Abstract
Osteoporosis is a multi-factorial common disease, which is caused by combination of genetic as well as environmental factors. Among several factors, osteoclast acidification pathways during bone resorption might play an important role. Carbonic anhydrases, consisting of a gene family, are essential for pH regulation by the osteoclast. Clinically, use of carbonic anhydrase inhibitors has been known to be associated with a bone-sparing effect as judged by spine bone mineral density (BMD). Here, we investigated single nucleotide polymorphisms (SNPs) in carbonic anhydrase genes that are expressed in bone tissues, i.e., CA8 and CA10, for possible association with femoral and lumbar BMD among 337 Japanese women with osteoporosis participated in BioBank Japan project. Significant correlation was observed between CA8 SNP, rs6984526, and femoral BMD (P = 0.00029); homozygous carriers of the major (C) allele (n = 166) had the highest BMD (0.754 ± 0.006 g/cm2, mean ± SD), while heterozygous carriers (n = 135) were intermediate (0.741 ± 0.07 g/cm2) and homozygous T-allele carriers (n = 31) had the lowest BMD (0.691 ± 0.012 g/cm2). CA8 SNP as well displayed significant association with lumbar BMD in recessive model (P = 0.00017). In addition, CA10 SNP, rs2106329, also displayed strong association with femoral BMD (P = 0.00002). The results suggest that the variations of CA8 and CA10 loci may be important determinants of osteoporosis in Japanese women.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteoporosis, one of the most prevalent disease condition in the elderly, is defined as a skeletal disorder characterized by compromised bone strength predisposing a person to an increased risk of fracture [1]. The diagnosis of this disease is done with low bone mineral density (BMD) and the presence of fragility fractures [2]. Like many other common diseases, multiple factors including genetic variations determine predisposition for onset or progression of osteoporosis, as has been indicated by genetic-epidemiological studies [3, 4]. Numerous studies on genetic risks for osteoporosis have been investigated to date, mainly by association studies and linkage analysis for BMD as a quantitative trait [5].
Carbonic anhydrases form a family of enzymes that catalyze the rapid conversion of carbon dioxide to bicarbonate and protons. It has been reported that continuous subcutaneous infusion of a carbonic anhydrase inhibitor acetazolamide partially prevented denervation-induced bone loss in a rat model of disuse osteoporosis [6]. In fact, it was further confirmed in postmenopausal white female with glaucoma that long-term use of carbonic anhydrase inhibitors, acetazolamide or methazolamide, was associated with a bone-sparing effect as judged by spinal BMD [7].
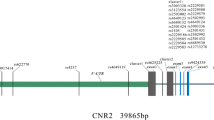
These reports suggested a role for the carbonic anhydrases in human bone resorption and, thus, prompted us to undertake genetic association study between single nucleotide polymorphisms (SNPs) on carbonic anhydrase genes. We focused on carbonic anhydrase 8 and 10 genes (CA8 and CA10) that are expressed in bone tissues. We chose representative intronic SNPs on each gene, i.e., rs6984526 (in CA8) and rs2106329 (in CA10) with minor allele frequencies above 0.1 in Japanese, and, examined correlation with femoral BMD as well as lumbar BMD among 337 Japanese women with osteoporosis participated in BioBank Japan project.
Materials and methods
Subjects
Female osteoporosis case subjects were recruited from the BioBank Japan project according to the Japanese diagnostic criteria for primary osteoporosis [2]. We selected 337 osteoporosis subjects that had measurements of both lumbar spine and femoral neck BMD. No participant had medical complications or was undergoing treatment for conditions known to affect bone and lipid metabolism, such as pituitary diseases, hyperthyroidism, primary hyperparathyroidism, liver disease, renal failure, adrenal diseases or rheumatic diseases, and none was receiving estrogen replacement therapy. DNA samples were obtained from peripheral blood and extracted using a standard procedure. We obtained written informed consent from all study subjects, and procedures were approved by the ethics committees of the Institute of Medical Science of the University of Tokyo and other participating clinical institutions.
Measurement of BMD
Lumbar spine BMD (anteroposterior; L2–L4) and femoral neck BMD were measured by dual-energy X-ray absorptiometry (DXA; Lunar DPX-L, Madison, Lunar, WI, USA) and expressed in g/cm2. The precision of DPX-L was 0.7% in CV. To calculate adjusted BMD [8], the measured BMD was normalized for differences in age and body mass index (BMI) by multiple regression analysis, using the Instat 3 software package (GraphPad Software, San Diego, USA). The adjustment equation for the study samples was as follows:
Genotyping procedure
Genotypes of SNPs, rs6984526 (in CA8) and rs2106329 (in CA10) were determined using Invader assay [9] according to the manufacturer’s instructions. Details of each SNP were archived in the dbSNP at http://www.ncbi.nlm.nih.gov/SNP. SNPs in the CA8 and CA10 genes with low minor allele frequency (<10%) in Japanese in HapMap database were excluded from the analysis for statistical power calculation.
Statistical analysis
Quantitative association between genotypes and adjusted BMD values (g/cm2) was analyzed via one-way analysis of variance (ANOVA) with regression analysis as post hoc test. Three genotypic categories of each SNP were converted into incremental values, 0, 1, and 2 corresponding to the number of chromosomes possessing minor allele nucleotide. Significant association was defined when the given P-value of the ANOVA F-test was less than 5% (P < 0.05) [10]. To ascertain the Hardy–Weinberg equilibrium among genotypes of the subjects, chi-square test was used (P > 0.05).
Results
Mean values and standard deviations (SD) of the 337 Japanese osteoporosis women were 72.7 ± 7.3 years for age, 149.2 ± 5.3 cm for body height, 47.5 ± 7.5 kg for weight, 21.3 ± 3.3 for BMI, 0.869 ± 0.157 g/cm2 for lumbar BMD and 0.743 ± 0.081 g/cm2 for femoral BMD.
We focused on genetic variations in CA8 and CA10 genes, both intronic SNPs, that displayed expression in bone tissue. SNPs with high minor allele frequencies (>10%) in Japanese population, rs6984526 (in CA8) and rs2106329 (in CA10), were genotyped among 337 Japanese osteoporosis women who had measurements of both femoral and lumbar BMD in BioBank Japan project. When the subjects were genotypically categorized into three groups for each SNP, no deviation of genotype frequencies from Hardy–Weinberg equilibrium was detected in the measured SNPs.
ANOVA with linear regression analysis detected a significant correlation between the genotypes of CA8 SNP, rs6984526, and femoral BMD (P = 0.00029); homozygous carriers of the major (C) allele (n = 166) had the highest BMD (0.754 ± 0.006 g/cm2, mean ± SD), while heterozygous carriers (n = 135) were intermediate (0.741 ± 0.07 g/cm2) and homozygous T-allele carriers (n = 31) had the lowest BMD (0.691 ± 0.012 g/cm2); Fig. 1a). When the subjects were separated into two groups, the T homozygote (n = 31) and those who bear at least one C-allele (C/T and C/C), the former subjects had significantly lower femoral BMD than the latter (BMD; 0.691 ± 0.012 g/cm2 versus 0.748 ± 0.005 g/cm2, P = 0.00017; Fig. 1b).
Femoral BMD and lumbar BMD among genotypic groups of CA8 SNP, rs6984526. Values are expressed as means ± SD. a Femoral BMD among three genotypic group of CA8 SNP, rs6984526. b Femoral BMD in recessive model for CA8 SNP, rs6984526. c Lumbar BMD among three genotypic group of CA8 SNP, rs6984526. d Lumbar BMD in recessive model for CA8 SNP, rs6984526
Genetic variation at CA8 SNP, rs6984526, also displayed association with lumbar BMD (P = 0.021); homozygous carriers of the major (C) allele (n = 166) had the highest BMD (0.877 ± 0.012 g/cm2, mean ± SD), while heterozygous carriers (n = 135) were intermediate (0.872 ± 0.014 g/cm2) and homozygous T-allele carriers (n = 31) had the lowest BMD (0.794 ± 0.016 g/cm2; Fig. 1c). When the subjects were separated into two groups, the T homozygote (n = 31) and those who bear at least one C-allele (C/T and C/C), the former subjects had significantly lower lumbar BMD than the latter (0.794 ± 0.016 g/cm2 versus 0.875 ± 0.009 g/cm2, P = 0.0057; Fig. 1d).
With respect to another candidate gene, CA10, an intronic SNP rs2106329, displayed strong association with femoral BMD (P = 0.00002). In this case, the homozygous A-allele carriers (n = 96) had the highest BMD (0.774 ± 0.008 g/cm2), while heterozygous carriers (n = 152) (0.728 ± 0.007 g/cm2) and homozygous G-allele carriers (n = 88) had low BMD (0.734 ± 0.008 g/cm2; Fig. 2a). When the subjects were separated into two groups, the A homozygote (n = 96) and those who posses at least one G-allele (A/G and G/G), the former subjects had significantly higher femoral BMD than the latter (BMD; 0.774 ± 0.008 g/cm2 versus 0.731 ± 0.005 g/cm2, P < 0.00001; Fig. 2b). Correlation between lumbar BMD and CA10 SNP, rs2106329, was insignificant.
These results suggest that genetic variations of CA8 and CA10 loci are related to pathogenesis of osteoporosis. We hypothesized that T-allele homozygote for CA8 SNP, rs6984526, is an important risk factor for osteoporosis, whereas A-allele homozygote for CA10, SNP rs2106329, is a protective factor for osteoporosis in adult Japanese women.
Discussion
In the work reported here, for the first time, we have detected a significant association between genotypes of SNPs on CA8 and CA10 genes with adjusted levels of femoral as well as lumbar BMD among osteoporosis patients of adult Japanese women.
The carbonic anhydrase gene family includes multiple members, which are major players in many physiological processes, including renal and male reproductive tract acidification, bone resorption, respiration, gluconeogenesis, signal transduction, and formation of gastric acid. Carbonic anhydrase isozymes have different kinetic properties, and they are present in various tissues and in various cell compartments [11].
Osteoclasts resorb bone by attaching to the surface and then secreting protons into an extracellular compartment formed between osteoclast and bone surface. This secretion is necessary for bone mineral solubilization and the digestion of organic bone matrix by acid proteases. Carbonic anhydrases are thought to be involved in this process by catalyzing the rapid conversion of carbon dioxide to bicarbonate and protons while maintaining normal intracellular pH in the osteoclast [12]. Carbonic anhydrase inhibitor use was associated with a bone-sparing effect as judged by spinal BMD [7], further supporting a proposed role for carbonic anhydrases in human bone resorption. The CA8 and CA10 genes have been thought to play a role in the central nervous system, especially in brain development [13]. The present study suggests an additional role of the CA8 and CA10 genes in the process of bone resorption; however, their role in bone metabolism is yet to be clarified in further study.
A limitation of this study would be the way of subject recruitment. The subjects studied here were all diagnosed as osteoporosis, since the BioBank Japan project only collected case samples with 47 different diseases and; therefore, no control samples were available from the bank. Further studies are required to extrapolate the results to the general population.
In summary, we showed significant association of the intronic variants in the CA8 and CA10 genes with femoral as well as lumbar BMD of osteoporosis population of adult Japanese women. A possibility cannot be ruled out, however, that these SNP markers may themselves be in linkage disequilibrium with other unmeasured and functional variants at or near the CA 8 or CA10 genes that are the true mechanistic basis for the associations. Functional studies will be required to rule out these possibilities. Longitudinal studies in large cohorts, as well as functional studies, may clarify the molecular events by which the CA8 and CA10 variations bring about alterations in bone metabolism.
References
NIH Consensus Development Panel on Osteoporosis Prevention, Diagnosis, Therapy (2001) JAMA 285:785–795
Orimo H, Hayashi Y, Fukunaga M, Sone T, Fujiwara M, Shiraki M, Kushida K, Miyamoto S, Soen S, Nishimura J, Oh-hashi Y, Hosoi T, Gorai I, Tanaka H, Igai T, Kishimoto H (2001) Diagnostic criteria for primary osteoporosis: year 2000 revision. J Bone Miner Metab 19:331–337
Peacock M, Turner CH, Econs MJ, Foroud T (2002) Genetics of osteoporosis. Endocr Rev 23:303–326
Albagha OM, Ralston SH (2003) Genetic determinants of susceptibility to osteoporosis. Endocrinol Metab Clin North Am 32:65–81
Liu YZ, Liu YJ, Robert RR, Deng HW (2003) Molecular studies of identification of genes for osteoporosis: the 2002 update. Endocrinology 177:147–196
Kenny AD (1985) Role of carbonic anhydrase in bone: plasma acetazolamide concentrations associated with inhibition of bone loss. Pharmacology 31:97–107
Pierce WM Jr, Nardin GF, Fuqua MF, Sabah-Maren E, Stern SH (1991) Effect of chronic carbonic anhydrase inhibitor therapy on bone mineral density in white women. J Bone Miner Res 6:347–354
Tsukamoto K, Orimo H, Hosoi T, Miyao M, Yoshida H, Watanabe S, Suzuki T, Emi M (2000) Association of bone mineral density with polymorphism of the human matrix Gla protein locus in elderly women. J Bone Miner Metab 28:27–30
Mein CA, Barratt BJ, Dunn MG, Siegmund T, Smith AN, Esposito L, Nutland S, Stevens HE, Wilson AJ, Phillips MS, Jarvis N, Law S, de Arruda M, Todd JA (2000) Evaluation of single nucleotide polymorphism typing with invader on PCR amplicons and its automation. Genome Res 10:330–343
Ota N, Nakajima T, Nakazawa I, Suzuki T, Hosoi T, Orimo H, Inoue S, Shirai Y, Emi M (2001) A nucleotide variant in the promoter region of the interleukin-6 gene associated with decreased bone mineral density. J Hum Genet 46:267–272
Breton S (2001) The cellular physiology of carbonic anhydrases. JOP 2:159–164
Rousselle AV, Heymann D (2002) Osteoclastic acidification pathways during bone resorption. Bone 30:533–540
Taniuchi K, Nishimori I, Takeuchi T, Fujikawa-Adachi K, Ohtsuki Y, Onishi S (2002) Developmental expression of carbonic anhydrase-related proteins VIII, X, and XI in the human brain. Neuroscience 112:93–99
Acknowledgments
We appreciate all the volunteers and participating institutions of BioBank Japan, University of Tokyo, Institute of Medical Science, Japan, for precious clinical data and samples. We also thank Prof. Yusuke Nakamura and Dr. Michiaki Kubo in RIKEN SNP Research Center, Tokyo, Japan, for their generous support in accomplishment of this work. The BioBank Japan Project on the implementation of personalized medicine was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT). This work was supported by a grant from BioBank Japan (No 03023120).
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Mori, S., Kou, I., Sato, H. et al. Nucleotide variations in genes encoding carbonic anhydrase 8 and 10 associated with femoral bone mineral density in Japanese female with osteoporosis. J Bone Miner Metab 27, 213–216 (2009). https://doi.org/10.1007/s00774-008-0031-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00774-008-0031-9