Abstract
Southern rice black-streaked dwarf virus (SRBSDV), an insect- and plant-infecting reovirus of the genus Fijivirus, induced the formation of virus-containing tubules in infected plant and insect vector cells. Expression of the nonstructural protein P7-1 of SRBSDV in insect cells by a recombinant baculovirus resulted in the formation of tubules with dimensions and appearance similar to those found in SRBSDV-infected cells. These tubules protruded from the cell surface, supporting the hypothesis that the P7-1 protein contains two putative transmembrane domains that are necessary for the formation of tubules. Furthermore, the self-interaction of SRBSDV P7-1 protein indicates that this protein has the capacity to form homodimers or oligomers to assemble the proposed helical symmetry structure of tubules. Taken together, our results indicate that SRBSDV P7-1 has the intrinsic ability of self-interaction to form tubules growing from the cell surface in the absence of other viral proteins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant-infecting reoviruses are found in the genera Phytoreovirus, Fijivirus and Oryzavirus of the family Reoviridae [1]. They are transmitted by leafhopper or planthopper vectors in a persistent-propagative manner. The formation of tubules with viral particles inside is a common feature of plant and insect cells infected with plant-infecting reoviruses [2, 3]. Rice black-streaked dwarf virus (RBSDV) is a fijivirus that replicates both in plants and in an invertebrate insect vector [1]. The viral genome consists of 10 segmented, double-stranded RNAs (dsRNAs), designated S1 through S10, in the order they appear in the electrophoretic migration pattern on SDS-polyacrylamide gels. The nonstructural protein P7-1 of RBSDV is localized to virus-containing tubules [4], suggesting that RBSDV P7-1 is one of the constituents of the tubules. Recently, a tentative species in the genus Fijivirus, named Southern rice black-streaked dwarf virus (SRBSDV), has been proposed by two groups [5, 6]. In the past several years, SRBSDV has spread rapidly throughout southern China and northern Vietnam and has caused severe damage to rice in some regions [7]. SRBSDV is transmitted by white-backed planthoppers, Sogatella furcifera (Hemiptera: Delphacidae), in a persistent-propagative manner. SRBSDV is most closely related to RBSDV, since both viruses have about 81% amino acid identity in P7-1, which is encoded by genomic segment S7 [5–7]. Furthermore, SRBSDV infection also induces the formation of tubules in rice plants and insect vector cells [5; Fig. 1A]. However, the significance and origin of the tubules remain unknown. In the present study, we found that P7-1 of SRBSDV has the intrinsic ability to form tubules growing from the cell surface in the absence of other viral proteins.
Materials and methods
Cloning of the P7-1 gene and plasmid construction
Total RNAs were extracted from rice tissues infected with the isolate of SRBSDV from Fujian Province, China, using an RNeasy Plant Mini Kit (QIAGEN) following manufacturer’s instructions. Reverse transcription was then performed using Superscript III reverse transcriptase (Invitrogen) and the appropriate reverse primers. The primer sequences used for cloning and mutagenesis in this study are listed in Supplemental Table 1. The reverse primer for SRBSDV P7-1 was fused in frame with the sequence of Strep-tag II. Gene sequences were amplified by PCR using Phusion DNA polymerase (NEB). The resulting cDNA fragments were purified and transferred by recombination into the entry vector pDONR221 (Invitrogen) using BP clonase II (Invitrogen), following the manufacturer’s protocol. The insert of the resulting pDONR clone was verified by sequencing. The pDONR221 vector containing P7-1-Strep-tag II was transferred by recombination into the Gateway baculovirus vector pDEST™8 to generate plasmid pDEST-P7-1-Strep-tag II. The pDONR221 vector containing SRBSDV P7-1 was recombined into binary destination vector pEarleygate101 [8, 9] to generate plasmid P7-1-yellow fluorescent protein (P7-1-YFP). The vectors used in bimolecular fluorescence complementation (BiFC) assay were described previously [10]. The pDONR221 vector containing SRBSDV P7-1 was recombined into BiFC vectors YN and YC to generate plasmids P7-1-YN and P7-1-YC. The yeast two-hybrid system used in this study was obtained from the DUALmembrane system (Dualsystems Biotech) and is suitable for identification of the interactions of transmembrane proteins. The fragment containing SRBSDV P7-1 was amplified by PCR and inserted into the DUALmembrane system bait vector pBT3-STE and the prey vector pPR3-N using the Sfi I restriction site to make the constructs SRBSDV-STE-P7-1 and SRBSDV-N-P7-1, respectively. The insert of the resulting DUALmembrane system clone was verified by sequencing.
Protein secondary structures were predicted based on their primary amino acid sequences using the CUBIC PredictProtein server (http://cubic.bioc.columbia.edu/pp/). Transmembrane helices (TM) in P7-1 were predicted using the programs PHDhtm [11], Tmpred [12] and TMHMM [13]. The P7-1 mutants in which the putative TM1 and TM2 were deleted individually were amplified by the fusion PCR protocol, as described previously [14, 15]. The PCR primer sequences and fusion PCR protocol are provided in Supplementary Table 1 and Fig. S1. The P7-1 mutant with the C-terminal 10 residues truncated was generated by PCR with the primers described in Supplementary Table 1. The resulting P7-1 fragments were recombined into plasmid pDONR221, and then into the Gateway baculovirus vector pDEST™8 to generate plasmids ΔTM1-Strep-tag II, ΔTM2-Strep-tag II and 1-348-Strep-tag II.
Expression of the P7-1 protein of SRBSDV by baculovirus
The recombinant baculovirus vectors P7-1-Strep-tag II, ΔTM1-Strep-tag II, ΔTM2-Strep-tag II and 1-348-Strep-tag II were introduced into DH10Bac (Invitrogen) for transposition into the bacmid. The recombinant bacmid was isolated and used to transfect Spodoptera frugiperda (Sf9) cells in the presence of CellFECTIN™ (Invitrogen). For cytological observations, Sf9 cells on coverslips were infected with the recombinant baculoviruses. The cells were further incubated for 72 h at 27°C before being processed for immunofluorescence or electron microscopy.
Purification of tubules
The purification of tubules from baculovirus-infected Sf9 cells was performed basically as described previously [16]. Briefly, 72 hours post-inoculation (p.i.), Sf9 cells were harvested, washed with phosphate-buffered saline (PBS), and resuspended in STE buffer (150 mM NaCl, 1 mM EDTA, 10 mM Tris-HCl [pH 7.5]) containing 0.5% Triton X-100. Cells were disrupted by homogenization, and the nuclei and cell debris were pelleted by centrifuging for 5 min at 1,500 g. The supernatant was loaded over a cushion of 3 ml of 40% sucrose in STE buffer and centrifuged for 2 h at 200,000 g. The resulting pellet was resuspended in a small volume of STE buffer, loaded onto a 10% to 40% gradient of sucrose in STE buffer, and then centrifuged for 1 h at 141,000 g. The pellet and 1.5 ml of each fraction of the gradient were collected, stained with 2% uranyl acetate, and then observed under an H-7650 electron microscope (Hitachi).
Immunofluorescence microscopy
At different times after inoculation, Sf9 cells on coverslips were fixed for 30 min at room temperature in 2% paraformaldehyde (Sigma) diluted in water. Cells were then washed with PBS and permeabilized in PBS containing 0.1% Triton X-100. Then, the cells were incubated with a monoclonal anti-Strep-tag II antibody (IBA), diluted in PBS containing 0.5% bovine serum albumin (BSA), for 60 min at 37°C. After three washes in PBS, fluorescein isothiocyanate (FITC) anti-mouse IgG (Sigma) diluted in PBS containing 0.5% BSA was added and incubated for 60 min at 37°C. Cells were visualized using a LEICA TCS SP5 fluorescence microscope. Mock-infected cells were included in each experiment as a negative control and were processed in the same manner as the infected cells.
Immunoelectron microscopy
For electron microscopy, Sf9 cells on coverslips or salivary glands of viruliferous white-backed planthoppers were fixed, dehydrated, and embedded as described previously [17]. For immunoelectron electron microscopy, sections on the grid were probed with monoclonal anti-Strep-tag II antibody followed by a 15-nm protein A-gold conjugate (Sigma) as described previously [17]. All samples were observed under an electron microscope (H-7650; Hitachi).
Transient expression in Nicotiana benthamiana
Four-week-old N. benthamiana plants grown in a greenhouse at 22°C to 24°C were used for Agrobacterium tumefaciens (strain GV3101)-mediated transient expression of P7-1-YN, P7-1-YC and P7-1-YFP fusion proteins, as described previously [18]. The relevant binary vectors for expressing P7-1-YN, P7-1-YC and P7-1-YFP fusion proteins were introduced into GV3101 by electroporation and infiltrated into leaf tissues using a 1-ml syringe by gentle pressure through the stomata on the lower epidermal surface. For leaf infiltration, agrobacteria harboring the relevant binary vectors were grown overnight in LB plus the appropriate antibiotics, collected by low-speed centrifugation, and then resuspended in 1 ml of 10 mM MgCl2 containing 100 μM acetosyringone.
Yeast two-hybrid assay
Yeast transformation was performed using the lithium-acetate-based protocol for preparing and transforming yeast competent cells following the instructions of the DUALmembrane starter kit user manual (Dualsystems Biotech). Briefly, yeast strain NMY51 was co-transformed with the bait vector SRBSDV-STE-P7-1 and the prey vector SRBSDV-N-P7-1. Simultaneously, NMY51 was co-transformed with the plasmid pTSU2-APP and pNubG-Fe65 or pPR3-N as a positive and a negative control, respectively. All transformants were grown on SD-trp-leu-his-ade plates for 3-4 days at 30°C. In addition, authentic transformants were confirmed by their color in the HTX-galactosidase assay, as described in the DUALmebrane starter kit user manual.
Results
P7-1 induces tubule formation in vivo
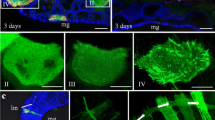
Ultrathin sections of salivary glands of viruliferous white-backed planthoppers showed tubules of about 85 nm in diameter with viral particles inside in the cytoplasm of infected cells (Fig. 1A). To investigate whether P7-1 has an inherent ability to form these tubules, Sf9 cells were infected with a recombinant baculovirus expressing P7-1-Strep-tag II and incubated for different periods of time. By immunoblot analysis, P7-1-Strep-tag II was first detected at 24 h p.i., and its expression reached maximum level at 72 h p.i. (data not shown). By immunofluorescence microscopy, P7-1-Strep-tag II was shown to aggregate to form numerous tubule-like structures at 72 h p.i. in all of the cells that were tested (Fig. 1B). These tubule-like structures extended from the cell surface (Fig. 1B). No specific fluorescence was detected in uninfected cells (data not shown).
Subcellular localization of P7-1 in baculovirus-infected Sf9 cells at 72 h p.i. (A) Ultrathin section of salivary glands of viruliferous insect vector showing tubules of about 85 nm in diameter with viral particles inside in the cytoplasm of infected cells. Bar, 100 nm. (B) Immunofluorescence staining of P7-1 showing aggregation of numerous tubule-like structures in the cytoplasm or protruding from the cell surface. Bar, 10 μm. (C) Electron micrograph showing the presence of tubules (of about 85 nm in diameter) located in the cytoplasm (arrow) or protruding from Sf9 cell surface (arrowheads). Bar, 100 nm. (D) Immunogold electron microscopy showing that the tubules specifically react with a monoclonal anti-Strep-tag II antibody in the cytoplasm (arrow) or protruding from the Sf9 cell surface (arrowhead). Bar, 100 nm. (E) Electron micrograph of a purified tubule. Bar, 100 nm. The image in B combines the images of P7-1-Strep-tag II fluorescence and that of differential interference contrast (DIC).
Electron microscopy revealed that tubules of about 85 nm in diameter were located in the cytoplasm or protruded from the Sf9 cell surface (Fig. 1C). These tubules had an appearance (e.g., diameter and length) similar to the tubules produced by wild-type virus (Fig. 1A). Furthermore, immunogold electron microscopy showed that P7-1 was localized in these tubules (Fig. 1D). Obviously, the results described above indicated that expression of P7-1 alone, in the absence of the virus replication process, is sufficient for tubule formation in Sf9 cells. Therefore, it is reasonable to conclude that P7-1 can self-aggregate to form tubules in SRBSDV-infected host cells. No reaction with cellular structures was observed by incubation of uninfected cells with monoclonal anti-Strep-tag II antibody, or with viral structures, by incubation of infected cells with preimmune rabbit serum (data not shown).
To purify the tubules, infected cell extracts were prepared and subjected to sucrose density gradient centrifugation. A portion of each fraction was analyzed by immunoblotting (data not shown). The peak fractions containing P7-1 protein were pooled and pelleted. Electron microscopy confirmed that the preparation contained tubules of about 85 nm in diameter (Fig. 1E).
The putative transmembrane domains of P7-1 is required for tubule formation in vivo
Based on secondary structure prediction using the CUBIC PredictProtein server (http://cubic.bioc.columbia.edu/pp/), P7-1 is composed of three regions, including a large N-terminal region that is predominantly composed of α-helices that are separated by short β-strand structures (Part I, residues 1–316), a region containing only β-strands (Part II, residues 323–340), and a small C-terminal tail (residues 334-358) with no apparent α-helical or β-strand structures (Fig. 2).
The amino acid sequences of the SRBSDV P7-1 and RDV Pns10 [17] proteins were submitted to the CUBIC PredictProtein server (http://cubic.bioc. columbia.edu/pp/) for secondary structure prediction. Secondary structure elements (e.g. α-helix 1–10 [a1–10] and β-strand A–N [bA-N]) are labeled above the sequence in red and blue. The two putative transmembrane domains (TM1 and TM2) of P7-1 are underlined in blue under the sequences. The hydrophobic residues Leu (L), IIe (I) and Val (V) are boxed.
Computer analysis of the P7-1 amino acid sequence further revealed the presence of two putative α-helical transmembrane domains of 18 and 17 amino acid residues, respectively. The two predicted transmembrane segments were designated TM1 and TM2 (Fig. 3A). We generated two P7-1 deletion mutants (ΔTM1 and ΔTM2) in which the TM1 and TM2 were deleted, respectively, and found that both deletion mutants were diffusely distributed in the cytoplasm, different from the distribution of P7-1-Strp-tag II in the tubules (Fig. 3A, B). Thus, deletion of either the TM1 or TM2 domain of P7-1 abolished the ability to form tubules. However, another P7-1 mutant with the C-terminal 10 residues deleted retained the ability to form the tubule-like structures growing from the cell surface (Fig. 3A, B). These data indicate that the putative transmembrane domains are necessary for P7-1 to form tubules in Sf9 cells.
(A) Schematic illustrations of SRBSDV P7-1 protein, which is 358 amino acids in length and includes two putative transmembrane (TM) domains. Portions of the putative TM1 and TM2 are colored red. Construct names were indicated at the left. Blocks represent the region of P7-1 contained in the construct. Discontinuous lines represent the deleted region. Numbers indicate the relative amino acid positions in P7-1. The subcellular distribution of protein derived from each construct is summarized at the right. (B) Subcellular localization of various deletion mutants of SRBSDV P7-1 in Sf9 cells. Sf9 cells were infected with baculovirus encoding P7-1-Strep-tag II, ∆TM1-Strep-tag II, ∆TM2-Strep-tag II or 1-348-Strep-tag II. Cells were fixed 3 days p.i., stained with monoclonal anti-Strep-tag II and FITC anti-mouse IgG as a secondary antibody, and visualized by confocal fluorescence microscopy. Bars, 10 μm.
P7-1 has the capacity of self-assembly to form tubules
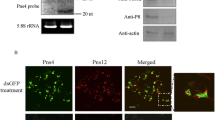
To determine whether there is direct interaction between P7-1 molecules to form homodimers or oligomers, a BiFC assay was carried out. P7-1-YN and P7-1-YC were coexpressed in N. bethamiana leaf cells, and indeed, there was strong BiFC fluorescence found in the inclusion bodies in the cytoplasm 48 h post-infiltration (Fig. 4A), whereas no BiFC fluorescence was visible in the negative control (data not shown). This result suggested that there was self-interaction of P7-1 in planta. To investigate whether P7-1 could form tubule-like structures in a non-host plant, N. benthamiana was used for transient expression via agoinfiltration [18]. We found that P7-1-YFP formed inclusion bodies rather than tubule-like structures in the cytoplasm of N. benthamiana leaf cells 48 h post-infiltration (Fig. 4B).
The direct self-interaction of SRBSDV P7-1. (A) BiFC array showing the self-interaction of P7-1 in planta. A strong BiFC fluorescence was observed in inclusion bodies in the cytoplasm of leaf cells of N. benthamiana coexpressing P7-1-YN and P7-1-YC 48 h post-agro-infiltration. Chl, chlorophyll autofluorescence. Bar, 10 μm. (B) P7-1-YFP formed inclusion bodies in the cytoplasm in the leaf cells of N. benthamiana 48 h post-infiltration. Bar, 10 μm. (C) Yeast two-hybrid assay showing the self-interaction of P7-1 protein. Transformants were grown on SD-trp-leu-his-ade plates for 3 days. (D) Transformants appear colored in the HTX β-galactosidase assay. I, Positive transformant of pTSU2-APP/pNubG-Fe65. II, Transformants of pBT3-STE-P7-1/pPR3-N-P7-1. III, Negative transformant of pTSU2-APP/pPR3-N.
Because P7-1 is a membrane-protein, we took advantage of the DUALmembrane system, a variant of the yeast two-hybrid approach, to detect P7-1 self-interaction on the surface of cellular membranes. Yeast cells co-transformed with all possible pairwise combinations of plasmids containing the NubG and Cub fusion proteins were then assayed for histidine and adenine prototrophs. As a result, only yeast expressing the homologous combination of P7-1 showed growth (Fig. 4C, D). These results confirmed that the specific self-interaction is not an artifact of the BiFC assay, and thus SRBSDV P7-1 protein was found to interact specifically with itself.
Discussion
Expression of one of the SRBSDV nonstructural proteins (P7-1) fused with Strep-tag II (P7-1-Strep-tag II) in Sf9 cells can induce the formation of tubules with dimensions and appearance similar to those found in SRBSDV-infected plant and insect vector cells [5, Fig. 1]. These results demonstrate that P7-1 of SRBSDV is responsible for the formation of tubular structure and that the production of tubules is not specific to host plant or insect vector cells.
The predicted secondary structure of SRBSDV P7-1 shows a high degree of similarity to the predicted secondary structure of the nonstructural protein Pns10 of rice dwarf virus (RDV), a phytoreovirus [1, 17; Fig. 2]. The expression of Pns10 in Sf9 cells is known to induce the formation of tubules similar to those in RDV-infected hosts [17]. Therefore, SRBSDV P7-1 and RDV Pns10, both of which are major constituents of tubular structures, appear to have similar structures with a conserved assembly strategy. Katayama et al. [19] have previously shown, using tomography and computer image processing, that the tubules of RDV Pns10 have a helical symmetry structure. We show in the current study that the purified tubules of SRBSDV P7-1 also appear to have a similar helical symmetry structure, suggesting a similar strategy for the assembly of tubules by SRBSDV and RDV. Our observations that P7-1 can interact with itself in yeast two-hybrid and BiFC assays provide evidence that this protein has the capacity of forming homodimers or oligomers (Fig. 4), which may play a role in forming the helical structure of the tubules. This type of helical structure is likely formed by end-to-end interactions between P7-1 molecules. In support of this contention, our data show that YFP fusion with the C-terminus of P7-1 (P7-1-YFP) abrogates the correct assembly of P7-1 to form tubular structures in non-host N. bethamiana leaf cells, and it instead forms an inclusion body.
Our microscopic observations indicate that the tubules of SRBSDV P7-1 protrude from the plasma membrane of Sf9 cells, suggesting that P7-1 is a membrane-associated protein. Computer analysis of membrane-spanning domains within the amino acid sequence of P7-1 identifies two putative α-helical transmembrane domains of approximately 18 and 17 amino acid residues, respectively (Figs. 2, 3). Deletion of either of the two putative transmembrane domains can abolish the formation of tubules in Sf9 cells, and the deletion mutants exhibit a punctate distribution in the cytoplasm (Figs. 2, 3). By contrast, the deletion of the C-terminal 10 residues of P7-1, which did not form α-helical and β-strand structures, does not eliminate the ability to form tubules in Sf9 cells (Figs. 2, 3). These data suggest that proper membrane association is necessary for the assembly of P7-1 into tubules.
The morphology of the tubules with virions inside them in host cells infected with plant-infecting reoviruses is similar to the structures formed by the movement proteins of several other plant viruses, such as cowpea mosaic virus, which form tubules that are embedded within highly modified plasmodesmata that transport viral particles [20]. The tubules of plant-infecting reoviruses have never been found to be associated with the cell wall [2, 3], suggesting that these tubules are not related to the cell-to-cell movement of viral particles in plants. By using insect vector cells in monolayers, we recently determined the functional roles of the tubules induced by a phytoreovirus, RDV, and found that the virus employs the tubules formed by RDV Pns10 to move along the actin-based filopodia extending toward neighboring insect vector cells to enhance viral movement [17, 21, 22]. The tubules containing RDV particles are found in association with the actin-based microvilli of the midgut in viruliferous vector leafhoppers [23]. Therefore, RDV might utilize these tubules to spread among cells of vector insects. It will be interesting to examine whether the similar tubules induced by fijiviruses play a similar role in the spread of virus among their insect vector cells. In this regard, the observation that P7-1 tubules of SRBSDV protrude from the plasma membrane of Sf9 cells suggests that the virus-containing tubules induced by fijiviruses infection can move out of the infected insect vector cells, providing a possible route for viral spread among insect vector cells.
The appearance of tubular structures during viral morphogenesis has been reported in studies of other members of the family Reoviridae. The infection and replication of bluetongue virus (BTV), member of the genus Orbivirus of the family Reoviridae, is accompanied by the formation of tubules of about 52 nm in diameter that are composed of the nonstructural protein NS1 [16]. The tubules composed of P7-1 of SRBSDV are morphologically similar to the NS1 tubules of BTV. However, SRBSDV tubules contain viral particles within them and have the ability to protrude from the plasma membrane of non-host Sf9 cells [16]. By contrast, BTV tubules do not contain viral particles within them and the expression of NS1 results in formation of tubules within the cytoplasm, rather than protruding from the plasma membrane of Sf9 cells. The NS1 protein of BTV has been reported to play a direct role in the cellular pathogenesis of BTV [24]. Therefore, the tubules induced by plant-infecting reoviruses and by BTV play different roles in viral infection.
References
Milne RG, del Vas M, Harding RM, Marzachì R, Mertens PPC. Fijivirus, pp 534–542; Omura T, Mertens PPC. Phytoreovirus, pp 543–549; Upadhyaya NM, Mertens PPC. Oryzavirus, pp 550–555 (all 2005). In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) Virus taxonomy: Classification and nomenclature of viruses, Eighth report of the international committee on taxonomy of viruses. Elsevier, Academic Press; Amsterdam, Holland, pp 450–606
Shikata E (1969) Electron microscopic studies on rice viruses: the virus disease of the rice plant. Johns Hopkins University Press, Baltimore, pp 223–240
Hibino H (1996) Biology and epidemiology of rice viruses. Annu Rev Phytopathol 34:249–274
Isogai M, Uyeda I, Lee BC (1998) Detection and assignment of proteins encoded by rice black streaked dwarf fijivirus S7, S8, S9 and S10. J Gen Virol 79:1487–1494
Zhou G, Wen J, Cai D, Li P, Xu D, Zhang S (2008) Southern rice black-streaked dwarf virus: a new proposed Fijivirus species in the family Reoviridae. Chin Sci Bull 53:3677–3685
Zhang HM, Yang J, Chen JP, Adams MJ (2008) A black-streaked dwarf disease on rice in China is caused by a novel Fijivirus. Arch Virol 153:1893–189
Wang Q, Yang J, Zhou G, Zhang HM, Chen JP, Adams MJ (2010) The complete genome sequence of two isolates of southern rice black-streaked dwarf virus, a new member of the Genus Fijivirus. J Phytopathol 158:733–737
Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45:616–629
Wei T, Huang TS, McNeil J, Laliberté JF, Hong J, Nelson RS, Wang A (2010) Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication. J Virol 84:799–809
Wei T, Zhang C, Hong J, Xiong R, Kasschau KD, Zhou X, Carrington JC, Wang A (2010) Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. Plos Pathog 6:e1000962
Rost B, Casadio R, Fariselli P (1996) Refining neural network predictions for helical transmembrane proteins by dynamic programming. Proc Int Conf Intell Syst Mol Biol 4:192–200
Hofmann K, Stoffel W (1993) TMbase—a database of membrane spanning protein segments. Biol Chem 347:166
Sonnhammer EL, von Heijne G, Krogh A (1998) A hidden markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 6:175–182
Atanassov II, Atanassov II, Etchells JP, Turner SR (2009) A simple, flexible and efficient PCR-fusion/Gateway cloning procedure for gene fusion, site-directed mutagenesis, short sequence insertion and domain deletions and swaps. Plant Methods 5:14
Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR (2006) Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc 1:3111–3120
Monastyrskaya K, Booth T, Nel L, Roy P (1994) Mutation of either of two cysteine residues or deletion of the amino or carboxy terminus of nonstructural protein NS1 of bluetongue virus abrogates virus-specified tubule formation in insect cells. J Virol 68:2169–2178
Wei T, Kikuchi A, Moriyasu Y, Suzuki N, Shimizu T, Hagiwara K, Chen H, Takahashi M, Ichiki-Uehara T, Omura T (2006) The spread of Rice dwarf virus among cells of its insect vector exploits virus-induced tubular structures. J Virol 80:8593–8602
Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1:2019–2025
Katayama S, Wei T, Omura T, Takagi J, Iwasaki K (2007) Three-dimensional architecture of virus-packed tubule. J Electron Microsc 56:77–81
Ritzenthaler C, Hofmann C (2007) Tubule-guided movement of plant viruses. Plant Cell Monogr 7:64–83
Wei T, Shimizu T, Omura T (2008) Endomembranes and myosin mediate assembly into tubules of Pns10 of Rice dwarf virus and intercellular spreading of the virus in cultured insect vector cells. Virology 372:349–356
Pu Y, Kikuchi A, Moriyasu Y et al (2011) Rice dwarf viruses with dysfunctional genomes generated in plants are filtered out in vector insects—implications for the virus origin. J Virol 85:2975–2979
Nasu S (1965) Electron microscopy studies on transovarial passage of rice dwarf virus. Jpn J Appl Entomol Zool 9:225–237
Owens RJ, Limn C, Roy P (2004) Role of an arbovirus nonstructural protein in cellular pathogenesis and virus release. J Virol 78:6649–6656
Acknowledgments
This work was supported by grants from the programs for the National Basic Research Program of China (No. 2010CB126203), Specialized Research Fund for the Ministry of Agriculture (No. 201003031), New Century Excellent Talents in University (No. NCET-09-0011), the National Natural Science Foundation of China (No. 31070130) and the Natural Science Foundation of Fujian Province (No. 2010J01075).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Y. Liu and D. Jia contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Schematic diagram of the use of fusion PCR to synthesize the fragments for creating mutant P7-1 in which the TM1 or TM2 segment was deleted. (A) Segment 1-107 and segment 127-358 were amplified with primers P1 and P2, and primers P3 and P4, respectively. P2 and P3 are complementary PCR primers consisting of two fused sections (shown in red and blue) that match regions flanking the deleted domain. The two fragments of PCR products were mixed and fused together by PCR using primers P1 and P4, creating mutant P7-1, in which the TM1 was deleted. (B) Segment 1-285 and segment 304-358 were amplified with primers P1 and P5, and primers P6 and P4, respectively. P5 and P6 are complementary PCR primers consisting of two fused sections (shown in red and blue) that match regions flanking the deleted domain. The two fragments of the PCR products were mixed and fused together by PCR using primers P1 and P4, creating mutant P7-1, in which TM2 was deleted. (TIFF 134 kb)
Rights and permissions
About this article
Cite this article
Liu, Y., Jia, D., Chen, H. et al. The P7-1 protein of southern rice black-streaked dwarf virus, a fijivirus, induces the formation of tubular structures in insect cells. Arch Virol 156, 1729–1736 (2011). https://doi.org/10.1007/s00705-011-1041-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-011-1041-9