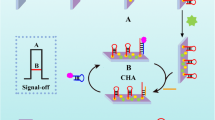
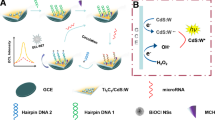
Abstract
An ultrasensitive photochemical biosensor based on the target miRNA-triggered catalytic hairpin assembly (CHA) reaction between Au nanoparticles (AuNPs)/C3N4 nanosheets and CdS quantum dots (QDs) was developed for the determination of miRNAs. Firstly, AuNPs/C3N4 nanosheets were immobilized onto a working glassy carbon electrode. Then, the hairpin probe 1 (H1) was loaded through Au–S bonding. Afterward, the unbound sites were blocked with 6-mercaptohexanol to avoid nonspecific adsorption. In the presence of the target miRNA, the CHA reaction between the H1 and hairpin probe 2-CdS QDs (H2-CdS QDs) could be triggered. As a result, the AuNPs/C3N4 nanosheet and CdS QDs were linked by the double helix structure H1-H2. Unlike the other CHA reactions, H2 used in this work is longer than H1 so that the AuNPs/C3N4 nanosheets could touch the CdS QDs. Given the matched energy band positions between the C3N4 nanosheet and CdS QDs, a strong photocurrent could be obtained after the CHA reaction was triggered by the target miRNA. In addition, p-type C3N4 nanosheets and n-type CdS QDs presented reduction photocurrents and oxidation photocurrents, respectively. Therefore, the photocurrents were vectors in this design that can eliminate the interference of nonspecific adsorption and avoid the generation of false-positive signals. Under the optimal conditions, the limit of detection was 92 aM. The constructed photoelectrochemical biosensor showed good reproducibility and selectivity in the analysis of serum samples, which indicates its great prospects in disease diagnostics and bioanalysis.
Graphical abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
MicroRNAs (miRNAs) are single-stranded, noncoding, and endogenous small RNA molecules [1]. These RNA molecules have been found to mediate gene silencing and translation repression, as well as trigger downstream signaling pathways [2]. Meanwhile, more evidence shows that the abnormal expression of miRNAs is associated with the occurrence, proliferation, invasion, and metastasis of tumors [3]. Therefore, miRNAs have been considered tumor biomarkers [4]. Previous studies have shown that the abnormal expression of miRNAs could be effectively detected by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) [5], microarray [6], Northern blotting [7], and deep sequencing [8] techniques. Although these methods present excellent accuracy in diagnosis, these processes are time-consuming and require the use of expensive equipment, thereby limiting their wide application. Therefore, it is imperative to explore sensing techniques with low cost, high sensitivity, and accuracy toward the detection of miRNAs.
Some emerging methods have been exploited to detect miRNAs, such as fluorescence [9], electrochemistry [10], electrogenerated chemiluminescence [11], photoelectrochemistry [12], surface-enhanced Raman spectroscopy [13], and colorimetry [14]. Among these methods, photoelectrochemistry (PEC), which integrates electrochemistry and optical excitation, has shown the advantages of simple instrumentation requirements, low noise, high sensitivity, and selectivity[15].
To obtain strong photocurrents in PEC biosensors, different semiconductor materials have been complexed by researchers. This is because the matched energy band positions between two semiconductors can promote the electron–hole separation efficiency [16, 17]. For example, Dong et al. constructed a photoelectrochemical platform based on a CdS@g-C3N4 composite for the bioanalysis of miRNA [18]. In addition, various composites, such as V2O5/CdS QDs [19], Ag2S@WS2 [20], MoS2-ReS2 [21], g-C3N4/Ti3C2 [22], and VS2 QDs-Bi2S3 [23], have been developed to enhance the photocurrent. However, most of the PEC biosensors developed based on these composites were signal “on–off” instead of “off–on.” In addition, there may be some false-positive signals due to the nonspecific adsorption. For instance, Victorious et al. developed a PEC biosensor based on the strong signal generated by the composites of TiO2 and AuNPs [24]. In their report, probe modified TiO2 was adopted as the substrate, and DNA-linked AuNPs could be captured by the probe. Meanwhile, the DNA was longer than the probe, showing that AuNPs could be located on the TiO2.
To achieve a large current change caused by miRNA, many efforts have been made to design signal amplification strategies. Some commonly used methods include strand displacement amplification (SDA) [25], catalytic hairpin assembly (CHA) reactions [26,27,28], rolling circle amplification (RCA) [29], and some endonuclease- [30] or exonuclease-mediated reactions [12]. Among these signal amplification methods, biological enzyme-mediated reactions are efficient but also expensive. Nevertheless, CHA-assisted signal amplification strategies are convenient, specific, low-cost, and efficient [31]. CHA is usually used to connect two different materials to achieve a certain signal enhancement. For example, Li et al. [32] used a miRNA-triggered CHA reaction and realized that plasmonic AuNPs with an appropriate interparticle distance could produce the optimal SERS signal.
Inspired by the above strategies, we tried to use a miRNA-triggered CHA reaction to assemble two semiconductor materials with matched energy band positions so that strong photocurrents could be obtained in the presence of a target. The previously reported CHA was aimed at assembling two materials on the two ends of double-stranded DNA. Here, we designed the length of the probe so that the two materials could be assembled with a smaller interparticle distance, and a stronger photocurrent signal could be achieved. As shown in Scheme 1, AuNPs/C3N4 nanosheets were utilized as the substrate, and hairpin probe 1 (H1) was attached to AuNPs by Au–S bonding. In the presence of the target miRNA and hairpin probe 2 (H2)-modified CdS QDs (H2-CdS QDs), CHA could be triggered and the CdS QDs were immobilized onto the working electrode. Due to the longer length of H2, the distance between the H2-modified CdS QDs and AuNPs/C3N4 nanosheets could be reduced. As a result, a stronger photocurrent could be obtained for the matched energy band positions between the C3N4 nanosheets and CdS QDs, which is typical of photocatalysts.
Experimental
Chemicals and materials
Dicyandiamide, chloroauric acid (HAuCl4), cadmium chloride (CdCl2), and 3-mercaptopropionic acid (MPA) were obtained from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Sodium sulfide (Na2S), N-hydroxy succinimide (NHS), 1-ethyl-3-(3-dimethyl amino propyl) carbodiimide hydrochloride (EDC), polyethyleneimine (PEI), 6-mercapto-1-hexanol (MCH), trisodium citrate dihydrate, and potassium ferricyanide (K3[Fe(CN)6]) were purchased from Aladdin Reagent Company (Shanghai, China). Human serum samples (H4522) were provided by Sigma-Aldrich. Besides, a breast cancer biomarker miRNA-21 was utilized as a representative miRNA. All the oligonucleotides used in this work were provided by Sangon Biotech Co., Ltd (Shanghai, China) and the corresponding sequences are displayed in Table S1.
Apparatus and measurements
The microstructures of the prepared samples were studied by scanning electron microscopy (SEM, FEI inspect F50, USA) and transmission electron microscope (TEM, Hitachi H800, Japan). Fourier-transform infrared (FT-IR) spectra were performed with a Nicolet 5700 spectrometer (Nicolet, USA). UV–vis absorption spectroscopy was carried out with a Shimadzu UV-2600 spectrophotometer (Japan).
Synthesis of H2-CdS QDs
H2 and MPA-capped CdS QDs were linked by amidation. Briefly, MPA-capped CdS QDs were synthesized according to Madrakian’s report [33]. Typically, CdCl2 (1 mmol) and MPA (1 mmol) were dissolved in 40 mL deionized water under stirring. Then, NaOH (2 M) solution was added to the above solution gradually to adjust the pH value of the solution to 11. Afterward, Na2S solution (0.1 M) was added gradually under stirring until the solution turned yellow. Thereafter, the solution was continuously stirred for 5 h and the MPA capped CdS QDs were formed. Furthermore, the MPA capped CdS QDs were precipitated by the addition of ethanol. The product was further washed with ethanol to remove the unreacted residues and dried at 60 ℃ for 12 h in a vacuum drying oven. Finally, the collected CdS QDs were dissolved in deionized water and stored at 4 ℃ in the dark for use.
Synthesis of H2-CdS QDs
H2 and MPA capped CdS QDs were linked by amidation. Briefly, MPA capped CdS QDs (1 mL, 1 mg mL−1) were mixed with 10 mL EDC solution (10 mg mL−1 in MES, pH 5.5) and stirred for 1 h to activate the carboxyl group in MPA. Afterward, 1 mL H2 solution (10 μM) and 10 mL NHS solution (5 mg mL−1 in MES, pH 5.5) were added to the above solution and reacted for 10 h at 25 ℃. Then, the formed H2-CdS QDs were washed with ethanol to remove the unreacted residues and dried at 60 ℃ for 10 h in a vacuum drying oven. Finally, the purified H2-CdS QDs were dissolved in deionized water and stored at 4 ℃ for use.
Preparation of AuNPs, C3N4 nanosheets, and AuNPs/C3N4 nanosheets
AuNPs, C3N4 nanosheets, and AuNPs/C3N4 nanosheets were synthesized according to our previous report [34]. The details are displayed in the supporting information.
Fabrication of the working electrode
Before fabricating the electrode, the glassy carbon electrode (GCE) was polished with 3 μm Al2O3 and washed with deionized water to obtain a mirror surface. Then, AuNPs/C3N4 nanosheet (10 μL) solution was dropped onto the GCE. Afterward, H1 (10 μL, 10 μM) was immobilized onto the AuNPs/C3N4 nanosheet–modified GCE through Au–S bonding. After that, MCH (10 μL, 1 mM) solution was used to seal the active sites on the AuNPs for 1 h, and then, the modified electrode was treated with 30 μL deionized water to remove excess mercaptoethanol. Then, the above-modified electrode was incubated with H2-CdS QDs (10 μL, 100 μM) and different concentrations of miRNA-21 for 2 h. Finally, the modified electrode was further treated with 30 μL deionized water to remove species bound by nonspecific adsorption.
Photoelectrochemical measurement
The PEC detection was performed on a CHI 660E electrochemical workstation (Shanghai Chenhua Instrument Co., Ltd., China) with a traditional three-electrode system consisting of the above-modified GCE as the working electrode, a Pt wire as the counter electrode, and a saturated calomel electrode (SCE) as the reference electrode. The photocurrents were recorded by obtaining I-T curves in PBS (pH = 6) under irradiation of a 150 W Xe lamp (LPX150 arc lamp, China) with an applied potential of 0.0 V vs SCE.
Results and discussion
Characterization of the AuNPs/C3N4 nanosheets and MPA-capped CdS QDs
First, the surface morphology of bulk C3N4 was explored by SEM. As displayed in Fig. 1A, bulk C3N4 was composed of a C3N4 nanosheet. Therefore, the C3N4 nanosheet could be obtained through ultrasonic peeling. The TEM image of the C3N4 nanosheet (Fig. 1B) indicates that a few layers of C3N4 nanosheets were obtained, while the AuNPs were assembled with C3N4 nanosheets. The size of the AuNPs was approximately 6–7 nm, and the AuNPs/C3N4 nanosheets were formed successfully, as shown in Fig. 1C. This could be explained by that the AuNPs with high surface energy are spontaneously assembling with the two-dimensional C3N4 nanosheet to reduce the surface energy. A TEM image of CdS QDs is shown in Fig. 1D, and its particle size could be estimated as 2–3 nm. In addition, the lattice spacing could be calculated to be approximately 0.23 nm, as observed from the HRTEM image (Fig. 1E). Furthermore, the selected area electron diffraction (SAED) pattern of CdS QDs is displayed in Fig. 1F.
(A) SEM image of bulk C3N4; TEM images of (B) C3N4 nanosheets (inset is the C3N4 nanosheets solution irradiated by ultraviolet light with a wavelength of 345 nm); (C) AuNPs/C3N4 nanosheets; (D) CdS QDs (inset is the CdS QDs solution irradiated by ultraviolet light with a wavelength of 345 nm); (E) HRTEM image of CdS QDs; and (F) SEAD pattern of CdS QDs
MPA-capped CdS QDs and H2-CdS QDs were characterized by FT-IR spectroscopy. As shown in Fig. 2, the FT-IR spectra of MPA-capped CdS QDs (curve a) displayed the significant vibrational peaks at 3435, 1560, 1400, and 1272 cm−1, which should be caused by the functional group of MPA. In addition, peaks at wavenumbers of 500–700 cm−1 were observed, which indicates the presence of Cd-S bonds [35]. After the MPA-capped CdS QDs were modified with H2 (curve b), the vibrational peak at 1640 cm−1 represented the amide bond [35]. In addition, the new vibrational peaks due to the presence of H2 indicate the functionalization of CdS QDs with H2 [36].
The UV–vis absorption spectra of the C3N4 nanosheets and MPA capped CdS QDs are shown in Fig. 3A. The C3N4 nanosheets and MPA-capped CdS QDs exhibit an absorption peak at 325 nm. Then, the corresponding Tauc plots shown in Fig. 3B were obtained according to the Tauc equation [37], (αhυ)2 = A(hυ − Eg), where α is the absorption coefficient, hυ is the photon energy, and Eg is the bandgap width. The semiconductor bandgap could be obtained by extending the tangent to the linear part of the graph through the horizontal axis. From Fig. 3B, the C3N4 nanosheets and MPA-capped CdS QDs depict noticeable absorption edges that correspond to the C3N4 (2.72 eV) and bulk CdS (2.4 eV) intrinsic absorptions, respectively [38].
Mott-Schottky plots of C3N4 nanosheets and CdS QDs were obtained through impedance spectroscopy analysis. According to the Mott-Schottky plots, the flat-band potential of the C3N4 nanosheets and CdS QDs could be determined, and the valance or conduction band potential could be reflected. Furthermore, the negative and positive slopes of the linear part of the curves indicate the p-type and n-type semiconductor behaviors, respectively [39]. As shown in Fig. 4A, the plot of the C3N4 nanosheets exhibits a negative slope, confirming its p-type semiconductor behavior with holes acting as the majority charge carrier. Figure 4B shows that CdS QDs exhibit a positive slope, indicating their n-type behavior, with electron acting as the majority charge carrier. Therefore, when the electrode was modified with C3N4 nanosheets or CdS QDs, different types of the peak currents could be obtained in the PEC test.
Electrochemical characterization of the constructed biosensor
To explore the stepwise assembly of the modified electrodes, cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS) were conducted in the electrolyte containing [Fe(CN)6]4−/3− (0.5 mM) and KCl (1 M) [40]. As shown in Fig. 5A, GCE exhibited a pair of [Fe(CN)6]4−/3− standard redox peaks (curve a). After the GCE was modified with AuNPs/C3N4 nanosheets (curve b), the peak currents of [Fe(CN)6]4−/3− decreased marginally, which can be explained by the C3N4 nanosheet with poor conductivity slowing electron transfer between the working electrode and solution. When H1 was immobilized onto AuNPs/C3N4 nanosheets (curve c), the redox peak currents of [Fe(CN)6]4−/3− further decreased, and the peak potential shifted. H1 was assembled successfully through Au–S bonding. Thereafter, the above-modified electrode was incubated with MCH (curve d) to seal nonspecific adsorption sites and the redox peak currents decreased again. Finally, miRNA-21 and H2-CdS QDs were incubated with the above electrode (curve e). As a result, the redox peak currents of [Fe(CN)6]4−/3− greatly decreased. This should be because of the poor conductivity of miRNA-21 and H2-CdS QDs and the stronger repulsion between [Fe(CN)6]4−/3− and the surface of the miRNA-21- and H2-CdS QDs-modified working electrode. Simultaneously, it could be demonstrated that the electrode was constructed successfully.
(A) Cyclic voltammograms and (B) EIS Nyquist plots (the insert is the equivalent circuit of the Nyquist plots) of different modified electrodes. (a) GCE, (b) GCE/AuNPs/C3N4 nanosheets, (c) GCE/AuNPs/C3N4 nanosheets/H1, (d) GCE/AuNPs/C3N4 nanosheets/H1/MCH, and (e) GCE/AuNPs/C3N4 nanosheets/H1/MCH/miRNA-21/H2-CdS QDs
The EIS results were consistent with the cyclic voltammograms and are shown in Fig. 5B. According to the Nyquist plots inserted, the electron-transfer resistance and solution resistance could be obtained [41]. Each step of the assembly of the working electrode inhibited the electron transfer between the electrode and solution. This indicates that the electrode was well modified and that the platform for detection was prepared successfully.
Feasibility of the proposed strategy for the miRNA diagnosis
To study the feasibility of the constructed miRNA detection platform, the working electrodes modified with or without target miRNA were compared in the PEC test. As shown in Fig. 6, the peak current was a reduction current when the working electrode was modified without target miRNA-21 (curve a). The C3N4 nanosheets were p-type semiconductors in the PBS (pH = 6), as demonstrated by the Mott-Schottky curve. In contrast, an oxidation peak current could be observed in the presence of target miRNA-21. This could be caused by the miRNA-21-triggered CHA reaction, and the n-type semiconductor CdS QDs were immobilized onto the working electrode. Therefore, the proposed biosensor could detect the target miRNA-21.
Optimization of the experimental conditions
To obtain the designed biosensor with optimal analytical performance, the concentration of H1 used for electrode modification and the incubation time of the CHA reaction were studied. First, H1 at different concentrations was dropped onto the AuNPs/C3N4 nanosheet-modified GCE. As shown in Fig. 7A, as the concentration of H1 increased, the impedance of the electrode gradually increased and gradually reached a plateau. Therefore, 10 μM H1 was utilized to modify the working electrode in the following experiments. In addition, the CHA reaction was affected by the incubation time [42]. Therefore, H1, miRNA-21, and H2-CdS QDs were incubated at different times to modify the working electrode. As shown in Fig. 7B, when the incubation time reached 2 h, the photocurrent was almost saturated, so the optimal incubation time was determined to be 2 h.
Analytical performance of the constructed PEC platform
After optimizing the experimental parameters of the proposed method, the performance of the miRNA analysis technique was investigated. Specifically, the working electrodes were modified with buffer with different concentrations of target miRNA-21 in the buffer. Then, the corresponding I-T curves were recorded at 0 V vs SCE with visible light. As shown in Fig. 8A, the peak current changed from a reduction peak current to an oxidation current when the electrode was constructed with target miRNA-21. In addition, the oxidation peak currents gradually became larger with increasing concentrations of miRNA-21. A good linear relationship between the logarithm of the miRNA-21 concentrations and the oxidation photocurrents is displayed in Fig. 8B. The corresponding linear regression equation was Ip (μA) = 0.03 logC + 0.0018 (where Ip represents the photocurrent and C is the concentration of the target miRNA-21 with R2 = 0.9892). The limit of detection (LOD) was calculated to be 92 aM (signal/noise = 3). Compared with other methods used for the determination of miRNA (Table S2), our designed platform in this strategy showed a lower LOD and wider linear range from 100 aM to 100 pM. Even though the photocurrent showed a downward trend as the target concentration increased, it only represented the change in the current state. This result is different from a previous report in the literature showing that the increased impedance decreased the current. In the absence of the target, some nonspecific substances may be adsorbed, which may cause the current to drop. However, no oxidation photocurrent could be generated when the CHA reaction was not triggered. Therefore, this strategy may better eliminate the interference of nonspecific adsorption and avoid the generation of false-positive signals.
(A) I-T responses of constructed biosensors incubated with different concentrations of miRNA-21: (a) blank, (b) 100 aM, (c) 1 fM, (d) 10 fM, (e) 100 fM, (f) 1 pM, (g) 10 pM, (h) 100 pM, (i) 1 nM, and (j) 10 nM in PBS. (B) Corresponding calibration plot for the photocurrents vs miRNA-21 concentrations. Error bars = RSD (n = 5)
Selectivity, reproducibility, and stability of the constructed biosensor
To explore the selectivity of the developed biosensor, different RNAs, such as single-base mismatched target, three-base mismatched target, and noncomplementary RNA with a concentration of 1 nM were utilized to replace the target miRNA-21 (100 pM) to modify the electrodes. The mixed sample was composed of all the interfering RNA and the target miRNA-21. As shown in Fig. 9A, weaker photocurrents were observed in all the interference experiment groups except for the sample group containing target miRNA-21. The CHA reactions were difficult to trigger by these interferences even though the concentrations of other RNAs were higher than the target miRNA-21 concentration and the photosensitizer could not be immobilized onto the electrode. Thus, the results indicated that the biosensor constructed by this strategy had a good selectivity.
(A) Photocurrent response of the biosensors incubated with different RNAs: (a) single-base mismatched target, (b) three-base mismatched target, (c) noncomplementary RNA, and (d) mixture of (a), (b), (c), and miRNA-21. (B) Time-dependent photocurrent response of the biosensor constructed with target miRNA-21 under periodic off–on illumination for 15 cycles
Furthermore, the stability of the biosensor was studied. The constructed biosensor of the target miRNA-21 was stored at 4 ℃ for 30 days. The photocurrents could retain 92% of the original value. In addition, the photocurrent response after 15 constant cycles was determined, with a relative standard deviation (RSD) of 3.13% (Fig. 9B). Therefore, the results demonstrated the good stability and repeatability of the constructed biosensor.
Detection in actual serum samples
To explore the applicability of the constructed platform in complex biological systems, standard addition experiments were carried out in real samples. MiRNA-21 at various concentrations was added to human serum samples to modify the electrodes, and the corresponding photocurrents are recorded in Table S3. The calculated recovery was distributed in the range of 95.45 to 103.95%, and the relative standard deviation was 3.12–4.88%. Therefore, the studies demonstrated that the constructed PEC biosensor has excellent stability and good sensitivity, which indicated the great feasibility of the designed biosensor in the determination of miRNA.
Conclusion
In conclusion, a photoelectrochemical biosensor was designed and constructed for the detection of miRNAs. The CHA reaction was triggered by the target miRNA, and the energy band positions matched AuNPs/C3N4 nanosheets and CdS QDs were assembled. In addition, the designed hairpin probe enabled CdS QDs to be located on the C3N4 nanosheet. In addition, p-type C3N4 nanosheets and n-type CdS QDs generated a reduction photocurrent and oxidation photocurrent, respectively, that could eliminate the interference of nonspecific adsorption and avoid the generation of false-positive signals. Furthermore, the designed biosensor showed good selectivity, reproducibility, and excellent stability in the analysis of serum samples, which indicates its good application prospects. In the future, we believe that the test time of this strategy could be further improved with the assistance of an enzyme or DNAzyme.
References
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–54
Peng Y, Croce CM (2016) The role of MicroRNAs in human cancer. Signal Transduct Target Ther 1:15004
Iorio MV, Croce CM (2012) MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 4:143–159
Lautner G, Gyurcsányi RE (2014) Electrochemical detection of miRNAs. Electroanalysis 26:1224–1235
Kimura Y, Ikeuchi M, Inoue Y, Ikuta K (2018) 3D microdevices that perform sample purification and multiplex qRT-PCR for early cancer detection with confirmation of specific RNAs. Sci Rep 8:17480
Premaratne G, Mubarak ZHA, Senavirathna L, Liu L, Krishnan S (2017) Measuring ultra-low levels of nucleotide biomarkers using quartz crystal microbalance and SPR microarray imaging methods: a comparative analysis. Sens Actuators, B Chem 253:368–375
Pall GS, Hamilton AJ (2008) Improved northern blot method for enhanced detection of small RNA. Nat Protoc 3:1077–1084
Yu Z-B, Han S-P, Bai Y-F, Zhu C, Pan Y, Guo X-R (2012) microRNA expression profiling in fetal single ventricle malformation identified by deep sequencing. Int J Mol Med 29:53–60
Luby BM, Zheng G (2017) Specific and direct amplified detection of microRNA with MicroRNA:Argonaute-2 Cleavage (miRACle) beacons. Angew Chem Int Ed 56:13704–13708
Cui L, Zhou J, Yang X-Y, Dong J, Wang X, Zhang C-Y (2020) Catalytic hairpin assembly-based electrochemical biosensor with tandem signal amplification for sensitive microRNA assay. Chem Commun 56:10191–10194
Kerr E, Farr R, Doeven EH, Nai YH, Alexander R, Guijt RM et al (2021) Amplification-free electrochemiluminescence molecular beacon-based microRNA sensing using a mobile phone for detection. Sensors Actuators B Chem 330:129261
Xia L-Y, Li M-J, Wang H-J, Yuan R, Chai Y-Q (2019) A novel “signal on” photoelectrochemical strategy based on dual functional hemin for microRNA assay. Chem Commun 55:9721–9724
Du X-Y, Wu S-H, Huang X-B, Sun J-J (2021) Ag nanocubes coupled with heating-enhanced DSN-assisted cycling amplification for surface-enhanced Raman spectroscopy detection of MicroRNA-21. ACS Appl Nano Mater 4:2565–2574
Juthani N, Doyle PS (2020) A platform for multiplexed colorimetric microRNA detection using shape-encoded hydrogel particles. Analyst 145:5134–5140
Mo F, Wu J, Chen M, Meng H, Han Q, Fu Y (2019) Enzyme-free “on-off-on” photoelectrochemical biosensor based on cascaded quadratic amplification strategy for miRNA 141 detection. Sens Actuators, B Chem 289:269–276
Wang Y, Su YR, Qiao L, Liu LX, Su Q, Zhu CQ et al (2011) Synthesis of one-dimensional TiO2/V2O5branched heterostructures and their visible light photocatalytic activity towards Rhodamine B. Nanotechnology 22:225702
Boruah PK, Szunerits S, Boukherroub R, Das MR (2018) Magnetic Fe3O4@V2O5/rGO nanocomposite as a recyclable photocatalyst for dye molecules degradation under direct sunlight irradiation. Chemosphere 191:503–513
Dong Y-X, Cao J-T, Wang B, Ma S-H, Liu Y-M (2017) Exciton–plasmon interactions between CdS@g-C3N4 heterojunction and Au@Ag nanoparticles coupled with DNAase-triggered signal amplification: toward highly sensitive photoelectrochemical bioanalysis of microRNA. ACS Sustain Chem Eng 5:10840–10848
Chen Y, Deng W, Tan Y, Xie Q (2020) CdS quantum-dots-decorated V2O5 nanosheets as chemically etchable active materials for sensitive photoelectrochemical immunoassay of carcinoembryonic antigen. ACS Appl Mater Interfaces 12:29066–29073
Wang Q, Yin H, Zhou Y, Wang J, Ai S (2021) Investigation of the inhibited biotoxicity of heavy metals towards 5- formylcytosine in rice by hydrochar based on photoelectrochemical biosensor. J Hazard Mater 414:125293
Liu L, Ma K, Xu X, Shangguan C, Lv J, Zhu S et al (2020) MoS2-ReS2 Heterojunctions from a bimetallic co-chamber feeding atomic layer deposition for ultrasensitive MiRNA-21 detection. ACS Appl Mater Interfaces 12:29074–29084
Yuan C, He Z, Chen Q, Wang X, Zhai C, Zhu M (2021) Selective and efficacious photoelectrochemical detection of ciprofloxacin based on the self-assembly of 2D/2D g-C3N4/Ti3C2 composites. Appl Surf Sci 539:148241
Zhang J, Shang M, Gao Y, Yan J, Song W (2020) High-performance VS2 QDs-based type II heterostructured photoanode for ultrasensitive aptasensing of lysozyme. Sensors Actuators B Chem 304:127411
Victorious A, Saha S, Pandey R, Soleymani L (2021) Enhancing the sensitivity of photoelectrochemical DNA biosensing using plasmonic DNA barcodes and differential signal readout. Angew Chem Int Ed 60:7316–7322
Yang K, Li J, Lamy de la Chapelle M, Huang G, Wang Y, Zhang J et al (2021) A terahertz metamaterial biosensor for sensitive detection of microRNAs based on gold-nanoparticles and strand displacement amplification. Biosens Bioelectron 175:112874
Fan Y, Liu Y, Zhou Q, Du H, Zhao X, Ye F et al (2021) Catalytic hairpin assembly indirectly covalent on Fe3O4@C nanoparticles with signal amplification for intracellular detection of miRNA. Talanta 223:121675
Ju T, Zhai X, Liu X, Han K (2021) A toehold-mediated strand displacement cascade-based DNA assay method via flow cytometry and magnetic separation. Anal Methods 13:1013–1018
Liu C, Zhang S, Li X, Xue Q, Jiang W (2019) Multi-code magnetic beads based on DNAzyme-mediated double-cycling amplification for a point-of-care assay of telomerase activity. Analyst 144:4241–4249
Hu Z, Xu F, Sun G, Zhang S, Zhang X (2020) Homogeneous multiplexed digital detection of microRNA with ligation-rolling circle amplification. Chem Commun 56:5409–5412
Yang L, Fang J, Li J, Ou X, Zhang L, Wang Y et al (2021) An integrated fluorescence biosensor for microRNA detection based on exponential amplification reaction-triggered three-dimensional bipedal DNA walkers. Anal Chim Acta 1143:157–165
Meng T, Jia H, An S, Wang H, Yang X, Zhang Y (2020) Pd nanoparticles-DNA layered nanoreticulation biosensor based on target-catalytic hairpin assembly for ultrasensitive and selective biosensing of microRNA-21. Sensors Actuators B Chem 323:128621
Li J, Koo KM, Wang Y, Trau M (2019) Native MicroRNA targets trigger self-assembly of nanozyme-patterned hollowed nanocuboids with optimal interparticle gaps for plasmonic-activated cancer detection. Small 15:1904689
Madrakian T, Maleki S, Afkhami A (2017) Surface decoration of cadmium-sulfide quantum dots with 3-mercaptopropionic acid as a fluorescence probe for determination of ciprofloxacin in real samples. Sens Actuators, B Chem 243:14–21
Ma X, Xu H, Qian K, Kandawa-Schulz M, Miao W, Wang Y (2020) Electrochemical detection of microRNAs based on AuNPs/CNNS nanocomposite with Duplex-specific nuclease assisted target recycling to improve the sensitivity. Talanta 208:120441
Butwong N, Noipa T, Burakham R, Srijaranai S, Ngeontae W (2011) Determination of arsenic based on quenching of CdS quantum dots fluorescence using the gas-diffusion flow injection method. Talanta 85:1063–1069
Masteri-Farahani M, Mosleh N (2019) Modified CdS quantum dots as selective turn-on fluorescent nanosensor for detection and determination of methamphetamine. J Mater Sci: Mater Electron 30:21170–21176
Gonullu MP, Ates H (2020) Investigation of the impact of annealing on the structural, optical and morphological evolution of mixture-phase ALD-TiO2 films containing brookite. Superlattice Microst 147:106699
Ahamad T, Majeed Khan MA, Kumar S, Ahamed M, Shahabuddin M, Alhazaa AN (2016) CdS quantum dots: growth, microstructural, optical and electrical characteristics. Appl Phys B 122:179
Singh AP, Arora P, Basu S, Mehta BR (2016) Graphitic carbon nitride based hydrogen treated disordered titanium dioxide core-shell nanocatalyst for enhanced photocatalytic and photoelectrochemical performance. Int J Hydrogen Energy 41:5617–5628
Xiong E, Zhang X, Liu Y, Zhou J, Yu P, Li X et al (2015) Ultrasensitive electrochemical detection of nucleic acids based on the dual-signaling electrochemical ratiometric method and exonuclease III-assisted target recycling amplification strategy. Anal Chem 87:7291–7296
Zhang L, Xiao X, Xu Y, Chen D, Chen J, Ma Y et al (2018) Electrochemical assay for continuous monitoring of dynamic DNA methylation process. Biosens Bioelectron 100:184–191
Ma X, Qian K, Ejeromedoghene O, Kandawa-Schulz M, Wang Y (2020) Electrochemical detection of microRNA based on SA-PPy/AuNPs nanocomposite with the signal amplification through catalytic hairpin assembly reaction and the spontaneous catalytic reaction of Fe3+/Cu2+. Electrochimica Acta 362:137168
Funding
This investigation was financially supported by the National Natural Science Foundations of China (No. 81571812) and A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (1107047002).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ma, X., Ma, Y., Ejeromedoghene, O. et al. Photoelectrochemical detection of microRNAs based on target-triggered self-assembly of energy band position–matched CdS QDs and C3N4 nanosheets. Microchim Acta 189, 65 (2022). https://doi.org/10.1007/s00604-022-05168-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-022-05168-x