Abstract
A photocathode is described for the determination of microRNA-21 by using CuInS2 as an active photocathode material. Exonuclease III assisted target recycling amplification was employed to enhance the detection sensitivity. The TATA-binding protein (TBP) was applied to enhance steric hindrance which decreases the photoelectrochemical intensity. This strategy is designed by combining the anti-interference photocathode material, enzyme assisted target recycling amplification and TBP induced signal off, showing remarkable amplification efficiency. Under the optimized conditions, the detection limit for microRNA-21 is as low as 0.47 fM, and a linear range was got from 1.0 × 10−15 M to 1.0 × 10−6 M.

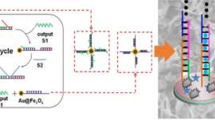
Schematic representation of sensitive photoelectrochemical detection of microRNA-21.CuInS2 is used as an active photocathode material. Combined Exonuclease III assisted target recycling amplification and TATA-binding protein decreased of photoelectrochemical intensity, the detection limit was 0.47 fM with good selectivity. (miR-21: microRNA-21; CS: chitosan).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The photoelectrochemical (PEC) process refers to photovoltaic conversion of photoelectrochemically active material from electron excitation to charge transfer under an applied light [1, 2]. Due to its separate source for photoirradiation and electrochemical detection, PEC assay has received substantial attention due to its low background signal and high sensitivity [3, 4].
From the way of electron transmission, PEC assay can be classified into photoanode assay and photocathode assay [5]. The n-Type semiconductors such as CdSe, CdS, ZnO and TiO2 [6, 7] are mainly used in photoanode assay. These materials rely on the ejection of electrons from the conduction-band to the electrode, which supply of electrons from electron donors in solution. However, the presence of sacrificial electron donor may compete with reductive ingredient (for example ascorbic acid, dopamine, H2O2 and thio compounds). In this case, the false positive signal can be appeared when the detection is carried out in the real samples [8,9,10]. The p-type semiconductors (such as CuS, PbS, BiOI and CuInS2) are always used as photoactive species for photocathode assay. The electrons transfer from the conduction-band to electron acceptor, and then the electrons are supplied from the electrode to the valence-band holes. In this process, electron acceptor (commonly use dissolved O2) instead of electron donor is required for cathodic photocurrent. Hence, the reductive component in the complex samples cannot interfere with the cathodic photocurrent. On the other hand, holes are used as carriers for electron acceptor in photocathode assay, which lead to lower photocurrent intensity compared with that of photoanode assay [11, 12]. Thus, highly active photocathode material has attracted much attention as enhancing the detection sensitivity. Among these p-type semiconductors, CuInS2 is highly attractive due to its small band gap and a large optical absorption coefficient [13, 14].
From the aspect signal format, the main established strategies for PEC assay can be classified as steric hindrance [15], biocatalytic precipitation [16], in situ generation of electron donor/acceptor [17], quenching effect [18], DNA conformational change [19] and introduction of photoelectric active materials [20]. Almost all these strategies utilize DNA for constructing PEC methods because DNA strands can bind with target molecular strongly and specifically. The interaction between DNA and protein plays important roles in controlling a variety of biological activities. Several works have been carried out on TATA-binding protein (TBP), which can bind to TATA sequence specifically [21,22,23]. With the advantage of the steric hindrance effect of TBP and easy amplification effect of DNA, a cathodic PEC method for detecting microRNA-21 was designed. In this strategy, CuInS2 is used as an active photocathode material and TBP is employed to enhance steric hindrance.
It has been reported that microRNA-21 is upregulated in many cancers, such as lung cancer, breast cancer and pancreatic cancer et al. [24]. However, it is difficult for the microRNA detection due to the instability, low content and short length of microRNA, as well as the similar sequence in microRNA families [25]. Various kinds of methods were designed for microRNA detection, which had difficulty meeting the requirements for high sensitivity [26,27,28]. To increase the detection sensitivity, exonuclease aided recycling was applied here. At the same time, target RNA was converted to DNA for further detection. The principle of PEC method was described in Scheme 1. Firstly, cathodic photoelectrochemical materials CuInS2 microflowers were dropped on the ITO glass, and heated to give a dense film. Then chitosan solution was added which provides amino groups for later modification and protects the CuInS2 on the ITO electrode. Subsequently, carboxyl group modified DNA2 was coupled on the electrode after reaction with chitosan through amide bond. If hairpin DNA1 is recognized by microRNA-21, the enzyme Exo III cuts off the blunt 3′ terminus of DNA1. This triggers the recycling of microRNA-21 and the release of DNA3 which can hybridize with DNA2 on the electrode. Finally, the DNA2-DNA3 duplex is recognized by TBP. The decrease of photocurrent caused by steric hindrance can be used to quantify microRNA-21.
Experimental section
Materials and reagents
ITO electrodes were provided by South China Science and Technology Co., Ltd. (Yiyang, China) (website: https://www.h-nxc.com), with sheet resistance ≤10 Ω/square. Copper Chloride (CuCl) and indium chloride (InCl3) were purchased from Sigma-Aldrich (website: https://www.sigmaaldrich.com/china-mainland.html). Thiourea (H2NCSNH2) and chitosan (CS) with degree of deacetylation >95.0% were the products of Aladdin Reagent Co., Ltd. (Shanghai, China) (website: http://www.aladdin-e.com). TATA-binding protein (TBP) and Exonuclease III (Exo III) were ordered from Sangon Biotech Co., Ltd. (Shanghai, China) (website: http://www.sangon.com). All other reagents used in the experiment were analytical grades and used directly. DNA oligonucleotides were gained from Sangon Biotech Co., Ltd. (Shanghai, China) (website: http://www.sangon.com) and the base sequences were as followed:
DNA1: 5’-CCTCGACTGAGGTATAAAAGTCCTCCTCAGTCTGATAAGCTA-3’;
DNA2: 5’COOH(CH2)-AG GACTTTTATACCTCAGTCGAGG-3’;
DNA3: 5’-CCTCGACTGAGGTAT AAAAGTCCT-3’;
microRNA -21: 5’-UAGCUUAUCAGACUGAUGUUGA-3’;
M1 microRNA -21: 5’-UAGCUUAUAAGACUGAUGUUGA-3’;
M2 microRNA -21: 5’-UAGCUUAUAACACUGAUGUUGA-3’;
M3 microRNA -21: 5’-UAGCUUAUAACCCUGAUGUUGA-3’;
miR-155: 5’-UUAAUGCUAAUCGUGAUAGGGGU-3’;
TBA: 5’-GGTTGGTGTGGTTGG-3’;
Apt-PDGF: 5’-TCAGGCTACGGCACGTAGAGCATCACCATGATCCTG-3’
Instruments
The electrochemical impedance spectroscopy (EIS) and photocurrent were performed on an electrochemical workstation (Zahner Zennium, Germany) using ITO conductive glass, saturated Ag/AgCl and Pt wire as working electrode, reference electrode and auxiliary electrode, respectively. The UV-vis absorption spectra were taken using a Cary 50 UV/vis-NIR spectrophotometer (Varian). Powder X-ray diffraction (XRD) patterns were recorded on an X-ray diffractometer with monochromatized Cu Kα radiation (Brook AXS, Germany). Scanning electron microscopy (SEM) image was taken with a JEM-7500F scanning electron microscope (JEOL Ltd., Japan).
Preparation of CuInS2 microflowers and modification of ITO electrode
CuInS2 microflowers were synthesized according to previous method with a little modification [29]. Briefly, to 100 mL Teflon-lined autoclave, 40 mL of the ethylene glycol solutions containing 0.03 M InCl3, 0.03 M CuCl, and 0.12 M H2NCSNH2 was added. The mixture was heated at 200 °C for 24 h, followed by washing with ethanol and deionized water and dried at 100 °C over night to give power products.
The ITO electrodes were ultrasonically cleaned in acetone, 2.0 M NaOH and deionized water successively for 20 min each, and then dried with nitrogen gas.
12.0 mg CuInS2 product was dissolved in 2.0 mL deionized water and ultrasonicated to obtain homogeneous suspension. Then 20.0 μL suspension was coated onto a bare ITO slices and dried at room temperature. The gained electrode was heated at 170 °C for 2.0 h followed by gradual cooling to room temperature. Subsequently, 20 μL of 0.1 wt% CS solution containing 1% acetic acid was spread onto the electrode. Finally, the electrode was dried at 50 °C, and washed with deionized water for three times to give ITO/CuInS2/CHIT electrode.
Detection of microRNA −21
Carboxyl functionalized DNA2 was assembled on ITO electrode surface through amidation coupling reactions. Firstly, 10 μL DNA2 was mixed with 90 μL PB buffer containing 7.0 mg EDC and 3.5 mg NHS. Then 25 μL mixture was spread on the electrode surface and incubated at 4 °C overnight. After washing with 10 mM PB, ITO/CuInS2/CHIT/DNA2 electrode was got.
At the same time, Exo III-assisted amplification was performed using 66 mM Tris-HCl (containing 0.66 mM MgCl2, pH = 8.0) as a reaction buffer. Specifically, 500 nM probe DNA1 was incubated with different concentration of target microRNA -21. 60 U Exo III was added to the mixture and incubate at 37 °C for 2 h. Then the enzyme was deactivated after heating at 75 °C for 10 min. Subsequently, 25 μL of mixture was dropped on the electrode surface and incubated for 2 h at 37 °C. Finally, 25 μL of 1000 ng mL−1 TBP was added onto the surface of the above electrode and incubated at 37 °C for 1.0 h in a humid condition. After each modification, the electrode was thoroughly washed for three times. The photocurrent measurements were performed in 0.1 M PB at a potential of 0 V. An excitation light source with a wavelength of 415 nm was switched on and off every 20 s.
Results and discussion
Characterization of materials
The gained CuInS2 was characterized by SEM. In Fig. 1a, spherical particles of microflowers morphology were observed with average size of 1.5–2.0 μm. Such rough surface caused by the layer structure of CuInS2 provides a lot of surface area for photogenerated electron exchange. As a result, PEC signal can be enhanced accordingly. The crystal structure of gained CuInS2 microflowers was further identified by XRD. As presented in Fig. 1b, five diffraction peaks are observed, which are assigned to the (112), (004), (204/220), (312), (332) crystal planes of CulnS2 [JCPDS Card No. 27–0159]. So, pure crystalline CulnS2 was successfully synthesized. These sharp peaks indicate the excellent crystallinity character of synthesized CulnS2.
The feasibility of the designed strategy
To illustrate the feasibility of the PEC assay, the photocurrent responses for the assemble processes were measured in 0.1 M PB (pH 7.4). As presented in Fig. 2, a remarkable photocathode response of ITO/CuInS2 is showed, due to the efficient charge transport and broad visible light absorption of the CuInS2 (curve a) [15]. After blocking with CS, which can protect the CuInS2 from dissolving to the detection solution as well as providing amino-group for further modification, the photocurrent intensity decreased dramatically (curve b). When carboxylated DNA2 was modified on the electrode (curve c) and hybridized with DNA3 (curve d), the photocurrent signal decreased slightly owning to the steric hindrance effect of DNA. After incubating with TATA-binding protein (TBP), the photocurrent intensity decreases obviously (curve e). This might be explained that TBP can bend the TATA sequence at an 80 degree angle through inserting amino acid side chains to base pairs of dsDNA sequence [22, 23], which blocked the transfer of electrons and led to a sharp decay in photocurrent intensity. Therefore, the characteristics of the photocurrent variation demonstrate the feasibility of the designed method.
Photocurrent responses of different ITO electrodes recorded in 0.10 M PB at a bias potential of 0 V for (a) ITO/CuInS2; b ITO/CuInS2/CS; c ITO/CuInS2/CS/DNA2; d ITO/CuInS2/CS/DNA2/DNA3; e ITO/CuInS2/CS/DNA2/DNA3/TBP incubated with 1.0 × 10−6 mol/L microRNA −21. The PEC tests were performed in 0.10 M PB (pH = 7.4) at a bias voltage of 0 V with 415 nm light excitation
The fabrication step of the PEC nanoprobe was also characterized by EIS. As exhibited in Fig. 3, the unmodified ITO shows a small semicircle (curve a). After modification of CuInS2 microflowers, the Ret value increased (curve b), indicating the poor conductivity of CuInS2. When CHIT was coated, the Ret value further increased due to the non-conductivity of chitosan (curve c). The assembly of the carboxyl functionalized DNA2 (curve d) and hybridization with DNA3 (curve e) inhibited the charge transfer furtherly, owning to the steric hindrance and the electrostatic repulsion between the negatively charged DNA2/DNA3 phosphoric acid backbone and the negatively charged Fe(CN)63−/4−. After recognition by TATA binding protein, the Ret value increased greatly (curve f), indicating the recognition between the protein and its aptamer as well as subsequent steric hindrance. Therefore, the characteristics of the ESI also demonstrate the successful preparation the electrode interface.
Nyquist plots of different electrodes in 0.1 mol/L KCl containing 5.0 mM K3Fe(CN)6/K4Fe(CN)6 (1:1): a bare ITO substrate; b ITO/CuInS2; c ITO/CuInS2/CS; d ITO/CuInS2/CS/DNA2; e ITO/CuInS2/CS/DNA2/DNA3; f ITO/CuInS2/CS/DNA2/DNA3/TBP incubated with 1.0 × 10−6 M microRNA-21. The frequency range for EIS measurements was from 0.01 to 100,000 Hz at an applied potential of 0.2 V
The characterization of enzymatic cleavage by polyacrylamide gel electrophoresis (PAGE)
Exo III has been widely used to recycle the target with signal amplification. Exo III recognizes a blunt 3′ terminus in double stranded nucleic acid substrate including duplex DNA-RNA and DNA-DNA [30,31,32,33]. The DNA strand in the duplex with 3′-flat or concave terminal is digested by Exo III to produce short oligonucleotide fragments, which released the target RNA with recycle for further improving sensitivity in various assay. To demonstrate the enzymatic cleavage process, polyacrylamide gel electrophoresis (PAGE) was applied. As shown in Fig.S1, lane a and lane b represent stem loop probe DNA1 and target microRNA −21, respectively. The bright band in lane c is attributed to the hybridization product of DNA1 with target microRNA −21. When incubating the DNA1/ microRNA −21 hybrid with Exo III, DNA1 was cleaved from blunt 3′ terminus to release the DNA3 and intact microRNA −141 simultaneously, which has similar number of bases (lane d). So, the image of PAGE shows the cycle amplification aided by Exo III digestion was performed successfully.
Sensitivity of the PEC assay
The concentrations of Exo III and TBP were optimized to improve the performance of designed PEC assay. The results (Fig. S2, supporting information) suggest an optimized concentration of 60 U for Exo III and 1000 ng·mL−1 for TBP. Under these optimum conditions, target microRNA 21 was detected quantitatively. As shown in Fig. 4, with increasing concentration of microRNA, the produced photoelectrochemical intensity of ITO/CuInS2/CHIT/DNA2/DNA3 decrease accordingly. The photocurrent response is proportional to the logarithm of microRNA concentrations across the range from 1.0 × 10−15 M to 1.0 × 10−6 M. The linear regression equation was expressed as I (nA) = −0.0937 lgC microRNA (M) -0.202, and the correlation coefficient (R2) was 0.999. The detection limit of microRNA −21 is estimated to be 0.47 fM (S/N = 3). A series of eleven repetitive measurements of 1.0 × 10−10 M microRNA 21 is used for estimating the precision, and the standard deviation (SD) is 0.27. Compared with some previous reports for microRNA −21 determination (Table 1), the designed PEC aptamer method shows a preferable detection performance. In contrast, an additional control experiment was performed without Exo III assisted amplification. DNA3 was employed as the cleaved product of the duplex strand by Exo III. The PEC signal was gained without Exo III by direct titration of DNA3 on the DNA2 modified electrode surface. The detection limit was approximately in the nM range (Fig. S3), which was nearly 6–7 orders of magnitude poorer than that of Exo III catalyzed amplification.
a Photocurrent responses of the designed assay incubated with different concentrations of microRNA-21: (from a to j 1.0 × 10−6, 1.0 × 10−7, 1.0 × 10−8, 1.0 × 10−9, 1.0 × 10−10, 1.0 × 10−11, 1.0 × 10−12, 1.0 × 10−13, 1.0 × 10−14, 1.0 × 10−15 M). b Calibration plot for microRNA-21 on the PEC assay. All photocurrent responses were measured in 0.10 M PB (pH = 7.4) at a bias voltage of 0 V with 415 nm light excitation. Error bars show the standard deviations of three replicate experiments
Specificity of the PEC assay
To investigate the selectivity of the designed PEC assay, other ssDNA or RNA including miR-155, thrombin aptamer (TBA) and platelet-derived growth factor aptamer (Apt-PDGF) were tested as an interfering substance. From the results shown in Fig. 5a, the relative photocurrent change for 1.0 μM microRNA −21 is obvious. However, as for TBA, miR-155 and Apt-PDGF in the same concentration, the relative photocurrents response was similar to that of the blank, indicating the excellent selectivity of the designed PEC method. When 1.0 μM microRNA −21 was mixed with equal amount of TBA, miR-155 and Apt-PDGF, the relative photocurrent change was almost the same as that of pure microRNA −21. The results demonstrate that other DNA and RNA do not interfere with the detection of target object. (I0 referred to the photocurrent of assembled electrode ITO/CuInS2/CHIT/DNA2, I referred to the final photocurrent response after addition different concentration of target microRNA −21.)
a Selectivity experiment for microRNA-21 detection against microRNA-155, TBA, Apt- PDGF, and mixture. b The specificity of our assay in discriminating other RNA strands with one, two or three nucleotide mutations (M1 miR-21, M2 miR-21 and M3 miR-21). c Effects of other proteins on the detection of 1000 ng mL−1 TBP in 0.10 M PB (pH = 7.4) at a bias voltage of 0 V with 415 nm light excitation. The bars represent 2000 ng mL−1 of CEA, Lyso, AFP, HAS, PSA, Thro and a mixture of the seven kinds of proteins, respectively. The concentrations of other samples are 1.0 μM. The error bars represent the standard deviations for three replicates
To investigate whether the mutation can be distinguished from the target microRNA, three different microRNAs were employed, including single-base mismatched microRNA-21 (M1 microRNA-21), two-base mismatched microRNA-21 (M2 microRNA-21) and three-base mismatched microRNA-21 (M3 microRNA-21). As displayed in Fig. 5b, the relative photocurrent change is occurred by the perfect target. The PEC signal outputs of M1 microRNA-21, M2 microRNA-21and M3 microRNA-21 are similar to the blank signal as well. These results verify that one or few nucleotides mutation is easily distinguished by our designed strategy.
In addition, the interference study was evaluated with other proteins. The modified electrode was incubated in sample solutions containing different interfering proteins such as carcinoembryonic (CEA), lysozyme (Lyso), α-fetoprotein (AFP), human serum albumin (HSA), prostate specific antigen (PSA) and thrombin (Thro). The mixed solution of these proteins was also detected under the same conditions. Figure 5c shows that none of other proteins exhibit an obvious signal change, expect for the TBP and mixed sample. This phenomenon is due to the high affinity of the TBP toward the TATA box.
Practical application in the human serum
The practical application of the designed method in the human serum was tested by standard addition method. Healthy human serum samples spiked with different concentrations of target microRNA −21 were tested. The gained recoveries are found to vary from 97.1% to 101.6% (Table 2), the RSDs are less than 4.3%, demonstrating the excellent potential of the designed strategy in complex samples.
Conclusion
A novel PEC method was designed for the detection of microRNA. Several advantages of our method have been demonstrated: 1) A photocathode assay using CuInS2 as a photoactive species is synthesized, which can perform well in complex sample without the interference of reductive ingredient; 2) Exo III assistant amplification led to the recycling of the target microRNA and enhanced the detection sensitivity; 3) In this method, DNA–protein bioconjugates were used to enhance steric hindrance. One major limitation of this strategy is that the DNA probes immobilized on the electrode exhibited weak PEC signal reduction, which reducing the range of PEC signal response to TBP. The photocurrent variation produced by this recognition process can be realized easily. What’s more, after carefully designing the recognition sequence, the designed PEC platform based on DNA amplification can also be used to detect other targets including DNA, protein and small molecule.
References
Zhao WW, Xu JJ, Chen HY (2015) Photoelectrochemical bioanalysis: the state of the art. Chem Soc Rev 46:729–741
Qu YQ, Duan XF (2013) Progress, challenge and perspective of heterogeneous photocatalysts. Chem Soc Rev 42:2568–2580
Zhang XR, Guo YS, Liu MS, Zhang SS (2013) Photoelectrochemically active species and photoelectrochemical biosensors. RSC Adv 3:2846–2857
Zhang N, Zhang L, Ruan YF, Zhao WW, Xu JJ, Chen HY (2017) Quantum-dots-based photoelectrochemical bioanalysis highlighted with recent examples. Biosens Bioelectron 94:207–218
Gill R, Zayats M, Willner I (2008) Semiconductor quantum dots for bioanalysis. Angew Chem 47:7602–7625
Yan K, Wang R, Zhang JD (2014) A photoelectrochemical biosensor for o-aminophenol based on assembling of CdSe and DNA on TiO2 film electrode. Biosens Bioelectron 53:301–304
Zhao WW, Shan S, Ma ZY, Wan LN, Xu JJ, Chen HY (2013) Acetylcholine esterase antibodies on BiOI nanoflakes/TiO2 nanoparticles electrode: a case of application for general photoelectrochemical enzymatic analysis. Anal Chem 85:11686–11690
Wang GL, Liu KL, Dong YM, Wu XM, Li ZJ, Zhang C (2014) A new approach to light up the application of semiconductor nanomaterials for photoelectrochemical biosensors: using self-operating photocathode as a highly selective enzyme sensor. Biosens Bioelectron 62:66–72
Wang GL, Shu JX, Dong YM, Wu XM, Zhao WW, Xu JJ, Chen HY (2015) Using G-quadruplex/hemin to "switch-on" the cathodic photocurrent of p-type PbS quantum dots: toward a versatile platform for photoelectrochemical aptasensing. Anal Chem 87:2892–2900
Li Y, Chen FT, Luan ZZ, Zhang XR (2018) A versatile cathodic "signal-on" photoelectrochemical platform based on a dual-signal amplification strategy. Biosens Bioelectron 119:63–69
Zhao WW, Dong XY, Wang J, Kong FY, Xu JJ, Chen HY (2012) Immunogold labeling-induced synergy effect for amplified photoelectrochemical immunoassay of prostate-specific antigen. Chem Commun 48:5253–5255
Kong C, Zhang G, Li Y, Li DW, Long YT (2013) Plasmon-enhanced photocurrent monitoring of the interaction between porphyrin covalently bonded to graphene oxide and adenosine nucleotides. RSC Adv 3:3503–3507
Fan CC, Shi XM, Zhang JR, Zhu JJ (2016) Cathode photoelectrochemical immunosensing platform integrating photocathode with photoanode. Anal Chem 88:10352–10356
Song Z, Fan GC, Li ZM, Gao FX, Luo XL (2018) Universal design of selectivity-enhanced photoelectrochemical enzyme sensor: integrating photoanode with biocathode. Anal Chem 90:10681–10687
Jiang XY, Zhang L, Liu YL, Yu XD, Liang YY, Qu P, Zhao WW, Xu JJ, Chen HY (2018) Hierarchical CuInS2-based heterostructure: application for photocathodic bioanalysis of sarcosine. Biosens Bioelectron 107:230–236
Zheng YN, Liang WB, Xiong CY, Yuan YL, Chai YQ, Yuan R (2016) Self-enhanced ultrasensitive photoelectrochemical biosensor based on nanocapsule packaging both donor-acceptor-type photoactive material and its sensitizer. Anal Chem 88:8698–8705
Zhao WW, Ma ZY, Yu PP, Dong XY, Xu JJ, Chen HY (2012) Highly sensitive photoelectrochemical immunoassay with enhanced amplification using horseradish peroxidase induced biocatalytic precipitation on a CdS quantum dots multilayer electrode. Anal Chem 84:917–923
Shen QM, Han L, Fan GH, Zhang JR, Jiang L, Zhu JJ (2015) “Signal-on” photoelectrochemical biosensor for sensitive detection of human T-cell lymphotropic virus type II DNA: dual signal amplification strategy integrating enzymatic amplification with terminal deoxynucleotidyl transferase-mediated extension. Anal Chem 87:4949–4956
Li MJ, Zheng YN, Liang WB, Yuan R, Chai YQ (2017) Using p-type PbS quantum dots to quench photocurrent of fullerene−AuNP@MoS2composite structure for ultrasensitive photoelectrochemical detection of ATP. ACS Appl Mater Interfaces 9:42111–44212
Zhang XR, Xu YP, Zhao YQ, Song W (2013) A new photoelectrochemical biosensors based on DNA conformational changes and isothermal circular strand-displacement polymerization reaction. Biosens Bioelectron 39:338–341
Shi XM, Fan GG, Shen QM, Zhu JJ (2016) Photoelectrochemical DNA biosensor based on dual-signal amplification strategy integrating inorganic−organic nanocomposites sensitization with λ-exonuclease-assisted target recycling. ACS Appl Mater Interfaces 83:5091–35098
Ma ZY, Ruan YF, Zhang N, Zhao WW, Xu JJ, Chen HY (2015) A new visible-light-driven photoelectrochemical biosensor for probing DNA–protein interactions. Chem Commun 51:8381–8384
Ma ZY, Ruan YF, Xu F, Zhao WW, Xu JJ, Chen HY (2016) Protein binding bends the gold nanoparticle capped DNA sequence: toward novel energy-transfer-based photoelectrochemical protein detection. Anal Chem 88:3864–3871
Oudeng G, Au M, Shi JY, Wen CY, Yang M (2018) One-step in situ detection of miRNA-21 expression in single cancer cells based on biofunctionalized MoS2 nanosheets. ACS Appl Mater Interfaces 10:350–360
Fang CS, Kim KS, Yu B, Jon S, Kim MS, Yang H (2017) Ultrasensitive electrochemical detection of miRNA-21 using a zinc finger protein specific to DNA-RNA hybrids. Anal Chem 89:2024–2031
Wang JX, Zhang LL, Lu LP, Kang TF (2019) Molecular beacon immobilized on graphene oxide for enzyme-free signal amplification in electrochemiluminescent determination of microRNA. Microchim Acta 186:2563–2569
Mahani M, Mousapour Z, Divsar F, Nomani A, Ju HX (2019) A carbon dot and molecular beacon based fluorometric sensor for the cancer marker microRNA-21. Microchim Acta 186:132
Tang YF, Liu MX, Zhao ZL, Li Q, Liang XH, Tian JN, Zhao SL (2019) Fluorometric determination of microRNA-122 by using ExoIII-aided recycling amplification and polythymine induced formation of copper nanoparticles. Microchim Acta 186:133
Liu NN, Yang ZK, Lou XD, Wei BM, Zhang JT, Gao PC, Hou RZ, Xia X (2015) Nanopore-based DNA-probe sequence-evolution method unveiling characteristics of protein-DNA binding phenomena in a nanoscale confined space. Anal Chem 87:4037–4041
Liao H, Zhou Y, Chai Y, Yuan R (2018) An ultrasensitive electrochemiluminescence biosensor for detection of MicroRNA by in-situ electrochemically generated copper nanoclusters as luminophore and TiO2 as coreaction accelerator. Biosens Bioelectron 114:10–14
Tang Y, Liu M, Xu L, Tian J, Yang X, Zhao Y, Zhao S (2018) A simple and rapid dual-cycle amplification strategy for microRNA based on graphene oxide and exonuclease III-assisted fluorescence recovery. Anal Methods 10:3777–3782
Huang RC, Chiu WJ, Li YJ, Huang CC (2014) Detection of microRNA in tumor cells using exonuclease III and graphene oxide-regulated signal amplification. ACS Appl Mater Interfaces 6:21780–21787
Min X, Zhang M, Huang F, Lou X, Xia F (2016) Live cell MicroRNA imaging using exonuclease III-aided recycling amplification based on aggregation-induced emission Luminogens. ACS Appl Mater Interfaces 8:8998–9003
Chen A, Gui GF, Zhuo Y, Chai YQ, Xiang Y, Yuan R (2015) Signal-off electrochemiluminescence biosensor based on Phi29 DNA polymerase mediated strand displacement amplification for microRNA detection. Anal Chem 87:6328–6334
Liao YH, Huang R, Ma ZK, Wu YX, Zhou XM, Xing D (2014) Target-triggered enzyme-free amplification strategy for sensitive detection of MicroRNA in tumor cells and tissues. Anal Chem 86:4596–4604
Yin BC, Liu YQ, Ye BC (2013) Sensitive detection of microRNA in complex biological samples via enzymatic signal amplification using DNA polymerase coupled with nicking endonuclease. Anal Chem 85:1487–11493
Lin MH, Wen YL, Li LY, Pei H, Liu G, Song HY, Zuo XL, Fan CH, Huang Q (2014) Target-responsive, DNA nanostructure-based E-DNA sensor for microRNA analysis. Anal Chem 86:2285–2288
Liu WP, Zhou XM, Xing D (2014) Rapid and reliable microRNA detection by stacking hybridization on electrochemiluminescent chip system. Biosens Bioelectron 58:388–394
Duan RX, Zuo XL, Wang ST, Quan XY, Chen DL, Chen ZF, Jiang L, Fan CH, Xia F (2013) Lab in a tube: ultrasensitive detection of MicroRNAs at the single-cell level and in breast cancer patients using quadratic isothermal amplification. J Am Chem Soc 135:4604–4607
Labib M, Khan N, Ghobadloo SM, Cheng J, Pezacki JP, Berezovski MV (2013) Three-mode electrochemical sensing of ultralow MicroRNA levels. J Am Chem Soc 135:3027–3038
Cong XX, Zhou MF, Hou T, Xu ZJ, Yin YZ, Wang XL, Yin M (2018) A sensitive photoelectrochemical aptasensor or miRNA-21 based on the sensitization effect of CdSe quantum dots. Electroanalysis 30:1140–1146
Wang B, Dong YX, Wang YL, Cao JT, Ma SH, Liu YM (2018) Quenching effect of exciton energy transfer from CdS:Mn to Au nanoparticles: a highly effcient photoelectrochemical strategy for microRNA-21 detection. Sensors Actuators B Chem 254:159–165
Acknowledgements
This work has been financially supported by the National Natural Science Foundation of China (Nos.: 21775080, 21705086); Key Research and Development Project of Shandong Province, China (No. 2017GSF221004).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 717 kb)
Rights and permissions
About this article
Cite this article
Liu, C., Zhao, L., Liang, D. et al. An CuInS2 photocathode for the sensitive photoelectrochemical determination of microRNA-21 based on DNA–protein interaction and exonuclease III assisted target recycling amplification. Microchim Acta 186, 692 (2019). https://doi.org/10.1007/s00604-019-3804-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3804-z