australia [26], Netherlands, and other six different populations [11]. One study involving 635 ALS patients reported that individuals carrying Pro34Ser variant of CHCHD10 were at higher risk of developing ALS than individuals without Pro34Ser variant [26]. The rest of the studies revealed no association between Pro34Ser variant and risk of ALS disease [8,9,10,11, 21,22,23,24,25,26].
Heterogeneity
Frequencies of the Pro34Ser allele exhibited heterogeneity based on a chi-squared test analysis (χ2 = 10.72; degrees of freedom, DF = 11, P = 0.467) and I2 value (0%). Therefore, allele data were analyzed using a fixed-effects model.
Meta-analysis
Pooled OR for risk of ALS in the presence of Pro34Ser variant relative to absence of Pro34Ser was 1.23 (95% CI 0.90–1.69; Fig. 2), and the test for overall effect, z = 1.28 (P = 0.201).
Analysis of sensitivity and publication bias
Analysis of sensitivity indicated that none of the independent studies significantly influenced our results (Fig. 3). Our funnel plot was visually symmetrical indicating that there was no significant publication bias (Fig. 4). Begg’s test revealed no significant risk of publication bias (P = 0.064).
Discussion
To our knowledge, this is the first meta-analysis assessing existing data on association of Pro34Ser variant of CHCHD10 with risk of ALS. Results from the present meta-analysis reveal that Pro34Ser variant of CHCHD10 gene is not considerably associated with high risk of developing ALS, particularly among Caucasian participants.
Results of our study are in line with previous findings suggesting that Pro34Ser variant in CHCHD10 is likely to be a benign polymorphism [15, 20, 25]. Dobson-Stone and others demonstrated Pro34Ser variant in a patient with FTD who harbors another established pathogenic mutation and its presence in 9/807 non-demented aged controls, six of whom were verified to be cognitively intact in their 70s–80s [15]. Meanwhile, non-pathogenicity was predicted in six out of eight functional predictive programs (PolyPhen-2, SIFT, FATHMM, MutationAssessor, PONP2, and Sibyl) [15]. The convergence of the above three evidence indicates that Pro34Ser is not a disease-causing mutation. In addition, Zhang and others analyzed 204 ALS and 158 FTD patients and established that the frequency of Pro34Ser variant substitution was comparable with the control samples (minor allele frequency = 0.0074), the 1000 genome project (minor allele frequency = 0.001), and the ExAC database (minor allele frequency = 0.003). This demonstrated that this variant is not pathogenic [20]. Moreover, time of median survival in four patients with Pro34Ser variant was 30 months, which corresponds with time of median survival of 29 months observed in total sporadic ALS cohorts that were tested. This contrasts with the gradual progression of the disease observed in most patients with motor neuron disease carrying a CHCHD10 mutation for which pathogenicity is supported by co-segregation data [23].
CHCHD10 genes are concentrated in exon 2 [9, 25] because this region is GC-rich (77.3% GC-content) and may not be effectively captured by oligonucleotide baits that are central to exome sequencing [22, 27]. Therefore, this variant is apparently absent or has low frequency in public databases and should not be solely relied on to elucidate its pathogenicity [15]. In addition, due to the low coverage of next-generation sequencing, the true frequency of Pro34Ser variation in the control group may indeed be higher, as reflected by Sanger sequencing data [22]. Although our meta-analysis does not confirm that Pro34Ser is a risk factor of ALS, there is need to perform large-scale studies to determine whether Pro34Ser variant is a risk allele for FTD-ALS with low penetrance.
Our meta-analysis provides the first comprehensive assessment of Pro34Ser variant in CHCHD10 and ALS risk. Our results should however be interpreted with caution because our study had several limitations. One of the limitations is that we did not perform a subgroup analysis based on gender and ethnicity due to limited data. Another limitation was that Begg’s test revealed that there was no significant risk in publication bias, although this risk is always present. Finally, included studies were only performed among Caucasian populations; thus, correlation between Pro34Ser variant and ALS risk in other races requires further investigation.
In conclusion, our results suggest that Pro34Ser variant of CHCHD10 is not associated with susceptibility to ALS, particularly in Caucasian populations. Future studies especially among African and Asian populations are required to validate and increase our understanding of the association between Pro34Ser variant and risk of ALS.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Kiernan MC, Vucic S, Cheah BC, Turner MR, Eisen A, Hardiman O, Burrell JR, Zoing MC (2011) Amyotrophic lateral sclerosis. Lancet 377(9769):942–955
Talbott EO, Malek AM, Lacomis D (2016) The epidemiology of amyotrophic lateral sclerosis. Handb Clin Neurol 138:225–238
Oskarsson B, Gendron TF, Staff NP (2018) Amyotrophic lateral sclerosis: an update for 2018. Mayo Clin Proc 93(11):1617–1628
Yang B, Jiang H, Wang F, Li S, Wu C, Bao J, Zhu Y, Xu Z, Liu B, Ren H, Yang X (2019) UNC13A variant rs12608932 is associated with increased risk of amyotrophic lateral sclerosis and reduced patient survival: a meta-analysis. Neurol Sci 40(11):2293–2302
Chia R, Chio A, Traynor BJ (2018) Novel genes associated with amyotrophic lateral sclerosis: diagnostic and clinical implications. Lancet Neurol 17(1):94–102
Genin EC, Plutino M, Bannwarth S, Villa E, Cisneros-Barroso E, Roy M, Ortega-Vila B, Fragaki K, Lespinasse F, Pinero-Martos E, Augé G, Moore D, Burté F, Lacas-Gervais S, Kageyama Y, Itoh K, Yu-Wai-Man P, Sesaki H, Ricci JE, Vives-Bauza C, Paquis-Flucklinger V (2016) CHCHD10 mutations promote loss of mitochondrial cristae junctions with impaired mitochondrial genome maintenance and inhibition of apoptosis. EMBO Mol Med 8(1):58–72
Bannwarth S, Ait-El-Mkadem S, Chaussenot A, Genin EC, Lacas-Gervais S, Fragaki K et al (2014) A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain 137(Pt 8):2329–2345
Chaussenot A, Le Ber I, Ait-El-Mkadem S, Camuzat A, de Septenville A, Bannwarth S et al (2014) Screening of CHCHD10 in a French cohort confirms the involvement of this gene in frontotemporal dementia with amyotrophic lateral sclerosis patients. Neurobiol Aging 35(12):2884.e1–2884.e4
Chio A, Mora G, Sabatelli M, Caponnetto C, Traynor BJ, Johnson JO et al (2015) CHCH10 mutations in an Italian cohort of familial and sporadic amyotrophic lateral sclerosis patients. Neurobiol Aging 36(4):1767.e3–1767.e6
Abdelkarim S, Morgan S, Plagnol V, Lu CH, Adamson G, Howard R, Malaspina A, Orrell R, Sharma N, Sidle K, Clarke J, Fox NC, Rossor MN, Warren JD, Clark CN, Rohrer JD, Fisher EMC, Mead S, Pittman A, Fratta P (2016) CHCHD10 Pro34Ser is not a highly penetrant pathogenic variant for amyotrophic lateral sclerosis and frontotemporal dementia. Brain 139(Pt 2):e9
Project Min EALSSC (2018) CHCHD10 variants in amyotrophic lateral sclerosis: where is the evidence? Ann Neurol 84(1):110–116
Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327(7414):557–560
Ning P, Yang X, Yang B, Zhao Q, Huang H, An R, Chen Y, Hu F, Xu Z, Xu Y (2018) Meta-analysis of the association between ZNF512B polymorphism rs2275294 and risk of amyotrophic lateral sclerosis. Neurol Sci 39(7):1261–1266
Egger M, Smith GD, Phillips AN (1997) Meta-analysis: principles and procedures. BMJ 315(7121):1533–1537
Dobson-Stone C, Shaw AD, Hallupp M, Bartley L, McCann H, Brooks WS, Loy CT, Schofield PR, Mather KA, Kochan NA, Sachdev PS, Halliday GM, Piguet O, Hodges JR, Kwok JBJ (2015) Is CHCHD10 Pro34Ser pathogenic for frontotemporal dementia and amyotrophic lateral sclerosis? Brain 138(Pt 10):e385
Bannwarth S, Ait-El-Mkadem S, Chaussenot A, Genin EC, Lacas-Gervais S, Fragaki K et al (2015) Reply: is CHCHD10 Pro34Ser pathogenic for frontotemporal dementia and amyotrophic lateral sclerosis? Brain 138(Pt 10):e386
Penttila S, Jokela M, Saukkonen AM, Toivanen J, Palmio J, Lahdesmaki J et al (2017) CHCHD10 mutations and motor neuron disease: the distribution in Finnish patients. J Neurol Neurosurg Psychiatry 88(3):272–277
Bannwarth S, Ait-El-Mkadem S, Chaussenot A, Genin EC, Lacas-Gervais S, Fragaki K et al (2014) Reply: are CHCHD10 mutations indeed associated with familial amyotrophic lateral sclerosis? Brain 137(Pt 12):e314
Bannwarth S, Ait-El-Mkadem S, Chaussenot A, Genin EC, Lacas-Gervais S, Fragaki K et al (2015) Reply: a distinct clinical phenotype in a German kindred with motor neuron disease carrying a CHCHD10 mutation. Brain 138(Pt 9):e377
Zhang M, Xi Z, Zinman L, Bruni AC, Maletta RG, Curcio SA et al (2015) Mutation analysis of CHCHD10 in different neurodegenerative diseases. Brain 138(Pt 9):e380
Ronchi D, Riboldi G, Del Bo R, Ticozzi N, Scarlato M, Galimberti D et al (2015) CHCHD10 mutations in Italian patients with sporadic amyotrophic lateral sclerosis. Brain 138(Pt 8):e372
Wong CH, Topp S, Gkazi AS, Troakes C, Miller JW, de Majo M et al (2015) The CHCHD10 P34S variant is not associated with ALS in a UK cohort of familial and sporadic patients. Neurobiol Aging 36(10):2908.e17–2908.e18
Marroquin N, Stranz S, Muller K, Wieland T, Ruf WP, Brockmann SJ et al (2016) Screening for CHCHD10 mutations in a large cohort of sporadic ALS patients: no evidence for pathogenicity of the p.P34S variant. Brain 139(Pt 2):e8
Dols-Icardo O, Nebot I, Gorostidi A, Ortega-Cubero S, Hernandez I, Rojas-Garcia R et al (2015) Analysis of the CHCHD10 gene in patients with frontotemporal dementia and amyotrophic lateral sclerosis from Spain. Brain 138(Pt 12):e400
Perrone F, Nguyen HP, Van Mossevelde S, Moisse M, Sieben A, Santens P et al (2017) Investigating the role of ALS genes CHCHD10 and TUBA4A in Belgian FTD-ALS spectrum patients. Neurobiol Aging 51:177.e9–177e16
McCann EP, Fifita JA, Grima N, Galper J, Mehta P, Freckleton SE et al (2020) Genetic and immunopathological analysis of CHCHD10 in Australian amyotrophic lateral sclerosis and frontotemporal dementia and transgenic TDP-43 mice. J Neurol Neurosurg Psychiatry 91(2):162–171
Johnson JO, Glynn SM, Gibbs JR, Nalls MA, Sabatelli M, Restagno G, Drory VE, Chiò A, Rogaeva E, Traynor BJ (2014) Mutations in the CHCHD10 gene are a common cause of familial amyotrophic lateral sclerosis. Brain 137(Pt 12):e311
Acknowledgements
Writing and revision: Pingping Ning.
Funding
This work was supported by the Sichuan application basic research project (20YYJC0146).
Author information
Authors and Affiliations
Contributions
Conceptualization: Baiyuan Yang, Chenghui Yang, Junwei Ren, and Zhenfang Lin; Methodology: Baiyuan Yang, Chenghui Yang, and Chengqing Zhong; Formal analysis and investigation: Keting Liu, Liusha Zhao, Li Li, Han Wang, and Mingling Zhu; Writing-original draft preparation: Baiyuan Yang, Chenghui Yang, and Junwei Ren; Writing-review and editing: Baiyuan Yang and Zhenfang Lin; Supervision: Zhenfang Lin.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Code availability
The code used and/or analyzed during the current study is available from the corresponding author on reasonable request.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Heading
1. This manuscript provides what we believe to be the first report of the association between CHCHD10 Pro34Ser variant and risk of amyotrophic lateral sclerosis.
2. This manuscript demonstrated that Pro34Ser variant in CHCHD10 is not associated with risk of ALS, particularly in Caucasian participants.
Rights and permissions
About this article
Cite this article
Yang, B., Yang, C., Ren, J. et al. Meta-analysis of the association between CHCHD10 Pro34Ser variant and the risk of amyotrophic lateral sclerosis. Neurol Sci 42, 625–631 (2021). https://doi.org/10.1007/s10072-020-04579-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-020-04579-z