Abstract
Suaeda subg. Brezia (Chenopodiaceae/Amaranthaceae) comprises ~45 halophytic species distributed worldwide along coastlines and in saline inland habitats. Thirteen species are currently accepted from the Americas, but species delimitation is difficult due to the scarcity of distinguishing characters. Little is known yet about phylogenetic relationships and biogeography of American Brezia species. Here, we present molecular phylogenies based on DNA sequence data from the nuclear ribosomal internal transcribed spacer (ITS) and the chloroplast rpl32-trnL intergenic region. Our sampling comprised 157 accessions covering all 13 American Brezia species along with 38 accessions from 16 Eurasian taxa. Phylogenetic trees were generated using parsimony and Bayesian methods. Three monophyletic lineages were discerned in the ITS tree: the Suaeda maritima, S. prostrata and S. corniculata group. Most American species proved to belong to the S. corniculata group. Species boundaries were mostly not recovered or even contradicted by the ITS data, which could be a consequence of low sequence variation in terminal clades and/or reticulate evolution. The rpl32-trnL phylogeny was poorly resolved, with the majority of American species being part of a polytomy with few supported internal nodes. Several incongruities were found between the nuclear and chloroplast tree, revealing at least four instances of hybridization and chloroplast capture between distant lineages. Chromosome counts showed that all American species are polyploid with hexaploidy prevailing. We discuss our results in terms of species relationships, hybridization, polyploidy and biogeography with emphasis on the colonization from NE Asia and Europe, and the subsequent spread and diversification in the Americas.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Suaeda Forssk. ex J.F.Gmelin (Suaedoideae; Chenopodiaceae/Amaranthaceae) is a genus of halophytic plants that comprises approximately 80–100 species with a worldwide distribution. In the course of an integrated study based on morphological, anatomical and molecular characters of the Old World species of genus Suaeda, we previously established the subgenus Brezia (Moq.) Freitag & Schütze with the single section Brezia (Moq.) Volk. (=Heterosperma Iljin) (Schütze et al. 2003). Brezia proved to be clearly separated from its sister subgenus Suaeda by a deep split in phylogenetic trees that were derived from nuclear ITS and plastid atpB–rbcL and psbB–psbH sequences (Schütze 2011; Schütze et al. 2003). In morphological respect, Brezia species consistently differ from other members of the genus by a unique pistil configuration characterized by the insertion of usually two stigmas at the top of the gradually tapering apex of the ovary, in combination with green-lined axes, C3 leaf anatomy, predominantly annual life form, and a distinctly hygrohalophytic habit. Like most Chenopodiaceae, Suaeda species are preferably anemophilous or self-pollinating. Schütze (2011) altogether listed 38 species in subgenus Brezia, with 23 species occurring in temperate Eurasia, 10 in the Americas and the remainder scattered over S Africa, S and SE Asia and Oceania. Our knowledge of putatively new taxa allows for an estimate of ~45 species. Brezia species can be found growing in two different types of habitats: (1) intertidal marshes, estuaries and beaches from the Arctic to the Tropics (Fig. 1a, e) but with highest diversity in the temperate zones and (2) saline and alkaline wetlands that frequently occur under semiarid and arid climates in plains, in depressions, at the borders of saline or brackish lakes (Fig. 1 b–d) and in the outer belts of springs, again centered in temperate and subtropical zones.
American species of Suaeda subg. Brezia: habitats, habit and details. a Coastal salt marsh of Suaeda aff. esteroa (light green) and Sarcocornia pacifica (Standl.) A.J.Scott (dark green), Baja California, Bahía de Las Animas, 02/01/1998; b Alkaline flat with S. occidentalis, Distichlis spicata (L.) Greene and Nitrophila occidentalis Wats. growing on washes between bushes of Sarcobatus vermiculatus (Hook.) Torr., Buffalo Valley, Nevada, 03/10/2010; c S. calceoliformis belt behind Salicornia rubra A.Nelson belt, southern edge of Great Salt Lake, Utah, 30/10/2010; d S. jacoensis on an alkaline plain of N Mexico, Nuevo Léon, 19/08/2014, 24°21′05.7″N/100°11′50.1″W; e S. patagonica on rocky shore, Antarctica Chileña near Pto. Natales, 25/02/2011. f S. rolandii, greenhouse cultivation in Kassel, with H. Freitag, 04/11/2011; g S. calceoliformis 1, prostrate annual form from open alkaline flat at Newark Lake, Nevada 01/10/2010; h S. pulvinata, prostrate perennial plant on temporarily flooded plain at Totolcingo Lake, C Mexico, 03/10/2012. i S. calceoliformis 2, erect plants, from an ecocline near the Great Salt Lake, Utah, 30/09/2010; j S. calceoliformis (S. minutiflora) in flower, from cultivation in Kassel, 16/01/2012; k S. calceoliformis, fruiting branch, near Great Salt Lake, Utah, 30/10/2010; l S. rolandii, fruiting branch, from greenhouse cultivation in Kassel, 26/10/2011; m S. occidentalis, Whorlwind Valley near Beowawe, Nevada, 03/10/2010; n S. calceoliformis (S. minutiflora), fruiting perianths, from cultivation in Kassel, 20/02/2012; o S. linearis, branch with fruiting perianths, Padre Island, Texas, 26/09/2010; p S. occidentalis, fruiting perianths from above, Whorlwind Valley near Beowawe, Nevada, 03/10/2010. Photos by R. Brandt (f, p), E. Dominguez (e), W. R. Ferren (a), H. Flores-Olvera (d, h), all others by H. Freitag
For the two American subcontinents, altogether 13 species of Suaeda sect. Brezia are currently accepted (Table 1). The distribution of the six N American taxa according to the Flora of North America is shown in Fig. 2. Delimitation of species and identification of individual specimens in the subgenus are often impeded by the scarcity of discriminating morphological characters and their plasticity related to different habitat conditions, in particular to nutrient and water supply and to the degree and type of salinity (see, e.g., Fig. 1i). Even characters that are usually quite reliable, such as life form, leaf parameters, structure of inflorescence, shape of the fruiting perianth and seed size are subject to considerable variation. This led to varying species circumscriptions which in some areas and some species groups have not been settled yet. Thus, previous and ongoing morphological studies in the Mexican Brezia species (Ferren and Whitmore 1983; Watson and Ferren 1991; Alvarado Reyes and Flores-Olvera 2013; Noguez-Hernández et al. 2013) have shown that the traditional classification does not correctly reflect the real species diversity, and descriptions of additional new taxa have already been announced from the area around the Gulf of California (Ferren and Roberts 2011).
Distribution of North American species of Suaeda subg. Brezia, from Ferren and Schenk (2003), by courtesy of the Flora of North America Association
Based on our own morphological, karyological and molecular work on Suaeda species from larger parts of Eurasia (e.g., Lomonosova and Freitag 2003, 2008; Freitag and Lomonosova 2006, 2013; Lomonosova et al. 2008) which was complemented by cultivation experiments, we suspected that certain Brezia species known from N America are in need of a critical recheck, despite the rather recent but partly contradicting revision by Hopkins and Blackwell (1977) and the account of Ferren and Schenk (2003). Hidden diversity was a priori suspected in species distributed through different climatic zones. In this respect, the most interesting candidates are S. calceoliformis and S. linearis. While the former species is reported from the Arctic coasts of the Beaufort Sea down to subtropical S California and from the Pacific coast throughout the western plains (Great Basin) to the Atlantic coast, the latter species is said to grow all along the coast from cool temperate SE Canada to tropical Yucatan (Fig. 2). The comparatively large number of taxonomical synonyms in S. calceoliformis and also in S. maritima is another potential indicator for hidden heterogeneity. Taken together, a profound taxonomic revision of the American Brezia species appears to be overdue.
So far, only few molecular data are available on the relationships among the American Brezia species and their affinities to species on other continents. Such data are however a necessary precondition for elucidating the phylogeny and the historical biogeography of the American taxa. Our previous genus-wide ITS analysis had shown that the Old World species of sect. Brezia are distributed among three lineages, i.e., the S. corniculata, S. prostrata and S. maritima group (Schütze et al. 2003). The two latter groups are closely related with each other, and were not distinguished in plastid phylogenies based on atpB–rbcL and psbB–psbH sequences that in general had a lower resolution than ITS. So far, only the S. corniculata group was found to be characterized also by a morphological apomorphy: the tepals being distinctly unequal, with at least one of them hooded or hooked (see Fig. 1n–p).
With S. patagonica s.l., the sampling of Schütze et al. (2003) included only a single collection from S America, which was found nested in the S. corniculata group. Since this early work, the ITS regions of six more American Brezia species have been sequenced, i.e., S. calceoliformis and S. occidentalis by Kapralov et al. (2006), and S. linearis, S. mexicana, S. esteroa and S. puertopenascoa by Steinmann, included in Schütze (2011). All species proved to be members of the S. corniculata group, but were poorly resolved in the ITS trees. To sum up, only a single ITS sequence each from seven of the 13 American species of subg. Brezia is known so far. This lack of knowledge delimits conclusions about the affinities among taxa, especially since unexpected molecular diversity was also found by extended sampling in well-circumscribed Brezia species from the Old World, e.g., in S. maritima, S. spicata and S. salsa (reviewed by Weising and Freitag 2007).
With regard to the origin of lineages, Schütze (2011) calculated from chronograms based on tree topologies of ITS and atpB–rbcL sequences that Brezia probably had evolved during the Oligocene at approximately 30 Mya somewhere between today’s E Mediterranean region and C Asia where the group also has its present center of diversity. The split into the three Old World lineages happened much later, presumably in the Late Miocene, whereas most of the diversification as represented by extant populations started in the Early Pleistocene. In his study, Schütze (2011) had used penalized likelihood with two calibration points of deeper Chenopodiaceae nodes taken from Kadereit et al. (2005b). His ITS dataset included 35 (24)Footnote 1 accessions from 27 (19) taxa of subg. Brezia and among them one sample each from 11 (7) species of the S. corniculata group, with all three American lineages represented by seven species. The estimates made by Schütze (2011) were later confirmed in a more representative, family-wide dated plastid phylogeny presented by Kadereit et al. (2012).
Polyploidy is widely distributed in Suaeda, but the karyological conditions of the American Brezia species are poorly known except for Canada, where Basset and Crompton (1978) constantly found tetraploids (2n = 36) in S. maritima, hexaploids (2n = 54) in S. calceoliformis, and dekaploids (2n = 90) in the newly described S. rolandii. Otherwise, only scattered data were reported for S. patagonica, S. edulis, S. linearis and S. calceoliformis (Table 2). More data are clearly needed because the results of Basset and Crompton (1978) already indicated that ploidy level is an important and potentially species-delimiting character in the American Brezia taxa. This is supported by our own experience in Eurasian Brezia species (Lomonosova 2011, and references therein) where the ploidy levels vary from diploidy (2n = 18) to dekaploidy (2n = 90). Ploidy levels were usually found to be constant within species, with a few notable exceptions (e.g., S. corniculata, S. kulundensis). By comparing nuclear and chloroplast phylogenies, the evolution of polyploids can in principle be traced back to their ancestors, as was shown in certain Asian Brezia species by Lomonosova et al. (2008) and Schütze (2011).
In the present study, we addressed the following questions: (1) How does the actual genetic diversity among the American species correspond to their current taxonomic classification? Are currently delimited species monophyletic, or can we detect hidden heterogeneity and/or new species? (2) Can we find evidence for reticulate evolution? (3) Where are the American species placed in the phylogenetic frame of the subgenus? (4) How many times were the Americas colonized, where did the colonizers come from, and how did they spread and diversify?
To answer these questions, we performed an extensive sampling of the American Brezia species along with presumably related species from other continents. Molecular phylogenies using the traditional nuclear ITS marker and the rpl32-trnL gene as a chloroplast DNA (cpDNA) marker were generated, and complementary chromosome counts were carried out which proved to be significant for understanding the evolution in the subgenus. In addition, most taxa were studied in ample herbarium material, in the field and in the greenhouse.
Materials and methods
Plant material for molecular studies
For the molecular study, we sequenced 157 samples from all 13 currently accepted American Brezia species including several samples with doubtful affinities, and 38 samples from 16 selected Eurasian Brezia species. Four Eurasian species of Suaeda subg. Suaeda sect. Schoberia were chosen as outgroups based on our previous analyses (Schütze et al. 2003; Schütze 2011). All accessions are compiled on a spreadsheet in Online Resource 1. We consciously increased the sampling in taxonomically critical and widely distributed taxa such as S. calceoliformis (40) and S. linearis (26). To some species, we had only limited access. Therefore, S. densiflora and S. pulvinata are just represented by one sample each. From several taxa, more herbarium specimens were sampled than presented here, but either failed in DNA extraction, or never delivered readable sequences. For provenances of the samples included in the molecular analyses see Fig. 3. The selection of the Eurasian species was based on the results of Schütze (see above) and our own worldwide dataset comprising about 350 sequences of Suaeda subg. Brezia (unpublished data). As we found that the vast majority of the American Brezia species belongs to the S. corniculata group, we included all species of that group known worldwide.
The majority of samples from America were gathered by ourselves from herbarium specimens, mainly in US American institutions, to (1) reduce the risk of getting wrongly identified material, (2) get material from locations close to the areas where the types had been collected, and (3) screen for potentially interesting morphological variations. The most helpful herbaria were BKL, CAS, GR, LPB, NY, RENO, RSA, SD, TEX, UCSB, and UTC (Thiers 2012). Some additional samples were taken from loans, particularly from ALA, DAO, IEB, and from material kindly provided by a number of colleagues. About one quarter of the samples was collected by RB and HF during an expedition along the northeastern, southern and western coasts of the US as well as in dry inland areas in September and October 2010. During fieldwork, the leaves were quick-dried with silica gel in parallel to the collection of herbarium specimens which are deposited in KAS. For a few samples, plants were grown from seeds collected during the expedition, and fresh leaves from these plants were used for DNA isolation.
Most accessions from the Eurasian species were sequenced from herbarium material, preferably from more recently collected specimens kept in KAS and NS. For details of the vouchers see Online Resource 1. Species identification was sometimes difficult, especially along the northeastern US coast where several species grow sympatrically in only slightly differing habitats. Responsibility for the names given to the American samples is by HF, and for the Eurasian samples jointly by HF and ML. The geographical coverage of the distributional areas is sufficiently even and almost complete, whereas sampling density differs considerably depending on the availability of samples. The only gap concerning the known distribution is Cuba from where we did not get readable sequences. The lack of samples from large strips along the Pacific coast of Canada and the United States correlates with the rare occurrence of Brezia species in these areas.
Plant material for karyological, morphological and taxonomic studies
All plants used for chromosome counting were grown from seeds in greenhouses in Kassel and/or in Novosibirsk. The seeds were mostly collected during the field trip to N America (see above), some were provided by W. R. Ferren (from Sandy Hook, New Jersey, USA), and seeds of S. patagonica (from S Chile) were sent by E. Dominguez. Regrettably, due to limited availability of seeds and to the comparatively short time span they remain able to germinate, not all American species could be grown in the greenhouse. All relevant karyological data from literature were also included into the evaluation.
Morphological and taxonomic information was gained from the relevant literature. Most types have been seen, at least as scans, and many herbarium specimens beyond those cited as vouchers in Online Resource 1 were studied in American herbaria and in the specimens made available by loans. During the expedition, complementary observations concerning morphological variation and ecology were made in the field. Other important information came from cultivation experiments in the greenhouse, where progeny of plants originating from seed collected in the field and/or received from W. R. Ferren were grown under most suitable and identical conditions (see, e.g., Fig. 1f) thus allowing to judge about the impact of adverse environmental factors in certain natural habitats.
DNA isolation, amplification and sequencing
Total DNAs were isolated from 50 to 100 mg of fresh leaves, or 20–50 mg of silica-dried or herbarium leaves of individual plants using a modified cetyltrimethylammonium (CTAB) procedure (Krapp 2013). DNA concentrations were determined electrophoretically versus known amounts of λ DNA as standards. For PCR, DNA samples were adjusted to a concentration of approximately 20 ng/μL.
For PCR amplification of the nuclear ribosomal ITS region and the chloroplast rpL32-trnL intergenic region, we used primer combinations described by Blattner (1999) and Shaw et al. (2007), respectively. For some difficult samples, the rpL32-trnL spacer was amplified in two parts, using newly designed internal primers in addition to the standard primers (int_fwd: 5′-AAC ATA GTA CTT TTG GTT GAA AAC CC-3′; int-rev: 5′-GAA AGT TTA ACG ATT TTC TTA TTT GCC-3′). All primers carried an M13-tail at their 5′ end to facilitate subsequent sequencing (see below). All PCRs were performed in 30 μL volumes using a Biometra T-Gradient Cycler. Each reaction contained approximately 20 ng of genomic template DNA, 1.5 mM MgCl2, 0.25 µM of each forward and reverse primer, 0.2 mM of each dNTP, 0.5 µg/µL bovine serum albumin and 0.1 U/µL Mango Taq DNA polymerase (Bioline, Taunton, USA) in a buffer supplied by the enzyme manufacturer. ITS amplification was initiated with a denaturation step at 95 °C for 3 min, followed by a touchdown PCR for 10 cycles, each consisting of 95 °C for 30 s, 60–55 °C (see below) for 30 s, and 72 °C for 30 s. Starting at 60 °C, the annealing temperature was reduced by 0.5 °C per cycle and then left constant at 55 °C for another 30 cycles. Final extension was at 72 °C for 7 min. For the chloroplast rpL32-trnL intergenic region, initial denaturation was performed at 80 °C for 5 min, followed by 30 PCR cycles, each consisting of denaturing at 95 °C for 1 min, annealing at 45 °C for 1 min and elongation at 65 °C for 2 min. Final extension was at 72 °C for 5 min. To check for the presence of distinct, single bands, aliquots of PCR products were electrophoresed on 1.5 % agarose gels and stained with ethidium bromide. Some samples failed to amplify with one or the other DNA region.
Double-stranded PCR products (10–30 ng per reaction) were sent to a commercial company (LGC Genomics, Berlin, Germany) for sequencing by the dideoxynucleotide chain termination method. M13 forward (5′-TGT AAA ACG ACG GCC AGT-3′) and M13 reverse sequencing primers (5′-CAG GAA ACA GCT ATG ACC-3′) were purchased from Metabion (Martinsried, Germany). All sequences were submitted to GenBank. Accession numbers are given in Online Resource 1.
Phylogenetic analyses
Consensus sequences were aligned with the help of PhyDE (Müller et al. 2011) followed by manual adjustments. Indels were excluded from all analyses. The alignments used to produce the phylogenies are provided as commented nexus files in Online Resources 2 and 3. All phylogenetic reconstructions were performed separately for the rpl32-trnL and the ITS sequence alignment. No combined analysis of nuclear and plastid datasets was attempted due to the considerable degree of incongruence. Maximum parsimony (MP) analyses were performed using PAUP* 4.0b (Swofford 2003). Consensus trees were generated from a heuristic search with 100 stepwise random addition replicates using tree bisection and reconnection (TBR) branch swapping with steepest-descent modification and MulTrees option activated. To evaluate the extent of homoplasy in the dataset, the consistency (CI) and retention (RI) indices were calculated. Statistical support values were estimated running 100 bootstrap pseudoreplicates with one random addition replicate followed by TBR branch swapping.
For Bayesian inference analyses, the datasets were tested for the best-fit model of evolution with MrModeltest v. 2.3 (Nylander 2004) using the Akaike Information Criterion. GTR + G + I represented the best fitting model for both datasets. Bayesian analyses were done using MrBayes 3.2 (Huelsenbeck and Ronquist 2001). Two independent runs of Markov chain Monte Carlo were initiated, with 1,000,000 generations, sampling trees every 100th generation. Each run consisted of three heated chains using a heating parameter of 0.2 and one cold chain. After plotting the likelihood-by-generation values, the first 10 % of the runs were discarded as burn-in, and Bayesian consensus trees with posterior probability values of nodal support were constructed.
Biogeography
Regarding the biogeographical aspects, we tried with ancient area reconstructions using the methods implemented in RASP (Yu et al. 2010, 2015) but failed in getting plausible results, maybe in part due to the peculiar kind of our data. In face of the general problems associated with purely statistical biogeographical evaluations of phylogenetic trees, in particular the lacking information about extinct species (see Cusimano and Renner 2014) we doubt that other standard methods of ancient area analyses, as MESQUITE or LAGRANGE, would yield more reliable results. For outlining historical biogeography, we instead complemented the direct, intuitively gained inference from the phylogenetic trees by information from our karyological data, from our knowledge of the species’ biology—in particular their ecology—, and from general experience in other groups of Chenopodiaceae.
Karyological methods
Chromosome counts of root tip meristems were carried out following the protocol of Smirnov (1968). In short, root tips obtained from germinated seeds or from cultivated plants were pretreated in 0.2 % colchicine for 2 h, fixed in ethanol–acetic acid (3:1) and subsequently stained with 1 % acetic hematoxylin. Observation of chromosomes, photo documentation and subsequent drawings of the mitotic metaphases were made with the help of an Axioscop-40 light microscope and the built-in video camera AxioCam MRc 5 (Zeiss, Germany).
Results
Alignment and sequence statistics
The nuclear ITS1—5.8SrDNA—ITS2 region and the plastid intergenic spacer rpl32-trnL were sequenced for a large taxon set with special focus on the American species of Suaeda sect. Brezia. The ITS dataset comprised 170 sequences (127 from the Americas), of which 157 sequences were generated for this study. The rpl32-trnL dataset consisted of 161 sequences (123 from the Americas) all of which were newly generated. The alignment of ITS contains 598 characters (size range 430–581 bp) showing 26.2 % variable sites of which 21.2 % were parsimony informative (33.6 and 28.6 % when outgroup taxa were included). The alignment length of the rpl32-trnL intergenic spacer was 1055 bp (size range 814–965 bp), with 8.8 and 6.0 % of the sites being polymorphic and parsimony informative, respectively (18.9 and 15.6 % when outgroup taxa were included).
Phylogenetic analysis of the ITS tree
The consensus tree resulting from Bayesian analyses of the ITS dataset is shown in Fig. 4. A similar topology was obtained by maximum parsimony (MP) analysis (not shown). Posterior probabilities and bootstrap support values obtained by Bayesian and MP analyses are indicated above and below the branches, respectively. One remarkable aspect of the ITS tree(s) is the apparent lack of correspondence between the positions of several samples and the names given to them, although much effort was dedicated to accurate species identification. For convenience, genetically differing samples that carry the same species name and might represent hidden species are distinguished by a suffix, like S. linearis 1, 2 or 3, or S. aff. calceoliformis.
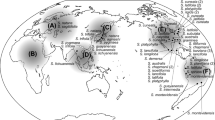
Molecular phylogeny of American and selected Eurasian species of Suaeda subg. Brezia derived from a Bayesian analysis of internal transcribed spacer (ITS) sequences. Posterior probabilities are shown above branches; bootstrap values from a maximum parsimony analysis are shown below branches. ID, species name, provenance and chromosome numbers are given for each accession (for details see Online Resource 1). Chromosome numbers in brackets refer to samples from nearby localities. After submission of the present manuscript the species S. „iranica“ was described as S. khalijefarsica Akhani. Shaded areas indicate American samples; for color coding see Fig. 3. A Austria, BOL Bolivia, CHN China, CDN Canada, DK Denmark, E Spain, F France, GB Great Britain, KZ Kazakhstan, IR Iran, MEX Mexico, MGL Mongolia, RA Argentina, RCH Chile, TJ Tajikistan, UA Ukraine, USA United States of America, UZ Uzbekistan
The backbone topology of the ITS tree confirms our previous findings (Schütze et al. 2003; Schütze 2011) concerning the well-supported monophyly of the subgenus Brezia and its basal dichotomy into the S. corniculata group and a second lineage that splits into the S. prostrata and the S. maritima groups. The S. prostrata group only contains two sequences from the Americas, one belonging to the S American S. densiflora which is presumed to be closely related to the Eurasian S. prostrata, and the second being almost identical with the available sequences of the W Mediterranean S. spicata, for the first time collected in S California. The S. maritima group is represented in N America by 13 accessions of S. maritima and by two accessions of the endemic S. rolandii. The N American S. maritima sequences do not differ significantly from those generated from S. maritima plants collected along the Atlantic coast of Europe.
The S. corniculata group is represented in our tree by all species known worldwide. It is much more diversified and, in contrast to the other two groups, appears to be clearly centered in the Americas. The ITS tree reveals a basal subdivision, with a well-supported (PP 0.99, BS 100) small group of geographically restricted Asian species being sister to a clade that contains all American species and the remaining Eurasian species. This very coherent group (PP 1, BS 100) which we here refer to as the S. arctica lineage includes S. arctica, S. tschujensis and two taxa that have not been formally described yet (S. “jacutica”, S. “tibetica”). The large clade containing the remainder of the S. corniculata group is split into another dichotomy, separating a weakly supported (PP 0.96, BS 54), purely N American group of 17 accessions that were morphologically identified as S. calceoliformis and S. mexicana from the remainder. For convenience, the first clade is here referred to as S. calceoliformis 1 lineage, shaded blue in Fig. 4 and marked by blue symbols in the map (Fig. 3). Internal groupings in the S. calceoliformis 1 lineage are only weakly supported.
The next split is a trichotomy. The first unit is a single sequence of an individual similar to S. calceoliformis (named here S. aff. calceoliformis) collected in inland S California. The second group (PP 0.69, BS 50) includes in an unresolved polytomy all samples from the three remaining Eurasian species of the group, namely S. corniculata, S. pannonica and S. tuvinica (here informally referred to as S. pannonica assemblage) together with a well-supported (PP1, BS 100) monophyletic group of American S. edulis and S. calceoliformis accessions. We refer to the latter as S. calceoliformis 2 lineage, shaded red in Fig. 4 and marked with red symbols in Fig. 3. The third, also weakly supported group of the trichotomy (PP 0.9, BS 69) comprises all remaining American taxa of the S. corniculata group, which we collectively refer to as S. linearis lineage, shaded green in Fig. 4 and represented by green symbols in Fig. 3.
The S. linearis lineage is split into a polytomy formed by several accessions of S. occidentalis and the only sequence of S. pulvinata together with four weakly to moderately supported subclades consisting of (1) two more sequences of S. occidentalis (PP 1, BS-), (2) all four sequences of the South American S. patagonica together with two sequences of S. puertopenascoa, a narrow-ranged endemic of the northern shores of the Gulf of California (PP 0.84, BS-), (3) all sequences from the Mexican endemic S. jacoensis together with three samples of S. mexicana 2 (PP 0.99, BS-), and (4) a lineage that comprises all sequences from S. linearis and S. esteroa s.l. (PP 1, BS 68). The latter subclade is composed of two sister groups. One of these contains eight sequences of S. linearis 1 along with the enigmatic S. linearis X (see “Discussion”) from the Atlantic coast (PP 0.96, BS 55), while the second, unsupported group again splits into a polytomy containing 11 sequences of S. linearis 2 from the northern Mexican Gulf coast, three accessions of S. esteroa s.l. from NW Mexico and adjacent California, three more accessions of S. esteroa s.l. from the Californian Gulf coast of Sonora and Sinaloa (PP 1, BS 77), six samples of S. linearis 3 from the Yucatan coast and a single accession from inland C Mexico that probably represents a new species (see “Discussion”), and a moderately supported lineage (PP 1, BS 66) embracing 13 more sequences of S. esteroa s.l. from the northern Californian Gulf coast and the Pacific coast from SW California to Baja California, including another well-supported sublineage from Baja California.
It should be noted that, in our extensive dataset from other continents, not a single accession was detected that is suspicious as having been derived from any American progenitor.
Phylogenetic analysis of the rpl32-trnL tree
In preliminary experiments, several non-coding cpDNA loci were amplified and sequenced from a small test set of plants, including atpB–rbcL and psbB–psbH that had been used by Schütze et al. (2003). Among these, the rpl32-trnL intergenic spacer suggested by Shaw et al. (2007) showed the most reliable performance and the highest diversity and was chosen for sequencing all samples. A 50 % majority rule consensus tree resulting from Bayesian analyses is shown in Fig. 5. A similar topology was obtained with an MP analysis (not shown, but the BS values were transferred to the respective branches of the Bayesian tree).
Molecular phylogeny of American and selected Eurasian species of Suaeda subg. Brezia derived from a Bayesian analysis of plastid intergenic spacer rpl32-trnL sequences. Posterior probabilities are shown above branches. ID, species name and provenance are given for each accession; shaded areas indicate American samples. For abbreviations see Fig. 4. After submission of the present manuscript the species S. „iranica“ was described as S. khalijefarsica Akhani
The most striking features of the cpDNA tree are: (1) Resolution and statistical support of the different lineages are generally much lower than in the ITS tree. (2) Several accessions and lineages are placed in different positions as compared with the ITS tree (for details see “Discussion”), with the outcome that none of the three major species groups recognized in sect. Brezia with ITS is monophyletic in the cpDNA tree. (3) In accordance with the ITS tree, the S. arctica lineage is sister to the moderately supported (PP 0.99, BS 69) remainder of the S. corniculata group that forms a polytomy of eight lineages. Seven of these correspond to the unresolved Eurasian S. pannonica assemblage of the ITS tree, whereas all but two American samples of the S. corniculata group are united in a single unsupported clade (shaded gray in Fig. 5). (4) The subdivision of the American species of the S. corniculata group into three lineages as suggested by the ITS tree is not seen in the cpDNA tree. Instead, only a few internal subclades are revealed among many unlinked accessions. These subclades are formed by S. patagonica (PP1, BS 90), the same S. jacoensis/S. mexicana 2 group (PP 1, BS 99) as in the ITS tree, three accessions of S. calceoliformis 1 and 2 (PP1, BS 68), and four accessions of S. calceoliformis 2 and S. edulis (PP 0.97, BS 54).
Chromosome counts
Our new counts from 31 N American and 12 Eurasian specimens are presented along with 36 previous counts in Table 2 on the background of a complete compilation of taxa dealt with in this article. Examples of metaphase plates are shown in Fig. 6. The coverage is still incomplete because we were not able to get seeds from S. jacoensis, S. mexicana, S. puertopenascoa, S. pulvinata, S. densiflora and the single American accession of S. spicata.
Mitotic metaphases of studied plants. a Suaeda “jacutica” nom. inedit. (2n = 18) [DNA-ID S440]; b Suaeda maritima (L.) Dumort. (2n = 36) [DNA-ID 2359]; c Suaeda occidentalis S.Watson (2n = 54) [DNA-ID 2216]; d Suaeda corniculata (C.A.Mey.) Bunge (2n = 54) [DNA-ID 1461]; e Suaeda calceoliformis (Hook.) Moq. (2n = 54) [DNA-ID 2221]; f Suaeda rolandii Bassett & Crompton (2n = 90) [DNA-ID 2197a]. Scale bar 5 μm; all photos by M. Lomonosova
Regarding the S. maritima group, we confirmed tetraploidy for the N American populations of S. maritima which is in accordance with the data from the Atlantic coast of Europe, and dekaploidy for S. rolandii. In the S. corniculata group, nearly all American specimens proved to be hexaploid, with the only exceptions being one tetraploid specimen found in the otherwise hexaploid S. patagonica, and the surprising discovery of dekaploidy in one plant from coastal Maine (referred to here as S. linearis X). The Eurasian species of the S. corniculata group cover the whole series from diploids (2n = 18) to dekaploids (2n = 90).
Discussion
Critical appraisal of the ITS and the rpl32-trnL phylogenies
In general, resolution of our ITS as well as cpDNA trees is limited, which at least in part has to be attributed to low sequence divergence. Given that the ITS tree is better resolved than the cpDNA tree, we cautiously consider it as the backbone of our phylogenetic inference. This is in line with previous studies on Suaedoideae (Schütze et al. 2003; Kapralov et al. 2006; Lee et al. 2007; Schütze 2011) and supported by general experience (e.g., Feliner and Rossello 2007, and references therein), which led to the almost universal usage of ITS sequencing for phylogenetic studies at the species level in the past. We are nevertheless aware of the limitations inherent to the ITS marker as summarized (and somewhat exaggerated) by Álvarez and Wendel (2003). As such they listed incomplete concerted evolution when different ITS repeats are merged within a single genome of (allo)polyploids, and the occurrence of paralogous sequences which both might confound phylogenetic reconstructions. Such phenomena might have caused the rather wide and very well-supported separation of two hexaploid S. calceoliformis lineages in the ITS tree. At least in some cases, pairs of samples belonging to either the S. calceoliformis 1 or 2 lineage, respectively, have been collected close to each other and are morphologically almost indistinguishable (e.g., ID 2338 and ID 2212 from Utah, and ID 2274 and ID 2340 from Saskatchewan).
Despite its lower resolution, the cpDNA tree is also important because it indicates the degree to which individual gene trees may differ from one another and hence from the underlying species tree (Doyle 1992; Maddison 1997). Though conflict between nuclear and plastid trees can be caused by different phenomena such as, e.g., coalescence and incomplete lineage sorting [see Pirie et al. 2009; Naciri and Linder (2015) discussed “seven veils”], in Suaeda probably more often they indicate reticulation by hybridization events. This applies in particular to taxa that have originated by reticulate evolution via alloploidy as shown by ourselves (Lomonosova et al. 2008) and by Schütze (2011) in Suaeda kulundensis and S. sibirica, two Asian counterparts of the American species of the S. corniculata group also included in our trees. Homoploid hybridization and introgression can have similar effects. To detect such phenomena, we carefully compared both trees with each other in the following paragraph.
Incongruities between the ITS and the rpl32-trnL trees
Major conflicts among ITS and cpDNA tree topologies are summarized in Fig. 7 where simplified versions of both trees are compared with each other. Whereas many of the incongruities regarding the position of individual accessions and lineages receive little statistical support, others are substantiated by stable and well-supported branches.
Comparison of ITS and rpl32-trnL trees. Only those accessions were included where both sequences were available. Shaded areas indicate American samples. For color coding of ITS lineages see Fig. 3. The hatched area in the rpl32-trnL tree contains intermingled American and Eurasian samples. Horizontal broken lines separate the three species groups. All other lines connect corresponding accessions
A major incongruity regarding the monophyly of the S. prostrata, S. maritima and S. corniculata groups was already noted and ascribed to hybridization by Schütze (2011), although other cpDNA markers (atpB–rbcL and psbB–psbH) were used in his study. In the ITS tree of Schütze (2011), the Asian species S. sibirica, S. kulundensis, S. crassifolia and S. heteroptera are placed in the S. maritima group, whereas they reside in more basal branches of the S. corniculata group in the cpDNA tree. We found the same distribution in our present analyses (compare Figs. 4, 5). For S. sibirica and S. kulundensis, (see Fig. 7). Lomonosova et al. (2008) confirmed the hybridization hypothesis by morphological and karyological arguments.
Among the American species, signatures for hybridization between members of widely separated species groups and combinations with “wrong” chloroplast DNA were found in several samples (see Fig. 7). One striking example concerns the Argentinean endemic S. densiflora (ID 2225) that morphologically and according to ITS data clearly belongs to the S. prostrata group but has a cpDNA sequence of the S. corniculata group. As no other species of these two groups are known from S America, the origin of this sequence combination is mysterious. Given that only one sample of S. densiflora was analyzed and no chromosome counts are available yet, this result needs to be validated by additional sampling.
Chloroplasts of the S. maritima group were found in plants that morphologically and according to ITS data belong to different lineages of the S. corniculata group (Fig. 7). Thus, two S. calceoliformis plants from Saskatchewan (ID 2274, 2340) that belong to either S. calceoliformis lineage 1 or 2 have S. maritima chloroplasts that only slightly differ from each other. These pairings are likewise difficult to explain. Whereas both S. calceoliformis 1 and 2 plants are present in the surroundings, the closest occurrence of S. maritima is far away at the Atlantic coast (see Fig. 3). Since cpDNA is transmitted by seed in most angiosperms and probably also in Suaeda, long-distance dispersal of S. maritima seeds to the inland may have occurred, followed by pollination of the seed-grown plant with S. calceoliformis pollen. Introgression of S. maritima cpDNA into S. calceoliformis was then eventually completed by repeated backcrossing with the paternal species. Most likely, both samples represent incidental and independent hybridization events. The only accession of Suaeda linearis X (ID 2200), from coastal Maine, is unique in its combination of ITS sequences of the S. linearis lineage with S. maritima chloroplasts (see also the subsequent paragraph on “Ploidy levels”).
Putative hybridization events between different lineages of the S. corniculata group are more difficult to detect because of the small sequence divergence. Here, we shortly comment on three examples traced in Fig. 7. First, Suaeda sp. nov. (ID 2400) from Texcoco in eastern central Mexico combines ITS sequences of S. linearis 3 (Yucatan) with cpDNA sequences close to but clearly distinct from S. mexicana 2 from northern central Mexico. This specimen also differs morphologically from its next relatives and probably represents a new species. Second, S. mexicana 1 (ID 2233) from Coahuila in northern central Mexico has ITS sequences close to those of the second S. mexicana 1 accession (ID 2282) from the same area and cpDNA sequences of the sympatric S. edulis. Unfortunately, we were unable to generate cpDNA sequence data from accession ID 2282, so we cannot judge whether this incongruity represents an incidental hybridization event without evolutionary consequences or a new species. Third, S. occidentalis, a morphologically well-circumscribed species, is represented in our sampling by five accessions collected in a coherent area and combining almost identical nuclear ITS sequences with more variable cpDNA sequences. Given that sequence variation in the cpDNA is usually much lower than in the nuclear ITS region, it is tempting to speculate that this divergence is likewise a consequence of cpDNA capture. We assume that much more hybridization has happened in the American species of Suaeda subg. Brezia but its traces can hardly be recognized in the “noise” of the regular sequence variation in both markers.
Ploidy levels
Our study has revealed all counted American species and populations of Suaeda subg. Brezia as polyploids (Table 2). Most likely, the same also applies to the populations which were not available for counting, with the potential exception of two species belonging to the S. prostrata group. As at least in the short run polyploidization is a unidirectional process (Scarpino et al. 2013, 2014), we can use—complementary to the molecular phylogenies—the increasing levels of polyploidy in the Suaeda groups for tracing their evolution. For convenient comparison, the polyploidy series of the Eurasian and the American representatives of the S. maritima and the S. corniculata groups of subg. Brezia are summarized and juxtaposed in Table 3. Furthermore, by combining karyological information with sequence data of nuclear and chloroplast markers, the hybrid parents or their close relatives can be detected provided they are genetically sufficiently distinct. In all groups, the presumably more ancestral diploids are restricted to Eurasia, though theoretically, at least in the S. corniculata group, they could have become extinct in the Americas.
S. maritima group In the S. maritima group, the tetraploid conditions constantly found in the American S. maritima underline the coherence with the European populations. Unexpectedly, the dekaploid S. rolandii that was interpreted by Basset and Crompton (1978) as an allopolyploid with the parents being S. maritima and S. calceoliformis proved to have not only ITS but also cpDNA sequences of S. maritima and therefore more likely it has to be considered as autopolyploid. We are aware that concerted evolution could also have caused this result, but in view of the young age as indicated by the missing or low sequence divergence from S. maritima this appears less likely. Even more surprising was our discovery of a dekaploid individual (“S. linearis X”, ID 2200) from the coast of Maine, which combined S. maritima cpDNA sequences with an ITS sequence of S. linearis (see Fig. 7). Though we collected only one solitary individual that morphologically resembled S. linearis rather than the likewise dekaploid S. rolandii, the unusually high ploidy level strongly suggests that it represents a new species. Both putative parental species occur in the same area. The two American dekaploids are most likely rather young. Interestingly, such perplexingly polyploid plants based on S. maritima have never been observed in Europe where the species occupies a much wider range, and the populations have had much more time for polyploidization.
S. corniculata group The diploid conditions found in all members of the Asian S. arctica lineage studied so far and only in them strongly support the molecular results concerning the ancestral position of that lineage which is also confirmed by molecular age estimates (see above). Tetraploidy, as the presumably next evolutionary step, occurs in some populations of the Eurasian S. corniculata and of S. patagonica from southernmost Chile only, while the majority of plants in both species show hexaploid chromosome complements.Footnote 2 From the coexistence of tetraploid and hexaploid populations in the same species and in two separate lineages, we conclude that hexaploidy was for some unknown reason favored in the group. At least the hexaploid populations of S. patagonica have most likely evolved by autopolyploidy because S. patagonica is the only extant representative of the subgenus in S America and it is most unlikely that another putative ancestor had ever reached Tierra del Fuego. However, some hexaploids may also have originated by hybridization between genetically slightly differing parental populations of the same ancestral tetraploids, thereby obscuring the separation between auto- and allopolyploids and by that eventually contributing to the actual problems in delimiting the American species of the S. corniculata group.
At first glance, tetraploid populations of S. patagonica seem to qualify as the progeny of all other American species of the S. corniculata group because there is no doubt about the plesiomorphic character of that chromosome number in the context of the related American species. However, this interpretation is conflicting with the ITS phylogeny where S. patagonica takes a basal position only in the S. linearis lineage. Considering the apparent ease of the change from tetra- to hexaploidy, it is however conceivable that the other two American lineages likewise started with tetraploid populations that later became extinct. Interestingly, the S. pannonica assemblage also includes tetraploid populations in the S. corniculata group besides the predominant hexaploids. Higher ploidy levels signaling terminal evolutionary positions in the group are known only from the octoploid S. pannonica (ID 2269, SE Europe) and from the unique (allo)dekaploid plant S. linearis X (ID 2200 from Maine, see preceding paragraph).
As general conclusion, we can confirm that like many other groups of higher plants, the American Suaeda populations have benefited from polyploidy “as a rampant evolutionary process that triggers drastic genome reorganization” (Comai 2005:836; see also Tayalé and Parisod 2013, and references therein. Whereas polyploidy in Suaeda subg. Brezia most probably contributed to the fitness of the populations and thereby to their success, our study does not give support to the recently postulated link between polyploidy and long-distance dispersal, as was deduced by Linder and Barker (2014) from their work on danthonioid grasses. In Suaeda subg. Brezia, long-distance dispersal always occurred incidentally as can be concluded from the case of S. patagonica: the longest distance was conquered by a colonizer equipped with the smallest genome. Also the first immigrants in N America were most likely diploids or tetraploids and only later higher ploidy levels were reached.
Provenance and spread of the American species of Suaeda subg. Brezia
The phylogenetic trees shown in Figs. 4 and 5 together with the molecular clock data of Schütze (2011) allow us to make some inferences on the timing of the different colonization events and the spatiotemporal evolution of Suaeda subg. Brezia in the Americas. Putative dispersal vectors as well as climatic history, ecological and morphological aspects have also to be taken into account. Our data strongly suggest Asia as the ancestral area not only of subg. Brezia but also of the S. maritima and S. corniculata groups, while the S. prostrata group which is not fully represented in the tree most likely originated in the Mediterranean area (Schütze 2011). From those places of origin, Brezia spread around the world. While Eurasia harbors 37 recognized taxa from all main groups, with centers of diversity—11 species each—in Asian Russia (Lomonosova and Freitag 2008) and Mongolia (Freitag and Lomonosova 2013), our new data show that the Americas hold the second rank with 11 recognized species, nine of them occurring in N and C America. However, the representation of the three main species groups in Eurasia and in the Americas is extremely unequal, with 16:2 in the S. maritima group, 6:2 in the S. prostrata group, and 5:10 in the S. corniculata group.
S. prostrata group The two American species of the S. prostrata group have certainly reached the continent separately. Suaeda spicata that we detected in one collection kept in the San Diego Herbarium is so similar to Mediterranean samples in morphology and in sequence data, that we do not question its S European provenance and recent anthropogenic dispersal. The origin of the Argentinean S. densiflora is obscure, but its unique genetic composition (see Fig. 7) suggests an earlier introduction. The species conquered a moderately large area that offers climatic and soil conditions similar to S Europe.
S. maritima group Two lines of evidence suggest a W European origin of the American members of the S. maritima group. First, all American populations are restricted to the N Atlantic coastline, whereas all related taxa of the S. maritima group show an Eurasian distribution. Second, the ITS and cpDNA sequences of S. maritima specimens from the opposing European and American coastlines are almost indistinguishable. The question remains when that migration happened and if it was a singular or a repeated event. At first glance an accidental introduction by the intensive shipping traffic appears to be plausible (Small 1933:469; Hultén 1971:188). According to Ball (pers. comm.), it is known that ships transporting horses and other farm animals have gathered hay from salt marshes around the ports [in England] at the beginning of a voyage and dumping the residue [with seeds] at or just outside their destination. However, most American authors consider S. maritima as being native, and some of our data also suggest an earlier colonization. As outlined above, some American populations were already involved in hybridization events with partners from remote inland areas where seeds of coastal S. maritima have a rare chance to arrive. Moreover, the tetraploid S. maritima populations in America already gave rise to two dekaploid taxa, i.e., S. rolandii and “S. linearis X” (see above). Otherwise, the sequence similarity of American and European S. maritima populations and the lack of bottleneck signatures suggest multiple recent introductions having followed an earlier colonization.
Two vectors could account for the postulated long-distance seed dispersal from Europe to N America (for a summarizing reevaluation see Milne 2006): (1) attachment to the feet of waterfowl (e.g., geese), and (2) oceanic currents which also might have contributed to the dispersal of S. maritima along European shores (Weising and Freitag 2007). By molecular studies, Kadereit et al. (2005a) showed that dispersal of salt water-resistant, viable seeds by floating on seawater played a dominant role in the extended migrations of coastal halophytes during and after glacial periods. Although the Gulf Stream follows the opposite direction from SW to NE, it appears conceivable that long-lasting easterly winds occasionally allow for floating of seeds at the sea surface from east to west. There are some other examples for amphi-Atlantic halophytes listed by Hultén (1958, 1971), as, e.g., Atriplex glabriuscula Edmondston, Glaux maritima L., Spergularia marina (L.) Griseb., Mertensia maritima (L.) Gray and Polygonum raji Bab. Hultén, however, considers these plants as remnants of formerly (before the Ice Ages) circumpolar ranges which at least in case of Suaeda maritima seems to be less likely. However, in warmer climates with a more northern distribution of S. maritima, the distance between the two disjunct areas must have been shorter. Taken together, early colonization via natural long-distance dispersal as well as later introductions by maritime trade may have contributed to the current distribution patterns.
After its successful establishment in N America, S. maritima apparently withstood the competition with the endemic Suaeda species of the S. corniculata group and conquered a long strip along the northeastern coast of N America. By autopolyploidy, allopolyploidy and introgression, S. maritima contributed to the biodiversity of Suaeda subg. Brezia in N America, but it did not change its habitat preferences and together with its derivates it occupies the same ecological niche as in Europe. It remains astonishing that despite of the much shorter distance obviously none of the numerous E Asian species of the S. maritima group ever invaded into the American continent, although it started to diversify much earlier than the S. corniculata group and contains many coastal taxa. Likewise the S African and Australian/Oceanian populations of the S. maritima group have never reached the Americas or have not survived there.
S. corniculata group Our molecular trees and karyological data consistently suggest that the American species of the S. corniculata group are derived from Asian colonizers that were closely related to the small S. arctica lineage that split from the S. maritima/S. prostrata clade in the Lower Pliocene at about 5.1–5.3 Mya. The extant species of the S. arctica lineage grow on arid high mountains of C Asia as well as in both coastal and inland areas of subarctic NE Asia where they adapted to microthermic habitats with a very short growing season. We consider it unlikely that colonization of the Americas started from such ecophysiologically specialized populations that might have originated only during the later Quaternary glacial cycles. It seems more conceivable that the ancestors of the American species grew under more favorable conditions prevailing in that period and became extinct afterwards.
The first colonizers were either diploids or tetraploids. If the molecular clock estimates are correct, the colonizers arrived in America in the Early Pleistocene (Calabrium) at about 1.6 Mya where they gave rise to the S. calceoliformis 1 lineage. They presumably arrived from NE Asia via long-distance dispersal, perhaps attached to the feet of waterfowl. We have no information about migratory bird routes in these times, but today a much used “East Asia/East Africa Flyway” that extends to Alaska is well known, in addition to the “Pacific Americas Flyway” used in the opposite direction and continuing from the Americas to the Chukotka Peninsula (Birdlife International 2014a, b). With high probability, we can exclude migration along the commonly cited Bering route used by numerous species including men (see, e.g., Hultén 1937/1972) because the temporary closing of the Bering Strait happened much later.
The phylogenetic trees seem to indicate the interior of Mexico as a “landing place” of the early colonizers and a later spread to N America. However, this scenario is not convincing because by its perennial and gypso-halophytic habit, S. mexicana 1 qualifies as a derived species compared to the other members of the group which have retained the annual and hygrohalophytic character of the more ancestral S. arctica lineage. Rather we consider S. mexicana 1 as a highly specialized relict that has derived from now extinct annuals of a more northern distribution. Also from a geographical viewpoint, a first arrival in N America seems to be more plausible.
The sequence and number of biogeographical events concerning the other lineages are ambiguous. If the various trees which show only weak or no branch support correctly reflect the phylogeny, at least one more colonization from Asia occurred. Both the S. calceoliformis 2 and the S. linearis lineages may have originated from a tetraploid invader belonging or related to the Eurasian S. pannonica assemblage. However, the uncertainties in tree topologies do not exclude the possibility that the S. pannonica assemblage originated in America and dates back to an early remigration event. Dating by molecular clock did not reveal substantial differences in the age of the three American lineages and the S. pannonica assemblage. However, the much higher diversity of the S. linearis lineage, presence of the plesiomorphic tetraploidy, geographical range and ecological versatility that considerably exceeds those of the two S. calceoliformis lineages cast doubts into the rough molecular estimates and strongly suggest a higher age as compared to the other American clades.
The well-supported and genetically homogeneous S. calceoliformis 2 lineage spread rather recently from interior N America (S. calceoliformis 2) to C Mexico (S. edulis). Likewise, the weakly supported but genetically heterogeneous S. linearis lineage probably evolved in interior N America where it is still represented by the basally branching S. occidentalis. From there, it dispersed to interior Mexico (S. pulvinata, S. mexicana 2, S. jacoensis), NW Mexico (S. puertopenascoa), S America (S. patagonica) and with the distinct S. linearis/S. esteroa subclade to the coastal marshes of N America and Mexico. Its separation into S. linearis and S. esteroa happened only in the middle Pleistocene (Ionian) at about 0.15 Mya. Along the coastlines of the Atlantic and the Mexican Gulf, more recently S. linearis split into three distinct geographical groups, and S. esteroa shows a similar pattern along the coasts of northwestern Mexico and California.
Additional remarks on the dispersal scenarios Dispersal over short, medium and long distances by water fowl has to be assumed as the most important vector for the further geographical spread of the American S. corniculata lineages. The occurrence of S. patagonica on Tierra del Fuego in southernmost S America and at the shores of saline lagoons of the Bolivian Altiplano are most striking examples of long-distance dispersal and can be correlated to the “Pacific Americas Flyway” (Birdlife International 2014b). It is also used by birds (e.g., geese) that inhabit the same habitats as the Suaeda species. Similar disjunctions on the American subcontinents were recently reviewed by Wen and Ickert-Bond (2009) and by Donoghue (2011). They likewise suggested bird dispersal as the most likely vector, though proof is rare. The same dispersal vectors probably apply to S. edulis in C Mexico that according to our molecular results is a rather recent derivate of the N American S. calceoliformis 2 lineage, and likewise to S. mexicana 1 from the S. calceoliformis 1 lineage. We also assume bird dispersal over shorter distances inside of N America where Suaeda habitats are restricted to island-like saline habitats, and for the first colonization of the coastal marshes along the Atlantic Ocean.
Along the coasts of the Pacific and the Atlantic as well as of the Gulfs of California and Mexico, short-distance dispersal by floating on seawater probably was the most effective vector, as along the European coasts (see above). The rigorous latitudinal shifts of the climatic zones during the climatic oscillations of the Pleistocene must have caused repeated migrations that enhanced diversification as can be deduced from the genetic diversity in S. linearis and S. esteroa in their essentially continuous distribution areas. While during cooling periods, the populations migrated southward, down to formerly (and today) tropical areas; in warmer intervals, the majority of the populations remigrated to the north except for individuals that had adapted to the changing temperature conditions. They built up new populations that were able to persist in subtropical and tropical environments where they even became components of mangrove ecosystems.
However, the distinct genetic discontinuities in the roughly coherent distribution areas of S. linearis and S. esteroa need to consider additional factors. Certainly, geographical barriers and differing systems of oceanic currents are important, not only as they exist today but also in glacial periods with a much lower sea level. This is obvious in S. linearis, where the most distinct genetic discontinuity was found between the populations of the Atlantic (S. linearis 1) and the Gulf coasts (S. linearis 2 and 3), which are separated from each other by the headland of S Florida. Similar barriers might have contributed to the unexpected diversity of the S. esteroa populations along the shores of NW Mexico as speculated by Ferren and Roberts (2011), but the rather scattered occurrence of salt marshes on Baja California is by itself suspicious of causing isolating effects.
Concordant discontinuities in population structure of salt marsh and intertidal animals of the N American Atlantic and Gulf coasts are well known since Avise (1992). Also the smaller but significant genetic subdivision (PP 0.95–0.99) in S. linearis 1 along the Atlantic coast between Maine and the Bahamas is paralleled in the invertebrate fauna of the salt marsh communities and was attributed to temporal latitudinal shifts (Díaz-Ferguson et al. 2010).
Besides effective dispersal vectors, we consider the genomic variability as most important precondition for the apparent success of the S. corniculata group in the Americas. It opened the path for physiological, ecological and morphological adaptations enabling its species to invade into new ecological niches though retaining their strictly halophytic constitution. From the extant species, it can be concluded that the Asian ancestors were hygrohalophytic annuals of coastal and inland habitats probably adapted to temperate conditions. While S. patagonica and some populations of S. calceoliformis had adjusted to subarctic conditions, other American species ecophysiologically adapted to habitats in warm temperate, subtropical and tropical climates. In some species, in particular, in S. esteroa, S. puertopenascoa, S. mexicana, S. jacoensis and S. pulvinata, this was associated with a transition from annual to perennial and eventually evergreen habit that allows for longer periods of photosynthesis and enhances competitiveness in a closed vegetation cover. A few species, namely S. occidentalis, S. mexicana and S. jacoensis, also acquired the ability to survive under much drier habitat conditions, thereby approaching to xero-halophytes.
Finally, it should be stated that two peculiar habitat characters might have greatly contributed to the success of the group under study: (1) salinity that strongly reduces the number of competing species, and (2) instable hydrological conditions that damage or kill the plants both in years with prolonged flooding and with permanent low water levels. The respective habitats around salty lakes, in depressions of arid areas and along seashore lagoons therefore usually carry more or less open plant communities. With their halophytic habit and opportunistic life strategy in being annual and therefore able to survive unfavorable years as dwarfed plants or seeds, most Suaeda species are perfectly adapted to those habitats. In coastal salt marshes with highly competitive perennials (e.g., Spartina sp. div., Sarcocornia pacifica [Standl.] A.J. Scott), annual Brezia species are either more or less restricted to disturbed (e.g., by erosion) microsites or they are lacking altogether, as along vast stretches of the Pacific coast.
Concluding remarks and outlook
The results obtained in the present study allow us to answer most but not all of the questions raised in the introduction: (1) ten out of 13 currently recognized Brezia species in the Americas belong to the S. corniculata group, two to the S. maritima group and one to the S prostrata group. The Americas turned out to represent the diversity center of the S. corniculata group, while the American species of the other groups just represent outposts. (2) Correspondence of the molecular phylogenies to the current taxonomic classification is weak. Only the species represented by one or few samples appear to be monophyletic, such as S. densiflora, S. puertopenascoa, S. pulvinata and S. rolandii. Some species are paraphyletic due to slight sequence divergence, such as S. edulis, S.jacoensis, S. occidentalis and S. patagonica. Two species proved to be clearly polyphyletic, indicating the presence of hidden or of confounded species. That applies to S. calceoliformis consisting of 3(4) different lineages, S. linearis and S. esteroa—which together form a monophylum in the ITS tree—each with at least two lineages, and to S. mexicana containing two lineages. (3) Several incongruities were found between the ITS and chloroplast tree, suggesting ancient lineage sorting and/or reticulate evolution. We found at least four instances of hybridization and chloroplast capture between distant lineages. (4) The Americas were colonized in five steps: The first and most successful two colonization events that founded the American S. corniculata lineages in N America started from NE Asia; later on a forerunner of S. densiflora arrived from the Mediterranean in Argentina, and S. maritima entered the northern Atlantic coast from W Europe. Only in most recent historical times, S. spicata not known from America yet was introduced into S California. Enhanced by recurrent polyploidization, commonly to hexaploidy but two times also to dekaploidy, and by gene flow, the invaders diversified and adapted to somewhat differing ecological niches. The current distribution pattern in the Americas is explained as having developed overland mainly via long- and short-distance dispersal by water fowl and along the seashores by additional or predominant drifting on seawater. Important driving forces behind the latter were the repeated latitudinal shifts of climatic zones from the late Pliocene onwards. No unambiguous “out of America” event was detected in our worldwide sampling but we cannot exclude an early remigration event to Asia that gave rise to the S. pannonica assemblage.
The molecular and karyological results delivered substantial insights into the phylogenetic and biogeographical relationships of the American taxa of Suaeda subg. Brezia, but only in few cases they fulfilled our expectations to be helpful in species delimitation. Nevertheless, they allow for suggestions towards an improved classification, together with results obtained from morphological studies in the field and on numerous herbarium specimens including most types. The initially suspected heterogeneity of the populations currently summarized under the names S. calceoliformis and S. linearis was confirmed and suggests on the one hand the reestablishment of several taxa discerned by earlier authors but lumped together in the more recent accounts since Basset and Crompton (1978). On the other hand, it also requires the description of new taxa. However, the available space does not allow an inclusion of these taxonomical aspects into the present article. We hope to complete our taxonomic account by more detailed studies in areas and herbaria that we could not visit so far. Additional work is particularly needed in populations of the two S. calceoliformis lineages, in the S. mexicana/S. jacoensis group and in the insufficiently understood taxa S. edulis, S. patagonica and S. densiflora.
Notes
1st number—ITS, 2nd number—atpB–rbcL.
Both numbers were also reported for S. calceoliformis by Ferren and Schenk (2003) but tetraploidy is most unlikely in this species because we did not find a respective record in any original publication, and all 23 counts known to us agree in 2n = 54.
References
Alvarado Reyes E, Flores-Olvera H (2013) Suaeda pulvinata (Chenopodiaceae), a new species from saline lakes of central Mexico. Willdenowia 43:300–314
Álvarez I, Wendel JF (2003) Ribosomal ITS sequences and plant phylogenetic inference. Molec Phylogen Evol 29:417–434
Avise JC (1992) Molecular population structure and the biogeographic history of a regional fauna: a case history with lessons for conservation biology. Oikos 63:62–76
Basset IJ, Crompton CW (1978) The genus Suaeda (Chenopodiaceae) in Canada. Canad J Bot 56:581–591
Birdlife International (2014a) East Asia/East Africa Flyway. http://www.birdlife.org/datazone/userfiles/file/sowb/flyways/6_East_Asia_East_Africa_Factsheet. Accessed 20 November 2014
Birdlife International (2014b) Pacific Americas Flyway. http://www.birdlife.org/datazone/userfiles/file/sowb/flyways/1_Pacific_Americas_Factsheet. Accessed 20 November 2014
Blattner FR (1999) Direct amplification of the entire ITS region from poorly preserved plant material using recombinant PCR. Biotechniques 27:1180–1186
Comai L (2005) The advantages and disadvantages of being polyploid. Nat Rev Genet 6:836–846
Cusimano N, Renner S (2014) Ultrametric trees or phylograms for ancestral state reconstruction: does it matter? Taxon 63:721–726
Díaz-Ferguson E, Robinson JD, Silliman B, Wares JP (2010) Comparative phylogeography of North American Atlantic salt marsh communities. Estuaries Coasts 33:828–839
Donoghue MJ (2011) Bipolar biogeography. Proc Natl Acad Sci USA 108:6341–6342
Doyle JJ (1992) Gene trees and species trees: molecular systematics as one-character taxonomy. Syst Bot 17:144–163
Ebrahimzadeh H, Ataei-Azimi A, Akhani H, Noori-Daloi MR (1994) Studies on the caryology of some species of the genus Suaeda (Chenopodiaceae) in Iran. J Sci Iran 5:81–88
Feliner G, Rossello JA (2007) Better the devil you know? Guidelines for insightful utilization of nrDNA in species-level evolutionary studies in plants. Molec Phylogen Evol 44:911–919
Ferren WR, Roberts F (2011) The genus Suaeda (Chenopodiaceae) and conservation of estuaries in the Baja California peninsula and Sonora, Mexico. Proc CNPS Conservation Conference. Sacramento, pp 56–70
Ferren WR, Schenk HJ (2003) Suaeda. In: Flora of North America Edit Committee (ed) Flora of North America 4. Missouri Bot Gard St. Louis, pp 390–398
Ferren WR, Whitmore SA (1983) Suaeda esteroa (Chenopodiaceae), a new species from estuaries of Southern California and Baja California. Madroño 30:181–190
Freitag H, Lomonosova MN (2006) Typification and identity of Suaeda crassifolia, S. prostrata and S. salsa, three often confused species of Suaeda sect. Brezia (Chenopodiaceae, Suaedoideae). Willdenowia 36:21–36
Freitag H, Lomonosova MN (2013) Suaeda. In: Virtual Guide to the Flora of Mongolia. http://www.greif.uni-greifswald.de/floragreif/?cat=13
Hopkins CO, Blackwell WH (1977) Synopsis of Suaeda (Chenopodiaceae) in North America. Sida 7:147–173
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17(8):754–755
Hultén E (1937/1972) Outline of the history of a biota during the Quaternary period: their evolution during and after the glacial period as indicated by the equiformal progressive areas of present plant species Thule, Stockholm, repr Cramer, Lehre
Hultén E (1958) The amphi-atlantic plants and their phytogeographical connections. Kungl Svenska Vetenskapsakademiens Handl Ser 4, vol. 7(1). Almquist & Wiksell, Stockholm
Hultén E (1971) The circumpolar plants II. Dicotyledons. Kungl Svensk Vetenskap Handl Ser 4, vol. 13(1). Almquist & Wiksell, Stockholm
Kadereit JW, Arafeh R, Somogyi G, Westberg E (2005a) Terrestrial growth and marine dispersal? Comparative phylogeography of five coastal plant species at a European scale. Taxon 54:861–876
Kadereit G, Gotzek D, Jacobs S, Freitag H (2005b) Origin and age of Australian Chenopodiaceae. Org Divers Evol 5:59–80
Kadereit G, Ackerly D, Pirie MD (2012) A broader model for C4 photosynthesis evolution in plants inferred from the goosefoot family (Chenopodiaceae s.s.). Proc R Soc London B 279:3304–3311
Kapralov MV, Akhani H, Voznesenskaya EV, Edwards GE, Franceschi VR, Roalson EH (2006) Phylogenetic relationships in the Salicornioideae/Suaedoideae/Salsoloideae s.l. (Chenopodiaceae) clade and a clarification of the phylogenetic position of Bienertia and Alexandra using multiple DNA sequence datasets. Syst Bot 31:571–585
Krahulcová A, Tomšovic P (1997) Ploidy levels in some European representatives of the Suaeda maritima group. Preslia 69:327–332 (in Czech)
Krapp F (2013) Phylogenie und Evolution der Gattung Dyckia (Bromeliaceae). Dissertation. University of Kassel, Germany
Lee JS, Park DS, Ihm BS, Lee WJ (2007) Taxonomic reappraisal on Suaeda australis (Chenopodiaceae) in Korea based on the morphological and molecular characteristics. J Pl Biol 50:605–614
Linder HP, Barker NP (2014) Does polyploidy facilitate long-distance dispersal? Ann Bot (Oxford) 113:1175–1183
Lomonosova MN (2011) Chromosome numbers, taxonomy and distribution of the subgenus Brezia (Suaeda, Chenopodiaceae). Turczaninowia 14:45–52 (in Russian)
Lomonosova M, Freitag H (2003) A new species of Suaeda (Chenopodiaceae) from the Altai. Willdenowia 33:139–147
Lomonosova M, Freitag H (2008) The genus Suaeda (Chenopodiaceae) in Asian Russia. Rastitel´nyj Mir Aziatskoj Rossii 2:12–19 (in Russian)
Lomonosova MN, Freitag H (2009) Chenopodiaceae. In: Marhold K (ed) IAPT/IOPB chromosome data 8, vol. 58. Taxon, pp 1284
Lomonosova MN, Shaulo DN (2010) Karyology of the Siberian representatives of the family Chenopodiaceae. Bot Zhurn (Sankt-Peterbourg) 95:422–426 (in Russian)
Lomonosova MN, Krasnikov AA, Krasnikova SA (2003) Chromosome numbers of the Chenopodiaceae family members of the Kazakhstan flora. Bot Zhurn (Sankt-Peterbourg) 88:134–135 (in Russian)
Lomonosova MN, Krasnikova SA, Krasnikov AA, Sukhorukov AP, Bananova VA, Pavlova NS (2005) Chromosome numbers of Chenopodiaceae species from Russia and Kazakhstan. Bot Zhurn (Sankt-Peterbourg) 90:1132–1134 (in Russian)
Lomonosova MN, Yusupova D, Akopyan JA (2007) Chromosome numbers of the Suaeda (Chenopodiaceae) representatives. Bot Zhurn (Sankt-Peterbourg) 9:1077–1078 (in Russian)
Lomonosova MN, Brandt R, Freitag H (2008) Suaeda corniculata (Chenopodiaceae) and related taxa from Eurasia. Willdenowia 38:81–109
Lorz A (1937) Cytological investigations on five chenopodiaceous genera with special emphasis on chromosome morphology and somatic doubling in Spinacia. Cytologia 8:241–276
Löve A, Löve D (1982) Reports. In: Löve A (ed) IOPB chromosome number reports LXXIV, vol. 31. Taxon, pp 120–126
Maddison WP (1997) Gene trees in species trees. Syst Biol 46:523–536
Milne RI (2006) Northern Hemisphere plant disjunctions: a window on Tertiary land bridges and climate change? Ann Bot (Oxford) 98:465–472
Moore DM (1981) Chromosome numbers of Fuegean angiosperms. Bol Soc Brot Ser 2(53):995–1012
Mulgura ME (1999) Catálogo de las plantas vasculares de la Républica Argentina 2. St. Louis
Müller J, Müller K, Quandt D (2011) PhyDE—Phylogenetic data editor. Version 0.9971. Program distributed by the author
Mulligan GA, Cody WJ (1973) Chenopodiaceae. In: Löve A (ed) IOPB Chromosome number reports XL, vol. 22. Taxon, pp 290
Naciri Y, Linder HP (2015) Species delimitation and relationships: the dance of the seven veils. Taxon 64:3–16
Noguez-Hernández R, Carballo-Carballo A, Flores-Olvera H (2013) Suaeda edulis (Chenopodiaceae), una nueva especie de lagos salinos del centro de México. Bot Sci (Mexico) 91:19–25
Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University
Pedrol J, Castroviejo S (1988) A proposito del tratamiento taxonomico y nomenclatural del genero Suaeda Forsskål ex Scop. (Chenopodiaceae) en “Flora Iberica”. Anales Jard Bot Madrid 45:93–102
Pirie MD, Humphreys AM, Barker NP, Linder HP (2009) Reticulation, data combination, and inferring evolutionary history: an example from Danthonioideae (Poaceae). Syst Biol 58:612–628
Probatova NC, Rubyka EG, Sokolovskaya AP (1998) Chromosome numbers in vascular plants from islands of Peter the Great Bay and Murayava-Amurskiy Peninsula (Primorsky Krai). Bot Zhurn (Sankt-Peterbourg) 83:125–130 (in Russian)
Scarpino SV, Hunt PJ, Garcia-De-Leon FJ, Juenger TE, Schart M, Kirpatrick M (2013) Evolution of a genetic incompatibility in the genus Xiphophorus. Molec Biol Evol 30:2302–2310
Scarpino SV, Levin DA, Meyers LA (2014) Polyploid formation shapes flowering plant diversity. Amer Naturalist 184:456–465
Schütze P (2011) Molekulare Systematik der Gattung Suaeda (Chenopodiaceae) und Evolution des C4-Photosynthesesyndroms. Dissertation, University of Kassel, Germany
Schütze P, Freitag H, Weising K (2003) An integrated molecular and morphological study of the subfamily Suaedoideae Ulbr. (Chenopodiaceae). Pl Syst Evol 239:257–286
Shaw J, Lickey EB, Schilling EE, Small RL (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Amer J Bot 95:275–288
Small JK (1933) Manual of the Southeastern Flora. University of N Carolina Press, Chapel Hill
Smirnov YA (1968) Accelerated method for studying somatic chromosomes in fruit trees. Tsitologiya 10:1132–1134 (in Russian)
Swofford DL (2003) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.0 Beta 10. Sinauer Associations, Sunderland, Massachusetts
Tayalé A, Parisod C (2013) Natural pathways to polyploidy in plants and consequences for genome reorganization. Cytogenet Genome Res 140:79–96
Thiers B (2012) Index Herbariorum: a global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium. http://sciweb.nybg.org/science2/IndexHerbariorum.asp and http://sweetgum.nybg.org/ih/[23.XI.2012]
Watson MC, Ferren WR (1991) A new species of Suaeda (Chenopodiaceae) from coastal Northwestern Sonora, Mexico. Madroño 38:30–36
Weising K, Freitag H (2007) Phylogeography of halophytes from European coastal and inland habitats. Zool Anz 246:279–292
Wen J, Ickert-Bond SM (2009) Evolution of the Madrean Tethyan disjunctions and the North and South American amphitropical disjunctions in plants. J Syst Evol 47:331–348
Yu Y, Harris AJ, He XJ (2010) S-DIVA (Statistical Dispersal-Vicariance Analysis): a tool for inferring biogeographic histories. Molec Phylogen Evol 56:848–850
Yu Y, Harris AJ, Chr Blair, He X (2015) RASP (Reconstruct ancestral state in phylogenies): a tool for historical biogeography. Molec Phylogen Evol 87:46–49
Zakhar’eva OI (1990) Suaeda olufsenii Pauls. In: Takhtajan A (ed) Numeri chromosomatum Magnoliophytorum Florae URSS 1. Aceraceae—Menyanthaceae. Nauka, Leningrad (in Russian)
Acknowledgments
With gratitude, we acknowledge the generous support of a large number of curators for providing herbarium specimens on loan or permitting removal of samples for molecular study in their institutions. This applies in particular to ALA, BKL, CAS, DAO, GR, IEB, LPB, NY, RENO, RSA, SD, TEX, UCSB, and UTC. Other colleagues were helpful by collecting and sending samples taken from herbarium material, as P. W. Ball (Toronto) and C. B. Villamil (Buenos Aires) or fresh material including seeds, as E. Dominguez (Punta Arenas), W. R. Ferren (Santa Barbara), S. Pfanzelt (Mainz), F. G. Schröder (Göttingen), N. Schütz (Stuttgart) and S. Zamudio (Pátzcuaro), E. Nikolin (Yakutsk); and M. Kucev (Barnaul) for providing each two ITS sequences of S. “jacutica” and S. arctica. We are also thankful to W. R. Ferren for joining and guiding our field trips in New Jersey and to J. Schenk (Fullerton), F. Roberts (San Louis Rey) and M. R. Sharifi (Long Beach) in California, as well as for pertinent discussions with the late S. E. Clemants (Brooklyn) and many others. Very kindly W. R. Ferren, E. Dominguez and H. Flores-Olvera also contributed by allocation of images, the latter also by unpublished chromosome counts. We thank the gardeners in Kassel University for their engagement in professionally cultivating American Suaeda plants. We also thank an anonymous reviewer and the editor for useful comments on earlier versions of the manuscript. The project was financially supported by the German Research Foundation (DFG) (Grant WE 1830/7-1 to K. Weising and H. Freitag) and by the Russian Foundation for Basic Research (Grant 12-04-00746 to M. Lomonosova).
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: Marcus Koch.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Brandt, R., Lomonosova, M., Weising, K. et al. Phylogeny and biogeography of Suaeda subg. Brezia (Chenopodiaceae/Amaranthaceae) in the Americas. Plant Syst Evol 301, 2351–2375 (2015). https://doi.org/10.1007/s00606-015-1233-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-015-1233-y