Abstract
In search for sea ice bacteria and their phages from the Baltic Sea ice, two ice samples were collected from land-fast ice in a south-west Finland coastal site in February and March 2011. Bacteria were isolated from the melted sea ice samples and phages were screened from the same samples for 43 purified isolates. Plaque-producing phages were found for 15 bacterial isolates at 3 °C. Ten phage isolates were successfully plaque purified and eight of them were chosen for particle purification to analyze their morphology and structural proteins. Phage 1/32 infecting an isolate affiliated to phylum Bacteroidetes (Flavobacterium sp.) is a siphovirus and six phages infecting isolates affiliated to γ-Proteobacteria (Shewanella sp.) hosts were myoviruses. Cross titrations between the hosts showed that all studied phages are host specific. Phage solutions, host growth and phage infection were tested in different temperatures revealing phage temperature tolerance up to 45 °C, whereas phage infection was in most of the cases retarded above 15 °C. This study is the first to report isolation and cultivation of ice bacteria and cold-active phages from the Baltic Sea ice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Baltic Sea is one of the largest brackish water areas in the world. Influence of freshwater input via rivers and limited water exchange with the North Sea can be seen in lower salinity (approximately 2–6 in the northern sub-basins; Granskog et al. 2006). Annually, a mean of 40 % of Baltic Sea area is covered by sea ice, duration varying from 6 months in the northernmost Gulf of Bothnia to sporadic ice occurrence in the southern Baltic Sea during severe winters (Granskog et al. 2006). Despite of the lower seawater salinity, Baltic Sea ice structure is similar to Polar sea ice with liquid brines concentrated into channels and pockets within the ice matrix (Granskog et al. 2006). The brine channel system and the lower surface of ice are colonized by ice-associated auto- and heterotrophic protists (Rintala et al. 2010; Majaneva et al. 2012) and prokaryotes (Norrman and Andersson 1994; Kaartokallio et al. 2008). Seasonal changes in incoming solar irradiance govern typical succession sequence of ice organism communities with algal bloom in ice in March, followed by heterotrophy-dominated post-bloom period before ice break-up in April or early May (Haecky et al. 1998; Kaartokallio 2004; Rintala et al. 2006).
Heterotrophic bacteria are the most abundant prokaryotes in Baltic Sea ice and likely depend on autochthonous dissolved organic matter (DOM) produced by ice algae (Kaartokallio 2004; Kuparinen et al. 2007; Stedmon et al. 2007) for their growth. According to 16S rDNA-based phylogenetic analyses, the most abundant heterotrophic bacterial groups found from the Baltic Sea ice are α- and γ-Proteobacteria classes and phylum Bacteroidetes (Kaartokallio et al. 2005, 2008). Bacterial community composition in the Baltic Sea ice in south-west (SW) coast of Finland has been shown to display seasonal succession pattern reflecting the duration of ice cover (Kaartokallio et al. 2008). Factors effecting bacterial mortality have not been studied in detail in Baltic Sea ice, but aside from grazing by flagellates and ciliates and in analogy to other aquatic systems, infection and lysis of bacteria by viruses (bacteriophages, phages) is assumed to be a major mortality factor (Proctor and Fuhrman 1990; Weinbauer 2004; Suttle 2005).
Sea ice-associated virus-like particle abundance and dynamics as well as phage–host systems have been occasionally studied in the Arctic (Yager et al. 2001; Boras et al. 2010; Collins and Deming 2011) and the Antarctic Sea ice (Gowing et al. 2004; Paterson and Laybourn-Parry 2012), but in the Baltic Sea there are no studies on sea ice-associated viruses to date. In sea ice, the number of viruses has typically been estimated by counting virus-like particles from melted sea ice samples (Yager et al. 2001; Maranger et al. 1994; Gowing et al. 2004). Maranger et al. (1994) reported one of the highest virus-to-bacteria ratios ever found from environmental samples from the Arctic Sea ice. This has been confirmed in other studies (Yager et al. 2001; Gowing et al. 2004) although much lower amounts have also been reported (Paterson and Laybourn-Parry 2012). There are only few accounts of isolation and cultivation of marine cold-active phages (Borriss et al. 2003; Wells and Deming 2006) i.e., phages that have active infection cycles at temperatures below 4 °C. Wells and Deming (2006) isolated and characterized three different phage–host systems from Arctic sea ice and melt ponds, and (Wells and Deming 2006) one phage–host system from Arctic seawater of the nepheloid layer. All four phages were shown to be either siphoviruses or myoviruses of the head–tail morphology. In both studies, viruses were very host-specific infecting hosts belonging to genera Shewanella, Flavobacterium and Colwellia, which are among the most common groups of bacteria found in the sea ice (Deming 2010).
So far the studies of heterotrophic bacteria in the Baltic Sea ice have been focused on the characterization of changes in biomass, activity and community composition using cultivation independent methods. The aim of this study was to isolate bacteria and their phages from the Baltic Sea ice, and cultivate them under laboratory conditions to gain detailed insights into their identity, infectivity and phage–host interactions. To that end, we characterized six myoviruses and one siphovirus that infected hosts belonging to two major sea ice bacterial groups.
Materials and methods
Sampling
Sea ice samples were collected from coastal land-fast level ice in the Baltic Sea near Tvärminne Zoological Station, University of Helsinki, in Hankoniemi, SW Finland (59°50.611′N, 23°15.098′E) in February and March 2011. First sampling was done in February, 10 weeks after freeze-up. Ice thickness during first sampling was 36 cm and air temperature −15 °C. Second sampling was done in March when ice thickness was 38 cm and air temperature was 2 °C. Ice was cut with handsaw (first sampling) or CRREL-type power auger (second sampling). Ice samples were melted in machine washed polyethylene buckets at 18 °C and immediately after that stored at 3 °C before use.
Growth media and conditions, phage buffer and storage of isolates
For isolation of bacteria three different media were used: basic ZoBell medium (5-g peptone, 1-g yeast extract, 800-ml aged seawater from the Baltic Sea, 200-ml ultrapure water, autoclaved at 121 °C, 20 min (ZoBell 1946; Middelboe et al. 2003), concentrated ZoBell medium (sea water was concentrated to half by boiling before adding nutrients) and low-nutrient ZoBell medium (0.05-g peptone, 0.01-g yeast extract, 800-ml aged seawater, 200-ml ultrapure water). Agar concentrations of 1.5 and 0.6 % were used for plates and top-layer agar, respectively. SM-buffer (50-mM Tris pH 7.5, 100-mM NaCl, 8-mM MgSO4, 0.01 % gelatin; Borriss et al. 2003) was used as the virus buffer. Purified bacteria and phages were stored at −80 °C freezer with 15 % glycerol. Bacteria were grown on a plate not more than 14 days to ensure purity and then cultured in liquid ZoBell medium aerobically at 3 °C.
Isolation of bacteria and phages
Isolation of bacteria was done by spreading 100 μl of melted ice sample on all three different plate types with duplicates and incubated at 3 or 10 °C. Different types of colonies were picked and isolates were purified by three consecutive rounds of colony purification.
For phage isolation, melted ice samples were filtered using 0.22-μm Millipore Durapore Membrane polyvinylidene difluoride (PVDF) filter (EMD Millipore Corporation, Billerica, USA) and concentrated approximately 200× using Amicon Ultra-15 concentration units (MWCO 100000 Da; Merck Millipore, Billerica, USA) by centrifugation (Eppendorf A-4-62, 3220×g, 10 min, 7 °C). Phages were isolated by plating 10, 100 or 200 μl of the filtered and concentrated sample with 100 μl of host suspension and 3 ml of top-layer agar on ZoBell plates. Single plaques were picked and resuspended in 500 μl of ZoBell media. Phages were purified through three similar, consecutive plaque purification steps.
Purification of phages
Plate lysates were prepared using semi-confluent plates. After plaque formation, the top-layer agar of each plate was collected and 2 ml of liquid ZoBell media per each plate was added. This lysate was incubated for 1.5 h at 3 °C in shaker. Cell debris and agar were removed by centrifugation (Sorvall SLA-1500 rotor, 9722×g for 40 min) and the supernatant was stored as the phage lysate. For purification, phages were concentrated by precipitation using 10 % (w/v) polyethylene glycol (PEG; 6000 MW Sigma-Aldrich, Steinheim, Germany; Yamamoto et al. 1970) in the presence of 0.5 M NaCl at 4 °C for 1 h. Phage precipitate was collected (Sorvall SLA-1500 rotor, 12305×g, 30 min), and the pellet was washed with SM-buffer. Precipitated phages were dissolved in the SM-buffer overnight at 4 °C. Viral aggregates were removed using Eppendorf 5415D table top centrifuge (9300×g, 10 min, 4 °C), and the non-aggregated viruses were subjected to linear 5–20 or 10–30 % (in SM-buffer) rate zonal sucrose gradient centrifugation (Sorvall TH 641 rotor, 153208×g, 20–40 min, 10 °C). The light-scattering zones containing the infective phage particles were collected (Sorvall T-865 rotor, 104087×g, 3 h, at 10 °C) and resuspended in SM-buffer.
Protein analyses
Protein concentration of the purified phage samples was measured by Bradford protein assay (Bradford 1976) using bovine serum albumin (BSA) as a standard. Structural proteins of purified phages were analyzed by 16 % sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using approximately 5 μg of phage proteins.
Transmission electron microscopy (TEM)
Morphology of the purified phages was analyzed by transmission electron microscopy (TEM) using negatively stained samples. Phages were allowed to adsorb to grids for 1 min and subsequently stained with 1 % potassium phosphotungstate pH 6.5, 3 % uranyl acetate pH 4.5 or 2 % uranyl acetate pH 4.5 for approximately 20 s. Micrographs were taken with Jeol 1200 EX II transmission electron microscope operating at 80 kV (Electron Microscopy Unit, Institute of Biotechnology, University of Helsinki).
Phage host range
In order to define the host range of the phages, 20 μl of different dilutions of the phage lysates were spotted on top of the host lawns. The results were verified using the plaque assay of suitable phage dilutions determined according to the titer of the phage lysates.
Influence of temperature on the growth of the hosts, infectivity of the phages and infection cycle
Bacterial growth at different temperatures was determined by plating 100 μl of bacterial suspension with 3 ml of top-layer agar on ZoBell plates and incubating them at 4, 15, 23, and 28 °C until a dense growth appeared on plates grown at 4 °C. Growth of the host strain as turbid lawn inside the soft agar was a positive result.
Temperature tolerance of six purified phages was tested by diluting the phage lysate to the concentration of 106 plaque forming units (pfu)/ml and incubating them at 4, 23, and 45 °C for 2 h. The phage suspensions where then further diluted to be able to determine the titer of each phage solution by plaque assay at 4 °C.
In order to determine the effect of temperature on the infection cycle, phage lysates were diluted so that we were able to determine the titer (tens of plaques on the titration plate). Phage dilutions (100 μl) were plated with host suspension (100 μl) and 3 ml of ZoBell soft agar as described above, and two replicates of each phage–host system were grown at 4, 15, 23, and 28 °C.
DNA extraction, amplification of 16S rRNA gene and sequencing
Bacterial cells were collected by centrifugation (Eppendorf 5424, 13000×g, 3 min) in late logarithmic growth phase after which genomic DNA was extracted with PowerSoil® DNA Isolation Kit (MO BIO Laboratories Inc., Carlsbad, CA, USA). Phage DNA was isolated from the purified phage preparation using proteinase K treatment and phenol–chloroform extraction.
The 16S rRNA genes were amplified by PCR using primers pA (5′ AGA GTT TGA TCC TGG CTC AG 3′; Edwards et al. 1989) and pH’ (5′ AAG GAG GTG ATC CAG CCG CA 3′; Edwards et al. 1989). The sequences of the 16S rRNA genes were determined using Sanger sequencing at the Institute of Biotechnology (University of Helsinki, Helsinki, Finland) using primers pD, pD’, pE and pF’ (Edwards et al. 1989). Sequences were assembled and primers removed manually with Gap4 program (version 4.11, Staden package version 2.0.0b9, Wellcome Trust Sanger Institute, Hinxton, UK; Staden et al. 2003). The final sequences of the 16S rRNA genes have been submitted in the NCBI GenBank database under accession numbers KF356204 (strain 4), KF356205 (strain 32), KF356206 (strain 40), KF356207 (strain 41), KF356208 (strain 44), and KF356209 (strain 49).
A Naïve Bayesian Classifier (version 2.5, 12.3.2013, 100 % confidence threshold) and Seqmatch (version 3, 3.3.2013, default settings; Cole et al. 2009) of Ribosomal Database Project database (Release 10, update 31, Michigan State University, USA; http://rdp.cme.msu.edu/; Wang et al. 2007) were used for the taxonomic identification of the host bacteria. Since Seqmatch could identify bacteria only at the genus level, sequences were also used to search for the closest matches using Blastn at the National Centre for Biotechnology Information (NCBI) database. In order to determine the overall similarity between the 16S rRNA gene sequences obtained in this study, they were aligned using the program LALIGN (Myers and Miller 1988).
Results
Isolation and characterization of ice bacteria and their phages
Ice bacteria were isolated from two ice samples as described in “Materials and methods”. Bacterial colonies with different morphology and color were selected from the plates for purification between 10 and 29 days after plating. Altogether 43 different types of colonies were picked, colony purified, and named in numerical order; 15 bacterial strains were found to serve as hosts for phages. Colonies isolated on different media were tested for growth on the basic ZoBell medium at 3 °C, and since all the colonies were able to grow in the tested conditions these were used as standard during the rest of the study.
Sequences of the 16S rRNA genes were determined for those six bacterial strains that served as hosts for further studied phage isolates (Table 1). The 16S rRNA gene sequence of strain 32 was closest to the corresponding sequences of strains belonging to species Flavobacterium gelidilacus LMG 21619 (99 % identity along 1467 nt). All the other host strains were closest to the genus Shewanella strains. At the species level, isolates 40 and 41 were found to be 97 % identical to Shewanella baltica strain BA175 and for strain 49 the closest match in the database was the rRNA gene sequence of Shewanella baltica strain OS195. Strains 44 and 4 were most similar to Shewanella frigidimarina NCIMB 400 (Table 1). By 16S rRNA gene sequence isolates 40 and 41 were identical.
All the 43 purified bacterial strains were used to screen for phages from the same melted sea ice samples by plaque assay at 3 °C. Plaques were formed 3–5 days after plating. Plaque-producing phages were found for 15 bacterial isolates and they were named according to the order they were isolated on a particular host strain. For example, phage 1/4 is the first plaque picked from the lawn of strain 4. Plaques produced by bacteriophage 1/32 were very small (<1 mm) and the other phages produced plaques that were usually hazy with clear centers. Although some of the phages were lost during the plaque purification, twelve different kinds of plaques were obtained for which plate lysates were made. At this step, we were not able to produce infective stock solutions of two of the isolated phages. The plate lysates of the rest of phages retained their infectivity at the same level for several months when stored at 3 °C.
Eight of the phages giving the highest titers were chosen for further characterization through purification. Concentrated phages were purified by linear rate zonal centrifugation to obtain the “1× purified” viral material as described in “Materials and methods”. Infectivity and the yield of the phages in different purification steps were determined and are shown in Table 2. Of the eight purified isolates, seven could be purified to the extent that we were able to determine their morphology by TEM and only the 1/44 preparation was too contaminated with other material to determine its morphology.
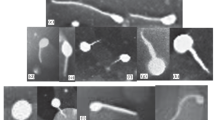
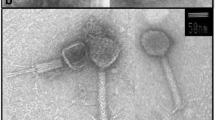
Negative stained samples examined by TEM showed that the bacteriophages were of the head–tail morphotype (Fig. 1). Phage 1/32 had long flexible, non-contractile tail characteristic for phages belonging to the family Siphoviridae. Phages 1/4, 1/40, 1/41, 2/41, 1/49, and 3/49 had rigid, contractile tails characteristic to Myoviridae family of phages. The head of the phages varied between 45 and 70 nm in diameter and the tail length between approximately 75–160 nm.
The genomic DNA of phage 1/4 was extracted and analyzed by restriction enzyme digest using KpnI (Fig. 2). Digest yielded fragments ranging in size from approximately 1.5 kb up to several fragments in the size range of 20 kb.
The structural proteins of purified phages were analyzed in 16 % SDS-PAGE (Fig. 3). In all of the bacteriophages that could be purified to the sufficient extent, one dominant structural protein, approximately 31–38 kDa in size, was seen together with a couple of less dominant and several minor proteins (Fig. 3). Protein patterns of phages 1/41 and 2/41 seemed to be identical and structural protein patterns of phages 1/4 and 1/40 differed slightly. All the other phages had unique patterns of structural proteins. Titrations of purified viruses with different hosts showed that all the studied phages were able to infect only their own host bacteria (data not shown). Since 1/4 and 1/40 could not infect strain 40 or strain 4, respectively, we concluded that these phages are different from each other.
Temperature sensitivity of phage–host systems
The effect of temperature on the phage–host systems was tested individually, i.e., phage suspension alone and bacterial isolate alone as well as during phage infection (Table 3). Influence of the temperature on the infectivity of the phages was tested by incubating phage lysates at 4, 23, and 45 °C and verifying the changes of infectivity by plaque assay at 4 °C. The infectivity of the phage solutions was slightly affected only with phage 1/40 where the infectivity was reduced to one tenth of the original infectivity at 45 °C.
Most of the Shewanella isolates tested alone did not grow in soft agar at temperatures above 23 °C (Table 3), and the growth of strains 40 and 44 was already retarded. The Flavobacterium isolate 32, on the contrary, grew even at 28 °C. The infection cycle (hosts were infected with phage and incubated at various temperatures) was not successful at temperatures above 15 °C except for phage 1/32 which produced plaques even at 23 °C. The infection cycle of 1/44 seemed to be influenced the most by increasing temperature. When infected hosts were grown at different temperatures, the hosts were growing faster and plaques appeared faster at the higher temperature that still allowed infection, i.e., for Shewanella phages at 15 °C and Flavobacterium phage 1/32 at 23 °C (data not shown).
Discussion
Bacteria inhabiting the Baltic Sea ice have been characterized before in terms of the biomass, species composition, and activity of biological processes such as biomass production and nitrogen transformations (Kaartokallio 2001, 2004; Petri and Imhoff 2001; Kaartokallio et al. 2005, 2007, 2008). However, before this study no published results exist of isolated and cultivated bacteria let alone their phages from the Baltic Sea ice.
Six hosts for which the viruses were characterized in this study were by 16S rRNA gene sequence shown to belong to genera Flavobacterium (1 strain) and Shewanella (5 strains). Strains belonging to these genera have also been found before from the Baltic Sea ice samples using cultivation independent methods such as denaturing gradient gel electrophoresis (DGGE) or temperature gradient gel electrophoresis (TGGE; Kaartokallio et al. 2005, 2008). For other sea ice systems it has been shown before that the cultivable fraction of ice bacteria corresponds well the indigenous sea ice bacterial populations determined by cultivation independent methods (Junge et al. 2002; Brinkmeyer et al. 2003).
15 different strains were found to serve as hosts for plaque-producing viruses, but during the plaque purification steps some viruses were lost. Also, for the plaque-purified viruses the viral titers in the plate lysates varied. In general, plate lysates of many isolates reached relatively high titers and infectivity of the plate lysates of different phages was retained the same for several months at 3 °C. Although we did not critically study the reasons for losing some of the phages, it is common that some phages have shorter time span for infectivity and some of these phages may be more sensitive to laboratory conditions used.
Many viruses isolated from an extreme environment are physically adapted well to the specific extreme conditions and major changes in them can completely abolish the infectivity of the virus (Kukkaro and Bamford 2009; Pietilä et al. 2013). In case of phages isolated from ice, temperature may be such a factor. However, in the light of our temperature sensitivity tests the studied phages were physically not very sensitive to high temperatures, since incubation even at 45 °C for 2 h reduced their infectivity maximally to one tenth of the initial infectivity (Table 3), but some of the hosts were not able to grow sufficiently enough at temperatures above 23 °C and successful infection cycle occurred only at 15 °C or below. Psychrophilic organisms are generally defined as those that can grow and reproduce at cold temperatures (at approximately 15 °C or below). For ice bacteria, however, it has been suggested that in addition to the ability to grow at low temperature, the ice bacteria also need to be able to grow at relatively high temperatures demanding flexibility from these organisms (Mock and Thomas 2005). Instead of psychrophile, the term “cold-active” has been used in association of ice bacteria and their phages and has been determined as those phage–host systems where the phage is able to produce plaques at 4 °C or below (Wells and Deming 2006). Since our phages produced plaques at 3 °C they can also be considered as cold-active. However, the plaques appeared faster at higher temperature. This most probably reflects the fact that the optimal temperature for growth of bacteria is usually close to the maximum temperature of growth. Borriss et al. (2007), when discussing genome structure of a cold-active phage, which infects a Bacteroidetes host, found affiliation to phages infecting mesophilic hosts in non-marine environments. Our results on high temperature tolerance of studied phages are concomitant with their results although no further conclusions can be drawn here.
Eight plaque-purified phage isolates reaching the highest titers in the plate lysates were chosen for viral purification and further analyses. Morphologically, all the seven phages of sufficient purity could be determined as typical eubacterial head–tail viruses representing the two most common families of Siphoviridae and Myoviridae. Analysis of the purified phage 1/4 genome using KpnI restriction digest showed that the genome is double-stranded DNA (dsDNA) perhaps more than 50 kb in size (Fig. 2). To date, all the known tailed bacteriophages contain a dsDNA genome. Isolates infecting hosts that belong to Shewanella sp. seemed to be myoviruses with contractile tails and only one phage, the flavovirus 1/32, is a siphovirus with flexible, but non-contractile tail. Although myoviruses are often having a wider host range (Sullivan and Waterbury 2003; Atanasova et al. 2012), all viruses isolated in this study seem to have a narrow host range. Specificity of ice phages has also been reported for the earlier studied ice phages (Borriss et al. 2003; Wells and Deming 2006). At first the high specificity of the phages isolated in this study seemed somewhat surprising, because according to the 16S rRNA gene sequence, the Shewanella sp. strains are very close to each other, if not identical, as is the case for strains 40 and 41. However, factors determining the phage sensitivity or resistance often reside in the non-core areas of the genome and are often dependent on the more variable cell surface structures (Bohannan and Lenski 2000; Avrani et al. 2011). Thus, our phages can reveal differences between the Shewanella sp. host strains in properties other than 16S rRNA gene sequence.
Analysis of the structural proteins suggests that 1/41 and 2/41 are highly similar if not identical phages. The structural protein patterns of 1/4 and 1/40 are also very similar with differences in the migration of two minor structural proteins (Fig. 3). As mentioned above, these two phage isolates infect only their own hosts that are not the same on the basis of cross-titration results. Thus, these phages are most probably closely related, but not identical. In general, further studies isolating new phages from the Baltic Sea ice will be required to critically assess the issue of phage diversity in this environment.
The seven new phages were isolated and purified in this study were more than double the number of cultivated cold-active phages from marine environments to date. They also represent the first phage isolates from the Baltic Sea ice. The four previously reported cold-active viruses have been isolated from Arctic sea water from melt ponds, the surface nepheloid layer or the sea ice itself (Borriss et al. 2003; Wells and Deming 2006). Currently, genomic sequences of only two ice phages have been published (Borriss et al. 2007; Colangelo-Lillis and Deming 2013).
We found viruses for bacteria belonging to the two major bacterial groups in sea ice, class γ-Proteobacteria and phylum Bacteroidetes. This implies a possibility for viral infection bearing a role in bacterial mortality in the Baltic Sea ice. Since the isolated hosts are different from each other and their phages host specific, it seems likely that the host–phage interactions in sea ice are very specific, implying minor effects at the community level due to single host–phage interaction dynamics. In order to be able to dissect the impact of the dynamics of host–phage interactions on the community level or further sea ice biogeochemical processes, much more work on the Baltic Sea ice phage populations is needed.
Abbreviations
- BSA:
-
Bovine serum albumin
- DGGE:
-
Denaturing gradient gel electrophoresis
- DOM:
-
Dissolved organic matter
- dsDNA:
-
Double-stranded DNA
- PEG:
-
Polyethylene glycol
- Pfu:
-
Plaque forming units
- PVDF:
-
Polyvinylidene difluoride
- SW:
-
South-west
- TEM:
-
Transmission electron microscopy
- TGGE:
-
Temperature gradient gel electrophoresis
References
Atanasova NS, Roine E, Oren A, Bamford DH, Oksanen HM (2012) Global network of specific virus–host interactions in hypersaline environments. Environ Microbiol 14(2):426–440. doi:10.1111/j.1462-2920.2011.02603.x
Avrani S, Wurtzel O, Sharon I, Sorek R, Lindell D (2011) Genomic island variability facilitates Prochlorococcus–virus coexistence. Nature 474(7353):604–608. doi:10.1038/Nature10172
Bohannan BJM, Lenski RE (2000) Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol Lett 3(4):362–377. doi:10.1046/j.1461-0248.2000.00161.x
Boras JA, Sala MM, Arrieta JM, Sa EL, Felipe J, Agusti S, Duarte CM, Vaque D (2010) Effect of ice melting on bacterial carbon fluxes channelled by viruses and protists in the Arctic Ocean. Polar Biol 33(12):1695–1707. doi:10.1007/s00300-010-0798-8
Borriss M, Helmke E, Hanschke R, Schweder T (2003) Isolation and characterization of marine psychrophilic phage–host systems from Arctic sea ice. Extremophiles 7(5):377–384. doi:10.1007/s00792-003-0334-7
Borriss M, Lombardot T, Glockner FO, Becher D, Albrecht D, Schweder T (2007) Genome and proteome characterization of the psychrophilic Flavobacterium bacteriophage 11b. Extremophiles 11(1):95–104. doi:10.1007/s00792-006-0014-5
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254 S0003269776699996[pii]
Brinkmeyer R, Knittel K, Jürgens J, Weyland H, Amann R, Helmke E (2003) Diversity and structure of bacterial communities in Arctic versus Antarctic pack ice. Appl Environ Microbiol 69(11):6610–6619
Colangelo-Lillis JR, Deming JW (2013) Genomic analysis of cold-active Colwelliaphage 9A and psychrophilic phage–host interactions. Extremophiles 17(1):99–114. doi:10.1007/s00792-012-0497-1
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi:10.1093/Nar/Gkn879
Collins RE, Deming JW (2011) Abundant dissolved genetic material in Arctic sea ice part II: viral dynamics during autumn freeze-up. Polar Biol 34(12):1831–1841. doi:10.1007/s00300-011-1008-z
Deming JW (2010) Sea ice bacteria and viruses. In: Thomas DN, Dieckmann GS (eds) Sea ice. Wiley-Blackwell, New York, pp 247–282
Edwards U, Rogall T, Blöcker H, Emde M, Böttger EC (1989) Isolation and direct complete nucleotide determination of entire genes—characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res 17(19):7843–7853. doi:10.1093/nar/17.19.7843
Gowing MM, Garrison DL, Gibson AH, Krupp JM, Jeffries MO, Fritsen CH (2004) Bacterial and viral abundance in Ross Sea summer pack ice communities. Mar Ecol Prog Ser 279:3–12. doi:10.3354/Meps279003
Granskog M, Kaartokallio H, Kuosa H, Thomas DN, Vainio J (2006) Sea ice in the Baltic Sea—a review. Estuar Coast Shelf S 70(1–2):145–160. doi:10.1016/j.ecss.2006.06.001
Haecky P, Jonsson S, Andersson A (1998) Influence of sea ice on the composition of the spring phytoplankton bloom in the northern Baltic Sea. Polar Biol 20(1):1–8. doi:10.1007/s003000050270
Junge K, Imhoff F, Staley T, Deming JW (2002) Phylogenetic diversity of numerically important arctic sea-ice bacteria cultured at subzero temperature. Microb Ecol 43(3):315–328. doi:10.1007/s00248-001-1026-4
Kaartokallio H (2001) Evidence for active microbial nitrogen transformations in sea ice (Gulf of Bothnia, Baltic Sea) in midwinter. Polar Biol 24(1):21–28. doi:10.1007/s003000000169
Kaartokallio H (2004) Food web components, and physical and chemical properties of Baltic Sea ice. Mar Ecol Prog Ser 273:49–63. doi:10.3354/Meps273049
Kaartokallio H, Laamanen M, Sivonen K (2005) Responses of Baltic Sea ice and open-water natural bacterial communities to salinity change. Appl Environ Microb 71(8):4364–4371. doi:10.1128/Aem.71.8.4364-4371.2005
Kaartokallio H, Kuosa H, Thomas DN, Granskog MA, Kivi K (2007) Biomass, composition and activity of organism assemblages along a salinity gradient in sea ice subjected to river discharge in the Baltic Sea. Polar Biol 30(2):183–197. doi:10.1007/s00300-006-0172-z
Kaartokallio H, Tuomainen J, Kuosa H, Kuparinen J, Martikainen PJ, Servomaa K (2008) Succession of sea-ice bacterial communities in the Baltic Sea fast ice. Polar Biol 31(7):783–793. doi:10.1007/s00300-008-0416-1
Kukkaro P, Bamford DH (2009) Virus–host interactions in environments with a wide range of ionic strengths. Environ Microbiol Rep 1(1):71–77. doi:10.1111/j.1758-2229.2008.00007.x
Kuparinen J, Kuosa H, Andersson A, Autio R, Granskog MA, Ikävalko J, Kaartokallio H, Karell K, Leskinen E, Piiparinen J, Rintala JM, Tuomainen J (2007) Role of sea-ice biota in nutrient and organic material cycles in the northern Baltic Sea. Ambio 36(2–3):149–154. doi:10.1579/0044-7447(2007)36[149:Rosbin]2.0.Co;2
Majaneva M, Rintala JM, Piisilä M, Fewer DP, Blomster J (2012) Comparison of wintertime eukaryotic community from sea ice and open water in the Baltic Sea, based on sequencing of the 18S rRNA gene. Polar Biol 35(6):875–889. doi:10.1007/s00300-011-1132-9
Maranger R, Bird DF, Juniper SK (1994) Viral and bacterial dynamics in Arctic sea ice during the spring algal bloom near resolute, N.W.T., Canada. Mar Ecol Prog Ser 111(1–2):121–127. doi:10.3354/Meps111121
Middelboe M, Riemann L, Steward GF, Hansen V, Nybroe O (2003) Virus-induced transfer of organic carbon between marine bacteria in a model community. Aquat Microb Ecol 33(1):1–10. doi:10.3354/Ame033001
Mock T, Thomas DN (2005) Recent advances in sea-ice microbiology. Environ Microbiol 7(5):605–619. doi:10.1111/j.1462-2920.2005.00781.x
Myers EW, Miller W (1988) Optimal alignments in linear space. Comput Appl Biosci 4(1):11–17. doi:10.1093/bioinformatics/4.1.11
Norrman B, Andersson A (1994) Development of ice biota in a temperate sea area (Gulf of Bothnia). Polar Biol 14(8):531–537
Paterson H, Laybourn-Parry J (2012) Antarctic sea ice viral dynamics over an annual cycle. Polar Biol 35(4):491–497. doi:10.1007/s00300-011-1093-z
Petri R, Imhoff JF (2001) Genetic analysis of sea-ice bacterial communities of the Western Baltic Sea using an improved double gradient method. Polar Biol 24(4):252–257. doi:10.1007/s003000000205
Pietilä MK, Laurinmäki P, Russell DA, Ko CC, Jacobs-Sera D, Butcher SJ, Bamford DH, Hendrix RW (2013) Insights into head-tailed viruses infecting extremely halophilic archaea. J Virol 87(6):3248–3260. doi:10.1128/JVI.03397-12
Proctor LM, Fuhrman JA (1990) Viral mortality of marine bacteria and cyanobacteria. Nature 343:60–62
Rintala JM, Piiparinen J, Ehn J, Autio R, Kuosa H (2006) Changes in phytoplankton biomass and nutrient quantities in sea ice as responses to light/dark manipulations during different phases of the Baltic winter 2003. Hydrobiologia 554:11–24. doi:10.1007/s10750-005-1002-y
Rintala JM, Piiparinen J, Uusikivi J (2010) Drift-ice and under-ice water communities in the Gulf of Bothnia (Baltic Sea). Polar Biol 33(2):179–191. doi:10.1007/s00300-009-0695-1
Staden R, Judge DP, Bonfield JK (2003) Managing sequencing projects in the GAP4 environment. In: Womble DD, Krawetz SA (eds) Introduction to bioinformatics: a theoretical and practical approach. Humana Press, New York
Stedmon CA, Thomas DN, Granskog M, Kaartokallio H, Papadimitriou S, Kuosa H (2007) Characteristics of dissolved organic matter in Baltic coastal sea ice: allochthonous or autochthonous origins? Environ Sci Technol 41(21):7273–7279. doi:10.1021/Es071210f
Sullivan MB, Waterbury JB, Chisholm SW (2003) Cyanophages infecting the oceanic cyanobacterium Prochlorococcus (vol 424, pg 1047. Nature 426(6966):584. doi:10.1038/Nature02147
Suttle CA (2005) Viruses in the sea. Nature 437(7057):356–361. doi:10.1038/nature04160
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naϊve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73(16):5261–5267. doi:10.1128/AEM.00062-07
Weinbauer MG (2004) Ecology of prokaryotic viruses. FEMS Microbiol Rev 28(2):127–181. doi:10.1016/j.femsre.2003.08.001
Wells LE, Deming JW (2006) Characterization of a cold-active bacteriophage on two psychrophilic marine hosts. Aquat Microb Ecol 45(1):15–29. doi:10.3354/Ame045015
Yager PL, Connelly TL, Mortazavi B, Wommack KE, Bano N, Bauer JE, Opsahl S, Hollibaugh JT (2001) Dynamic bacterial and viral response to an algal bloom at subzero temperatures. Limnol Oceanogr 46(4):790–801
Yamamoto KR, Alberts BM, Benzinge R, Lawhorne L, Treiber G (1970) Rapid bacteriophage sedimentation in presence of polyethylene glycol and its application to large-scale virus purification. Virology 40(3):734. doi:10.1016/0042-6822(70)90218-7
ZoBell CE (1946) Marine microbiology. A monograph on hydrobacteriology. Chronica Botanica Company, Waltham
Acknowledgments
We thank Sari Korhonen for technical assistance. This study was funded by Onni Talas foundation and Walter and Andrée de Nottbeck foundation (AML, JMR) and University of Helsinki three-year grant 2010–2012 (ER). All the field work was carried out at Tvärminne Zoological Station, University of Helsinki whilst the office and laboratory facilities for the isolation work were provided by The Finnish Environmental Institute (SYKE), Marine Research Centre. We thank Academy of Finland (grant 271413) and University of Helsinki for the support to EU ESFRI Instruct Centre for Virus Production and Purification used in this study. This study is dedicated to the memory of Prof. Kielo Haahtela.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Huang.
Rights and permissions
About this article
Cite this article
Luhtanen, AM., Eronen-Rasimus, E., Kaartokallio, H. et al. Isolation and characterization of phage–host systems from the Baltic Sea ice. Extremophiles 18, 121–130 (2014). https://doi.org/10.1007/s00792-013-0604-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-013-0604-y