Abstract
The multiple factors contributing to the pathogenesis of osteoporosis include genetic and environmental factors. Because decrease in bone mineral density (BMD) is the major clinical indicator and a useful quantitative trait, many association and linkage studies of BMD have been conducted. Although the series of studies showed apparently significant associations, the genes have not been found that can be utilized in clinical practice. Several genes identified in robust genome-wide association studies will be the new cutting edge in genetic studies of osteoporosis. Our recent reports of functional single nucleotide polymorphism in the tissue-nonspecific alkaline phosphatase gene and gamma-carboxylase gene are presented in this review to discuss the future prospects in the genetic research of osteoporosis from the point of view of genome–nutrition interaction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
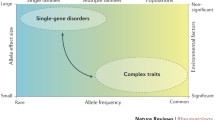
Osteoporosis brings about deterioration in activities of daily living (ADL) and quality of life (QOL) of the affected patients. Although fragility fractures resulting from osteoporosis continue to increase in the current aging society, it is assumed that this disease is still undertreated [1]. In the 1990s, osteoporosis was defined as a disease characterized by low bone mass and microarchitectural deterioration of bone tissue, leading to enhanced bone fragility and a consequent increase in fracture risk [2]. This definition reflects the importance of bone mass and microarchitecture in determining bone strength. Because there have not been practical measures of microarchitecture, bone mass or bone mineral density (BMD) has been used as a quantitative trait in searching the genes for osteoporosis. It seems reasonable that a vast series of association and linkage studies have been conducted with BMD, but one should keep in mind that BMD is one of the complex traits of osteoporosis and one of the surrogate markers for bone fragility.
Recently, osteoporosis was redefined as a skeletal disorder characterized by compromised bone strength predisposing a person to an increased risk of fracture [3]. In the previous definition of osteoporosis, low BMD was not considered as a sole factor of osteoporosis, but the new definition declares more clearly that bone strength is determined not only by BMD but also by factors other than BMD [3]. According to the new definition, the genes of osteoporosis should be a group of genes contributing to the multiple aspects of pathogenesis. Although case–control studies by defining the case with the diagnostic criteria of this disease are suggested, the diagnosis of osteoporosis might not be suitable as a “phenotype” in genetic studies because the diagnosis contains biologically heterogeneous components. In this review, genetic aspects of osteoporosis are discussed mainly using BMD as one of the measurable phenotypes of osteoporosis.
Genetic aspects of bone phenotypes
Predisposing factors of osteoporosis include both lifestyle factors and genetic factors. The first step in preventing osteoporosis should be the reduction of lifestyle-related risk factors. On the other hand, the genetic factors cannot be removed even when these are identified. However, it will be useful if one can learn that she or he has the genetically predisposing factor(s) and is thus motivated to avoid certain lifestyle risk factors.
Family history of fractures is included among the established risk factors for osteoporotic fractures [4], indicating the importance of the genetic background of osteoporosis. Twin studies also supported the heritability of BMD [5], which is the most valuable indicator of bone strength. On the other hand, it was reported that the possibility that the genetic determinants of BMD and those of fractures might be different [6]. Factors contributing to the variation in bone quality will be studied further from genetic aspects. For example, femoral neck cross-sectional geometry was successfully used as a clinical measure in quantitative trait locus analyses [7].
Approaches for the pathogenesis of diseases can be classified as deductive or inductive. Analyses about the roles of known substances or genes would be classified into deductive approaches and belong to the genetic approach. On the other hand, the recent availability of whole genome information has made the inductive approach possible, which is named a genome-wide association study (GWAS). In this mini-review, the recent genetic and genomic approaches for osteoporosis are reviewed, and our studies on the functional single-nucleotide polymorphisms (SNPs) related to osteoporosis are introduced.
Candidate gene approaches to the determinants of bone mineral density
Until now, BMD has been utilized most widely as a quantitative measure in genetic and genomic studies for osteoporosis. Quite a few association studies with BMD have been done with so-called candidate gene approaches [8]. Candidate genes have been chosen based on basic bone cell biology and clinical observations. In addition, the genome-wide linkage and association studies will show novel series of candidate genes that should be investigated further.
Association studies with the polymorphisms of these genes were done using genetic polymorphisms. Among the polymorphisms, SNPs were most commonly utilized. SNPs in the regulatory region (rSNPs) and those in coding regions (cSNPs) could be related to quantitative or qualitative variations of the gene expressions. In addition, other SNPs, for example, those in introns, could affect the gene expression or could be markers for genomic study. Microsatellite polymorphisms (e.g., dinucleotide repeat or triple repeat) are other kinds of polymorphisms that have also been utilized in osteoporosis research.
Association studies with candidate gene polymorphisms have been published by many groups including ours [9–40]. If you search the database using the key words “gene polymorphisms and bone mineral density,” 1,000 and more articles will be hit. The genes analyzed are classified into nuclear receptors and related molecules, collagen and other matrix proteins, receptor activator of nuclear factor-kappa B ligand (RANKL)/RANK system, cytokines and related molecules, hormones and related molecules, enzymes, cell cycle-related molecules, lipoprotein receptor-related peptides (LRPs) and Wnt signals, cell-surface molecules, transcription factors, and others (Table 1). However, the contribution of most genes to determining BMD is small and the result is not always reproducible [41, 42]. Lifestyle-related factors as confounding factors against genetic factors should be managed in the association studies. In addition, ethnic factors have to be considered appropriately [43].
The vitamin D receptor gene has been studied most extensively, but the implications of vitamin D receptor gene polymorphisms have not been established [44]. Recent extensive meta-analyses [45] showed that the effects of the vitamin D receptor gene polymorphisms seem modest, although the significant effects of the polymorphisms on BMD and osteoporotic fractures were proved.
Searches for functional SNPs affecting variation in bone metabolism
When polymorphisms of genes were significantly and reproducibly associated with bone phenotypes, biological relevancy should be confirmed, and the methods of clinical application should be considered following that process. In other words, it would be a rational method in the genetic approach for osteoporosis to examine the association of functional polymorphisms with bone phenotypes. Although the contribution of each polymorphism to BMD would be small, the significant effects of each polymorphism supported by functional studies will be a clue suggesting that the gene should play important roles in the pathogenesis of osteoporosis.
We reported two functional SNPs in two genes that are related to the variation in BMD of the elderly. The first one was an SNP in the tissue-nonspecific alkaline phosphatase (TNSALP) gene [46]. TNSALP resides in the plasma membrane of osteoblasts and supplies phosphate to the calcification site. We searched for nonsynonymous and functional SNPs in the exons of this gene. As a result, an SNP in exon 7 (787C > T), which replaces tyrosine at codon 246 to histidine, gives the biochemical differences between the products of each genotype. The K m value of 787 His was smaller than that of 778 Tyr, which means that persons with 787 His may supply phosphate to the calcification site more efficiently. Elderly Japanese women with 787 His had higher radial BMD than those with other genotypes. This study demonstrated the importance of phosphate metabolism in bone metabolism in the elderly. Additional in vitro experiments supported the biochemical variations resulting from this polymorphism [47, 48]. Further studies are underway to examine the clinical meaning of this variation, for example, the effects of this genotype on the relationship between phosphate intake and hormones in calcium metabolism and aging.
Another gene is vitamin K-dependent gamma-glutamyl carboxylase (GGCX) [49]. GGCX carboxylates vitamin K-dependent proteins including bone Gla protein (osteocalcin) and matrix Gla protein. Functional polymorphisms in the GGCX gene, if any, might explain the variation in bone metabolism and BMD. Also in this case, polymorphisms in the exons were screened in Japanese elderly women and a nonsynonymous SNPs was found: about 8762 G > A (Arg325Gln). When the kinetic parameters of GGCX325-Gln and GGCX325-Arg were compared in vitro, V max/K m was significantly higher for GGCX325-Gln than for GGCX325-Arg. Association study of this polymorphism with radial BMD of Japanese postmenopausal women showed that the body mass index (BMI)-adjusted Z score in the subpopulation older than 75 years was higher in those with 325 Gln than those with 325 Arg/Gln or 325 Arg. In this study, we first reported the different activities of GGCX between the common genotypes and their association with BMD. Vitamin K deficiency is known as a nutritional risk factor for osteoporotic fractures, and a regimen of vitamin K2 is utilized for osteoporosis treatment. The common allelic variation in the GGCX gene may explain the individual variation in the response to nutritional and/or pharmacological intervention with vitamin K. It would be rational to utilize the allele information in finding the level of vitamin K intake at which the effects of the genotype with lower enzymatic activity can be avoided. We have already reported that this GGCX gene polymorphism affects the correlation between the vitamin K status and gamma-carboxylation of osteocalcin in young males [50], and this kind of study is awaited in the group of elderly.
Implication from monogenic bone diseases
There are rare diseases involving bone that are caused by mutations of single genes and considered to be monogenic diseases. The causative genes of these diseases were identified by linkage analyses of the affected families. These genes would have important implications for the variations of bone phenotype also in the general population. A distinguished example is the gene for osteoporosis-pseudoglioma syndrome (OPPG) [51]. Positional cloning with the affected pedigrees showed that rare mutations in lipoprotein receptor-related peptide 5 (LRP5) gene cause the disease. In addition, another mutation in the same gene was demonstrated to cause a syndrome with high BMD [52]. It is also interesting that the LRP5 gene resides in the locus that has been among the loci related to BMD in the linkage studies [53]. Several groups including ours examined the relationship between the polymorphisms of LRP5 gene and BMD, and the results were reproducible [54–56]. In addition, the LRP5 gene was screened out by the recent GWAS, as mentioned below [57]. These results strongly suggest that variations in this gene would contribute to the variation of BMD in the general population.
Genes suggested by genome-wide association studies
Systematic search for the genes for osteoporosis has been done by genome-wide linkage studies with pedigrees, which have shown some hotspots linked to BMD, for example, those on chromosome 11 [53]. Further fine mappings were required to specify the genes contributing to the pathophysiology of osteoporosis and consequent analyses of their functions in bone biology. Recent advances in analyzing SNPs distributing to the whole genome area made it possible to conduct a GWAS (Table 2). One of the GWAS studies identified two SNPs, rs4355801 on chromosome 8 and rs3736228 on chromosome 11 [57]. The former is close to the osteoprotegerin gene and the latter nonsynonymous SNP is in the LRP5 gene, both of which are major components in bone biology. In the series of candidate gene approach, the significant correlation between the polymorphisms in LRP5 gene has been reproducible. The identification of the LRP5 gene in the GWAS study further strengthens the importance of this gene in the pathogenesis of osteoporosis. Osteoprotegerin was also identified to be correlated with BMD in another GWAS study [58].
Other examples of genes identified in GWAS studies are RANKL [58], estrogen receptor 1 (ESR1) [58], ADAM metallopeptidase with thrombospondin type 1 motif, 18 (ADAMTS18) [59], and transforming growth factor-beta receptor III (TGFBR3) [59].
Prevention of osteoporotic fractures is the major clinical goal of osteoporosis therapy, and the incidence of osteoporotic fractures should be an ideal phenotype used in the genetic studies searching the genes for osteoporosis. Recently, Kung et al. [60] reported the association of the JAG1 gene with osteoporotic fractures as well as BMD with GWAS study. They also demonstrated the possible molecular mechanism with which the genetic variation of this gene affects bone metabolism [60].
Discussion
Selection of candidate genes for polymorphism studies of osteoporosis is rather arbitrary. This situation cannot be avoided because we do not know how many genes are involved in the pathogenesis of osteoporosis or in the determination of BMD. Recent genome-wide studies with a large population size are successfully overcoming this issue, and several genes were identified for osteoporosis. These genes include the novel series of candidate genes whose implications should be studies. So far the new list of genes contains “previous” candidate genes that are well known in the field of bone biology.
BMD is a surrogate marker for bone fragility, and one should not consider the genes for low BMD as immediately being those for osteoporosis. Although BMD is still a useful quantitative measure in genetic studies for osteoporosis, other phenotypes, particularly the incidence of fractures, should be kept in mind. Further studies are required to utilize the products of genetic studies for the advancement of osteoporosis practice.
References
Kroth PJ, Murray MD, McDonald CJ (2004) Undertreatment of osteoporosis in women, based on detection of vertebral compression fractures on chest radiography. Am J Geriatr Pharmacother 2:112–118
Consensus Development Center (1993) Consensus Development Conference V, 1993. Diagnosis, prophylaxis and treatment of osteoporosis. Am J Med 94:646–650
NIH Consensus Development Panel (2001) NIH consensus development panel on osteoporosis prevention, diagnosis, and therapy. JAMA 285:785–793
Kanis JA, Johnell O, De Laet C, Jonsson B, Oden A, Ogelsby AK (2002) International variation in hip fracture probabilities: implication for risk assessment. J Bone Miner Res 17:1237–1244
Pocock NA, Eisman JA, Hopper JL, Yeates MG, Sambrook PN, Eberl S (1987) Genetic determinants of bone mass in adults. A twin study. J Clin Invest 80:706–710
Andrew T, Antioniades L, Scurrah KJ, Macgregor AJ, Spector TD (2005) Risk of wrist fracture in women is heritable and is influenced by genes that are largely independent of those influencing BMD. J Bone Miner Res 20:67–74
Xiong DH, Shen H, Xiao P, Gua YF, Long JR, Zhao LJ, Liu YZ, Deng HY, Li JL, Recker RR, Deng HW (2006) Genome-wide scan identified QTLs underlying femoral neck cross-sectional geometry that are novel studied risk factors of osteoporosis. J Bone Miner Res 21:424–437
Liu YJ, Shen H, Xiao P, Xiong DH, Li LH, Recker RR, Deng HW (2006) Molecular genetics of gene identification for osteoporosis: a 2004 update. J Bone Miner Res 21:1511–1535
Sano M, Inoue S, Hosoi T, Ouchi Y, Emi M, Shiraki M, Orimo H (1995) Association of estrogen receptor dinucleotide repeat polymorphism with osteoporosis. Biochem Biophys Res Commun 217:378–383
Kobayashi S, Inoue S, Hosoi T, Ouchi Y, Shiraki M, Orimo H (1996) Association of bone mineral density with polymorphism of the estrogen receptor gene. J Bone Miner Res 11:306–311
Shiraki M, Shiraki Y, Aoki C, Hosoi T, Inoue S, Kaneki M, Ouchi Y (1997) Association of bone mineral density with apolipoprotein E phenotype. J Bone Miner Res 1:1438–1445
Mizunuma H, Hosoi T, Okano H, Soda M, Tokizawa T, Kagami I, Miyamoto S, Ibuki I, Inoue S, Shiraki M, Ouchi Y (1997) Estrogen receptor gene polymorphism and bone mineral density at the lumbar spine of pre- and postmenopausal women. Bone (NY) 21:379–383
Hosoi T, Miyao M, Inoue S, Hoshino S, Shiraki M, Orimo H, Ouchi Y (1999) Association study of parathyroid hormone gene polymorphism and bone mineral density in Japanese postmenopausal women. Calcif Tissue Int 64:205–208
Miyao M, Hosoi T, Inoue S, Hoshino S, Shiraki M, Orimo H, Ouchi Y (1998) Polymorphism of insulin-like growth factor I gene and bone mineral density. Calcif Tissue Int 63:306–311
Ota N, Hunt SC, Nakajima T, Suzuki T, Hosoi T, Orimo H, Shirai Y, Emi M (1999) Linkage of interleukin 6 locus to human osteopenia by sibling pair analysis. Hum Genet 105:253–257
Ogawa S, Urano T, Hosoi T, Miyao M, Hoshino S, Fujita M, Shiraki M, Orimo H, Ouschi Y, Inoue S (1999) Association of bone mineral density with a polymorphism of the peroxisome proliferator-activated receptor gamma gene: PPAR gamma expression in osteoblasts. Biochem Biophys Res Commun 260:122–126
Nakajima T, Ota N, Shirai Y, Hata A, Yoshida H, Suzuki T, Hosoi T, Orimo H, Emi M (1999) Ethnic difference in contribution of Sp1 site variation of COL1A1 gene in genetic predisposition to osteoporosis. Calcif Tissue Int 65:352–353
Tsukamoto K, Orimo H, Hosoi T, Miyao M, Yoshida G, Watanabe S, Suzuki T, Emi M (2000) Association of bone mineral density with polymorphism of the human matrix Gla protein locus in elderly women. J Bone Miner Metab 18:27–30
Tsukamoto K, Orimo H, Hosoi T, Miyao M, Ota N, Nakajima T, Yoshida H, Watanabe S, Suzuki T, Emi M (2000) Association of bone mineral density with polymorphism of the human calcium-sensing receptor locus. Calcif Tissue Int 66:181–183
Urano T, Hosoi T, Shiraki M, Toyoshima H, Ouchi Y, Inoue S (2000) Possible involvement of the p57 (Kip2) gene in bone metabolism. Biochem Biophys Res Commun 269:422–426
Yamada Y, Harada A, Hosoi T, Miyauchi A, Ikeda K, Ohta H, Shiraki M (2000) Association of transforming growth factor beta1 genotype with therapeutic response to active vitamin D for postmenopausal osteoporosis. J Bone Miner Res 15:415–420
Ogawa S, Hosoi T, Shiraki M, Orimo H, Emi M, Muramatsu M, Ouchi Y, Inoue S (2000) Association of estrogen receptor beta gene polymorphism with bone mineral density. Biochem Biophys Res Commun 269:537–541
Miyao M, Morita H, Hosoi T, Kurihara H, Inoue S, Hoshino S, Shiraki M, Yazaki Y, Ouchi Y (2000) Association of methylenetetrahydrofolate reductase (MTHFR) polymorphism with bone mineral density in postmenopausal Japanese women. Calcif Tissue Int 66:190–194
Miyao M, Hosoi T, Emi M, Nakajima T, Inoue S, Hoshino S, Shiraki M, Orimo H, Ouchi Y (2000) Association of bone mineral density with a dinucleotide repeat polymorphism at the calcitonin (CT) locus. J Hum Genet 45:346–350
Ota N, Hunt SC, Nakajima T, Suzuki T, Hosoi T, Orimo H, Shirai Y, Emi M (2000) Linkage of human tumor necrosis factor-alpha to human osteoporosis by sib pair analysis. Genes Immun 1:260–264
Ota N, Nakajima T, Nakazawa I, Suzuki T, Hosoi T, Orimo H, Inoue S, Shirai Y, Emi M (2001) A nucleotide variant in the promoter region of the interleukin-6 gene associated with decreased bone mineral density. J Hum Genet 46:267–272
Ogata N, Shiraki M, Hosoi T, Koshizuka Y, Nakamura K, Kawaguchi H (2001) A polymorphic variant at the Werner helicase (WRN) gene is associated with bone density, but not spondylosis, in postmenopausal women. J Bone Miner Metab 19:296–301
Ogata N, Matsumura Y, Shiraki M, Kawano K, Koshizuka Y, Hosoi T, Nakamura K, Kuro-O M, Kawaguchi H (2002) Association of klotho gene polymorphism with bone density and spondylosis of the lumbar spine in postmenopausal women. Bone (NY) 31:37–42
Ohmori H, Makita Y, Funamizu M, Hiooka K, Hosoi T, Orimo H, Suzuki T, Ikari K, Nakajima T, Inoue I, Hata A (2002) Linkage and association analyses of the osteoprotegerin gene locus with human osteoporosis. J Hum Genet 47:400–406
Kawano K, Ogata N, Chiano M, Molloy H, Kleyn P, Spector TD, Uchida M, Hosoi T, Suzuki T, Orimo H, Inoue S, Nabeshima Y, Nakamura K, Kuro-o M, Kawaguchi H (2002) Klotho gene polymorphisms associated with bone density of aged postmenopausal women. J Bone Miner Res 17:1744–1751
Ota N, Nakajima T, Ezura Y, Iwasaki H, Suzuki T, Hosoi T, Orimo H, Inoue S, Ito H, Emi M (2002) Association of a single nucleotide variant in the human tumor necrosis factor-alpha promoter region with decreased bone mineral density. Ann Hum Biol 29:550–558
Ezura Y, Nakajima T, Kajita M, Ishida R, Inoue S, Yoshida H, Suzuki T, Shiraki M, Hosoi T, Orimo H, Emi M (2003) Association of molecular variants, haplotypes, and linkage disequilibrium within the human vitamin D-binding protein (DBP) gene with postmenopausal bone mineral density. J Bone Miner Res 18:1642–1649
Feng D, Ishibashi H, Yamamoto S, Hosoi T, Orimo H, Machida T, Koshihara Y (2003) Association between bone loss and promoter polymorphism in the Japanese women with hip fracture. J Bone Miner Metab 21:225–228
Omasu F, Ezura Y, Kajita M, Ishida R, Kodaira M, Yoshida H, Suzuki T, Hosoi T, Inoue S, Shiraki M, Orimo H, Emi M (2003) Association of genetic variation of the RIL gene, encoding a PDZ-LIM domain protein and localized in 5q31.1, with low bone mineral density in adult Japanese women. J Hum Genet 48:342–345
Ezura Y, Kajita M, Ishida R, Yoshida S, Yoshida H, Suzuki T, Hosoi T, Inoue S, Shiraki M, Orimo H, Emi M (2004) Association of multiple nucleotide variations in the pituitary glutaminyl cyclase gene (QPCT) with low radial BMD in adult women. J Bone Miner Res 19:1296–1301
Ishida R, Ezura Y, Emi M, Kajita M, Yoshida H, Suzuki T, Hosoi T, Inoue S, Shiraki M, Ito H, Orimo H (2003) Association of a promoter haplotype (-1542G/-525C) in the tumor necrosis factor receptor associated factor-interacting protein gene with low bone mineral density in Japanese women. Bone (NY) 33:237–241
Urano T, Shiraki M, Fujita M, Hosoi T, Orimo H, Ouchi Y, Inoue S (2005) Association of a single nucleotide polymorphism in the lipoxygenase ALOX15 5’-flanking region (-5229G/A) with bone mineral density. J Bone Miner Metab 23:226–230
Sudo Y, Ezura Y, Kajita M, Yoshida H, Suzuki T, Hosoi T, Inoue S, Shirak M, Ito H, Emi M (2005) Association of single nucleotide polymorphisms in the promoter region of the pro-opiomelanocortin gene (POMC) with low bone mineral density in adult women. J Hum Genet 50:235–240
Mori S, Kou I, Sato H, Emi M, Ito H, Hosoi T, Ikegawa S (2008) Association of genetic variations of genes encoding thrombospondin, type 1, domain-containing 4 and 7A with low bone mineral density in Japanese women with osteoporosis. J Hum Genet 53:694–697
Mori S, Kou I, Sato H, Emi M, Ito H, Hosoi T, Ikegawa S (2009) Nucleotide variations in genes encoding carbonic anhydrase 8 and 10 associated with femoral bone mineral density in Japanese females with osteoporosis. J Bone Miner Metab 27:213–216
Shen H, Liu YJ, Liu PY, Recker RR, Deng HW (2005) Non-replication in genetic studies of complex diseases: lessons learned from studies of osteoporosis and tentative remedies. J Bone Miner Res 20:365–376
Ionnidis JP, Trikalinos TA, Ntzani EE, Contopoulos-Ionnidis DG (2003) Genetic association in large versus small studies: an empirical assessment. Lancet 361:567–571
Deng HW (2001) Population admixture may appear to mask, change or reverse genetic effects of genes underlying complex traits. Genetics 159:1319–1323
Thakkinstian S, D’Este C, Eisman J, Nguen T, Attia J (2004) Meta-analysis of molecular association studies: vitamin D receptor gene polymorphisms and BMD as a case study. J Bone Miner Res 19:419–426
Uitterlinden AG, Ralston SH, Brandi ML, Carey AH, Grinberg D, EPOS Investigators; EPOLOS Investigators; FAMOS Investigators; LASA Investigators; Rotterdam Study Investigators; GENOMOS Study et al (2006) The association between common vitamin D receptor gene variations and osteoporosis: a participant-level meta-analysis. Ann Intern Med 145:255–264
Goseki-Sone M, Sogabe N, Fukushi-Irie M, Mizoi L, Orimo H, Suzuki T, Nakamura H, Orimo H, Hosoi T (2005) Functional analysis of the single-nucleotide polymorphism (787T>C) in tissue-nonspecific alkaline phosphatase gene associated with bone mineral density. J Bone Miner Res 20:773–778
Sogabe N, Oda K, Nakamura H, Orimo H, Watanabe H, Hosoi T, Goseki-Sone M (2008) Molecular effects of the tissue-nonspecific alkaline phosphatase gene polymorphism (787T>C) associated with bone mineral density. Biomed Res 29:213–219
Orimo H, Goseki-Sone M, Hosoi T, Shimada T (2008) Functional analysis of the mutant tissue-nonspecific alkaline phosphatase gene using U2OS osteoblast-like cells. Mol Genet Metab 94:375–381
Kinoshita H, Nakagawa K, Narusawa K, Goseki-Sone M, Fukushi-Irie M, Mizoi L, Yoshida H, Okano T, Nakamura T, Suzuki T, Inoue S, Orimo H, Ouchi Y, Hosoi T (2007) A functional single nucleotide polymorphism in the vitamin K-dependent gamma-glutamyl carboxylase gene (Arg325Glu) is associated with bone mineral density in elderly Japanese women. Bone (NY) 40:451–456
Sogabe N, Tsugawa N, Maruyama R, Kamao M, Kinoshita H, Okano T, Hosoi T, Goseki-Sone M (2007) Nutritional effects of gamma-glutamyl carboxylase gene polymorphism on the correlation between the vitamin K status and gamma-carboxylation of osteocalcin in young males. J Nutr Sci Vitaminol (Tokyo) 53:419–425
Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Osteoporosis-Pseudoglioma Syndrome Collaborative Group et al (2001) LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell 107:513–523
Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, Wu D, Insogna K, Lifton RP (2002) High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med 346:1513–1521
Koller DL, Econs MJ, Morin PA, Christian JC, Hui SL, Parry P, Curran ME, Rodriguez LA, Conneally PM, Joslyn G, Peacock M, Johnston CC, Foroud T (2000) Genome screen for QTLs contributing to normal variation in bone mineral density. J Clin Endocrinol Metab 85:3116–3120
Urano T, Shiraki M, Ezura Y, Fujita M, Sekine E, Hoshino S, Hosoi T, Orimo H, Emi M, Ouchi Y, Inoue S (2004) Association of a single-nucleotide polymorphism in low-density lipoprotein receptor-related protein 5 gene with bone mineral density. J Bone Miner Metab 22:341–345
Audrey Koay M, Brown MA (2005) Genetic disorders of the LRP5–Wnt signalling pathway affecting the skeleton. Trends Mol Med 11:129–137
Ezura Y, Nakajima T, Urano T, Sudo Y, Kajita M, Yoshida H, Suzuki T, Hosoi T, Inoue S, Shiraki M, Emi M (2007) Association of a single-nucleotide variation (A1330V) in the low-density lipoprotein receptor protein 5 gene (LRP5) with bone mineral density in adult Japanese women. Bone (NY) 40:997–1005
Xiong DH, Liu XG, Guo YF, Tan LJ, Wang L et al (2009) Genome-wide association study and follow-up replication studies identified ADAMTS18 an TGFBR3 as bone mass candidate genes in different ethnic groups. Am J Hum Genet 84:388–398
Richards JB, Rivadeneira F, Inouye M, Pastinen TM, Soranzo N et al (2008) Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet 371:1505–1512
Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, Jonsdottir T, Saemundsdottir J, Center JR, Nguyen TV, Bagger Y, Gulcher JR, Eisman JA, Christiansen C, Sigurdsson G, Kong A, Thorsteinsdottir U, Stefansson K (2008) Multiple genetic loci for bone mineral density and fractures. N Engl J Med 358:2355–2365
Kung AW, Xiao SM, Chemy S, Li GH, Gao Y et al (2010) Association of JAG1 with bone mineral density and osteoporotic fractures: a genome-wide association study and follow-up replication studies. Am J Hum Genet 86:229–239
Author information
Authors and Affiliations
Corresponding author
Additional information
T. Hosoi is a recipient of the 2002 JSBMR Distinguished Scientist Award.
About this article
Cite this article
Hosoi, T. Genetic aspects of osteoporosis. J Bone Miner Metab 28, 601–607 (2010). https://doi.org/10.1007/s00774-010-0217-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00774-010-0217-9