Abstract
Cadmium-telluride quantum dots (QDs) were conjugated to an antibody (Ab) against Citrus tristeza virus (CTV), while the coat protein (CP) of the CTV was immobilized on the surface of carbon nanoparticles (CNPs). Following immunobinding of the QD-Ab and the CP-loaded CNPs, the fluorescence of the CdTe QDs was quenched by the CNPs. This effect was exploited to design a detection assay for the CTV which was found more sensitive and specific than the existing enzyme linked immunosorbent assay (ELISA). The limit of detection was measured at about 220 ng⋅ mL‾1 of CTV. The Stern-Volmer plot of the CNPs-QD quencher pair showed a positive deviation from linearity which was ascribed to the presence of both static and dynamic quenching.

Cadmium-telluride quantum dots (QDs) were conjugated to an antibody (Ab) against Citrus tristeza virus (CTV), while the coat protein (CP) of the CTV was immobilized on the surface of carbon nanoparticles (CNPs). Following immunobinding of the QD-Ab and the CP-loaded CNPs, the fluorescence of the CdTe QDs was quenched by the CNPs. This effect was exploited to design a detection assay for the CTV which was found more sensitive and specific than the existing enzyme linked immunosorbent assay (ELISA).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Citrus tristeza virus (CTV), a member of the genus Closterovirus under the family Closteroviridae, is the causative agent of one of the most economically destructive crop diseases called tristeza [1]. Over the last decades, CTV has been disseminated to approximately all citrus-growing areas leading to significant losses in fruit yield and quality worldwide [2]. Early detection is significantly critical for effective control of the CTV, however, the conventional detection methods suffer from serious drawbacks [2–4]. Therefore, there is an urgent need for a simple, rapid, sensitive, and specific screening technique to detect CTV at very early stages of infection. In line with that, many attempts have been dedicated to find a more accurate detection method [2, 4, 5].
The accessibility of protein detection systems has highlighted them as revolutionary process to detect different analytes in a vast kind of samples [6, 7]. However, it should be noted that in these systems, proteins are generally immobilized on a solid surface which can negatively affect the physicochemical properties and binding tendency of the proteins [8]. Therefore, the effectiveness of these detection systems may be jeopardized when very accurate detection is targeted [8]. In order to overcome this challenge, protein detection systems in a homogeneous suspension were suggested in order to benefit the advantages such as easy handling, easy modification, and controlled synthesis [9].
On the other hand, to design high-throughput and simple proteins detection systems, fluorescent semiconductor nanocrystals, also known as quantum dots (QDs), were used to circumvent some of the practical limitations encountered by conventional proteins detection systems thanks to their predominant optical properties [10]. In fact, QDs have attracted an intensive attention in the detection of different kinds of biological molecules [11, 12] because of their unique chemical and physical features including photo-stability, expansive excitation spectra, narrow emission spectrum, and photobleaching resistance [10]. The introduction of carbon nanoparticles (CNPs) and their appealing nano-sized properties has also marked them as interesting candidates to be applied in diverse fields including biosensing, bioimaging probes, light emitting diode materials, and drug delivery [13].
Different chemical materials and nanoparticles have been used as acceptor in the fluorometric-based sensors including rhodamine [14], tamra [15], gold nanoparticles [16] graphene oxide [17], and carbon nanotubes [18].
Both dynamic and static quenching mechanisms may be involved in the quenching process and the quenching mechanism strongly varies with the fluorophore–quencher pair [19]. Until now, except for a few reports which have tried to explain the quenching kinetics mechanism of QDs, the quenching mechanism of the other fluorescent molecules in the vicinity of organic or inorganic nanoparticles has not entirely investigated. For example, the quenching kinetics mechanism of QDs and tetracycline was studied by Stern-Volmer relationship [20]. In another effort, Mohd Yazid and co workers (2013) investigated the quenching mechanism of CNPs and Sn(II) ions by using Stern-Volmer relationship. They concluded that the quenching mechanism of CNPs by Sn(II) ions was predominantly of the static type [21].
It is worth mentioning that studying the quenching kinetic mechanism of acceptor/donor pairs is imperative not only in physical sciences, but also in biological, chemical, and medical sciences [22]. More especially, due to the fact that fluorometric-based platforms are limited to a distance and subsequently the accuracy of the nanosensors is affected by the distance between the donor and acceptor; therefore, calculation of the effective quenching radius for thorough interpretation of the behavior of nanoparticles is vital.
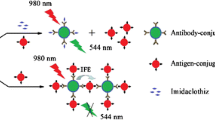
In continuation of the above-mentioned efforts, a fluorometric-based detection system for CTV using CNPs as acceptor was developed in the present study to investigate their capability in quenching QDs (as donor). More specifically, specific antibodies against CTV’s coat protein (CP) were labeled with QDs and the target proteins (CP) were labeled with CNPs. The vicinity of the QDs and CNPs led to the formation of an immuno-complex resulting in quenching interaction between the donor and the acceptor which subsequently decreased the fluorescence intensity of the QDs. While in the presence of the target proteins in the samples, the CNP-labeled CP was competitively displaced by free CPs causing the fluorescence intensity of the QDs to be recovered. This strategy enabled sensitive, rapid, and easy detection of the CTV. This fluorometric-based assay also possessed the attractive features of making use of nontoxic, and economic CNP nanoparticles.
Moreover, the quenching kinetics mechanism of the CNP-QDs pair was explored by using Stern-Volmer relationship and the effective quenching radius of the CNPs was also calculated. To the best of our knowledge, the capability of CNPs in quenching of QDs and the related quenching kinetic mechanism has not been reported.
Materials and method
Materials and apparatus
Tris-HCL buffer, 96-well microtiter plate and N-hydroxysuccinimide (NHS) were purchased from Thermo Fisher Scientific Inc. (USA, www.fishersci.com). Alkaline phosphatase-conjugated goat anti-rabbit IgG was purchased from Abcam (UK, http://www.abcam.com). The ELISA Kit was purchased from Bioreba Agro-Diagnostics company (Switzerland, http://www.bioreba.ch). Thiolglycolic acid (TGA), 1-ethyl-3-(3 dimethylaminopropyl)-carbodimide (EDC), Te powder and all other chemicals were bought from Sigma-Aldrich chemical company (St. Louis, Mo, http://www.sigmaaldrich.com).
All fluorometry studies were performed by using a Shimadzu, RF-5301PC Spectrofluorophotometer (Japan, http://www.shimadzu.com). All spectrophotometer analysis was performed using a Perkin-Elmer Lambda 35 UV-vis spectrometer (USA, http://www.perkinelmer.com). The synthesized nanoparticles were characterized by using a Hitachi H-7100 transmission electron microscope (TEM) (http://www.hitachi.com) and Thermo Nicolet, Smart Orbit Fourier transform infrared spectroscopy (FTIR) Nexus (Thermo Fisher Scientific Inc., http://www.thermoscientific.com) and the particle diameter size was determined using the ImageJ software (Version 1.46r, National Institute of Health, USA). The sonication was performed by using a Hwashin power sonic 420 (Hwashin Technology Co., Seoul, http://hwashintech.en.ec21.com) and filtration was carried out by a Filtres Fioroni filter paper (France, http://www.filtres-fioroni.com).
Field samples
The infected and healthy samples were collected from citrus fields located in Selangor and Pahang states, Malaysia. Young leaves from four different locations around the canopy were collected and stored at −20 °C until use.
CTV infection confirmation
The presence and confirmation of CTV infection was performed at the plant protection department of Sina seed and plant research institute by using RT-PCR technique.
Sap extraction from plant materials
Plant sap was extracted from healthy and infected citrus trees by crushing 1 g leaves in liquid nitrogen followed by suspension in 500 μl Tris-HCL buffer (pH 7.5). The sap samples were then stored at −20 °C until use.
Enzyme linked immunosorbent assay (ELISA)
In order to investigate the sensitivity and specificity of the nanobiosensor, the ELISA was performed as a standard test using a Bioreba ELISA Kit following the manufacture protocol with some modifications. Briefly, each well in a 96-well microtiter plate was coated with anti-CP antibody using carbonate coating buffer and incubated for 2 h at 37 °C. The plate was then washed with PBS three times. Then, the samples were added to the plate in three replicates and left overnight at 4 °C and washed with PBS as described earlier. Afterwards, diluted alkaline phosphatase-conjugated goat anti-rabbit IgG was added to the wells and the plate was incubated for 2 h at 37 °C. Lastly, the pNPP was added to each well as substrate and the plate was incubated for 30 min and the absorbance was read for each well.
Nanobiosensor fabrication
Synthesis of TGA capped CdTe QDs
In order to synthesize water soluble CdTe QDs, a freshly-prepared oxygen-free NaHTe aqueous solution was added into a solution containing 0.4 g CdCl2.2.5 H2O and 100 mL nitrogen-saturated double-distilled water (pH = 10) in the presence of 250 μL TGA as a stabilizing agent. The mixture solution was heated at 92 °C and stirred in a reflux system under nitrogen gas.
Labeling of antibody with QDs
To label the CTV specific antibodies with QDs, 200 μL of a freshly-prepared solution containing EDC (6.4 mgmL−1) and NHS (4.5 mgmL−1) was added to 400 μL QDs and incubated in a dark water bath at 37 °C for 1 h. Then, 60 μL antibodies (0.25 mgmL−1) were added drop-wise to the solution and the reaction was stopped by adding 540 μL tris buffer (6 mgmL−1 pH = 7.2). To separate the QD-labelled antibodies (QD-Ab), the mixture was centrifuged at 18,900×g for 10 min and the upper phase was diluted by 500 μL tris buffer and stored at 4 °C until use.
Synthesis of CNPs
Used facial tissues were washed and dried in an oven at 37 °C for 2 h. The dried facial tissues were then burned completely under air atmosphere. The obtained ash was dispersed in 50 ml double distilled water. Subsequently, 3.34 mL of HNO3 was added to the suspension and stirred for 30 min. The suspension was sonicated for 60 min, and filtered through a filter paper. Afterwards, the filtrate was centrifuged at 7000×g for 15 min and was then dialyzed against double distilled water to remove all excess impurities till a transparent solution was obtained.
Labeling of antigen with CNPs
In order to label the antigen (coat protein (CP)) with CNPs, the EDC/NHS chemistry was used. Briefly, 200 μL of a freshly-prepared solution containing EDC and NHS was added to 200 μL CNPs solution. Then, 45 μL of CP (0.375 mgmL−1) was added drop-wise to the solution and stirred gently at 4 °C for 2 h. Afterwards, the reaction was stopped by adding tris buffer to the solution. To separate the CNP-labeled antigen (CNP-CP) from the excess precursors, the solution was dialyzed against tris buffer for 16 h.
Nanobiosensor evaluation
Firstly, in order to investigate the capability of the nanobiosensor in detection of CTV, artificially-contaminated samples (0.1 to 1 μg mL−1 CP) and a known negative sample (as negative control) were tested in triplicate using a spectrofluorimeter. In addition, the specificity of the nanobiosensor was also tested by using four WBDL-infected samples. Afterwards, to determine the capability of the nanobiosensor in detection of field samples, 15 CTV-infected samples and 17 healthy samples were investigated. The detection procedure was conducted by adding 9 μL QD-Ab to 490 μL tris buffer (6 mgmL−1 pH = 7.2). Afterwards, 0.3 μL of CNP-CP was added to the solution and incubated for 3 min to ensure the formation of the QD-Ab/CNP-CP immuno-complex. Subsequently, the baseline curve was monitored to obtain the quenched baseline emission of the QD-Ab. Then, samples were added to the reaction mixture containing QD-Ab and CNP-CP and incubated for 5 min and the baseline curve was recorded again. The excitation wavelength of the QDs was set at 370 nm and the emission spectra were monitored between 450 to 750 nm to obtain the fluorescence intensity spectra.
Data analysis
The sensitivity and specificity of the nanobiosensor were calculated using the following formula (Eqs. 1 and 2).
Kinetic study of quenching mechanism
One of the most frequently used models for explaining quenching kinetic mechanism between donors and acceptors is the Stern–Volmer relationship [19, 20]. Therefore, herein, the kinetic quenching mechanism of the CNPs-QDs pair was explored by using this model. The Stern–Volmer equation regularly applied to describe the quenching kinetics mechanism is as follows [20]
where I0 is fluorescence intensity of QDs in the absence of quencher, I is fluorescence intensity of QDs in the presence of quencher, while KSV and [Q] are the Stern–Volmer quenching constant and concentration of quencher, respectively.
The Stern–Volmer equation can be extended as follows to investigate whether the ground-state complex plays a partial or pure role in the quenching mechanism of a donor/acceptor pair [19].
where KSV and Kg are dynamic/(S–V) and ground-state quenching constants of the complex, respectively.
Up to know, a number of quenching models based on the modified Stern–Volmer equation have been expanded to distinguish the static or instantaneous quenching route of donor/acceptor pairs [23] as follows:
Where W is computed as follows:
where V is the static quenching constant and exemplifies an active number element surrounding the fluorophore in its excited state.
The sphere of action quantity is given by the following equation (Eq. 7)
where N' and r are Avogadro’s number and the radius of sphere of action called kinetic distance, respectively.
Since the quantity of W depends on the [Q], the Stern–Volmer plot for a strong quencher usually deviates from linearity. Therefore, the Eq. 5 can be rewritten as follows:
Results and discussion
Up to now, different chemicals have been used as acceptor in the fluorometric-based sensors. The conventional fluorophores such as tamra and rhodamine shows some drawbacks include photobleaching, which is a reduction in fluorophore emission after repeated excitation. Emergence of nanoparticles such as gold nanoparticles have revolutionized fluorometric-based diagnostic techniques. However, the cost, toxicity, and strong background light scattering of these nanoparticles have restricted such nanoparticles to research applications only. Therefore, it is continuously desired to look for new energy donor-acceptor pairs, so as to circumvent the above-mentioned shortcomings and to acquire improved efficiency and analytical performances. CNPs owing to their tunable emissions as well as their unique electronic, mechanical, and thermal properties are considered as the next generation non-toxic green nanoparticles and have attracted a substantial deal of attention in bioassays [24, 25]. Therefore, the CNPs were used as acceptor to develop a fluorometric-based nanobiosensor for detecting CTV. To the best of our knowledge, the QD/CNP fluorometric-based nanobiosensor reported herein has not been employed in the detection of any pathogens.
Since ELISA has been used for the detection of CTV more than any other plant viruses [1]. Therefore, it was considered as a gold standard technique for detecting CTV and was compared with the developed nanosensor. The cut-off value of the ELISA was estimated considering six replications of the negative control at 0.577 ± 0.125 by using the following formula (Eq. 9):
where X is the average OD value of the negative samples and SD is the standard deviations.
Overall, 12 samples out of the 15 positive samples and 15 samples out of the 17 negative samples were successfully detected by ELISA (Table 1). In addition, two samples that were detected as positive by the nanobiosensor were identified as negative by the ELISA method (Table 1). Therefore, the sensitivity and specificity of the ELISA for detecting CTV were measured at 80% and 88%, respectively (Table 1).
Regardless of the false negative results observed by using the ELISA, this technique can effectively differentiate the CTV and the WBDL. In fact, the false negative and false positive results obtained by using the ELISA are ascribed to the concentration of the virus in the samples as well as the uneven distribution of the virus in different plant parts. The low concentration of viral particles in leaves and its critical effect on the detection procedures have been previously reported [26, 27].
CNPs and QDs were synthesized and used in fabricating the nanobiosensor. The TEM image of the synthesized CNPs and QDs showed that they were of spherical shape, and appropriate monodispersity (Fig. 1a, b). Particle size analysis showed that the synthesized CNPs and QDs had particle sizes of approximately 25 ± 8.1 and 4 ± 0.92 nm, respectively. Overall, the TEM images confirmed that the spherical water-soluble QDs and the water-soluble carboxyl-functionalized CNPs were appropriate to be used in the nanobiosensor.
Based on the FTIR analysis, the QDs exhibited characteristic stretching vibrations of O–H at 3429 cm−1, C-H at 2925 cm−1, 2858 cm−1, N-H stretching vibrations at 1511 cm−1, C–N at 1381 cm−1 and 1242 cm−1, C-O at 1739 cm−1, C-C at 1632 cm−1, and C-S stretching vibrations at 1050 cm−1. The CNPs exhibited characteristic absorption bands of O–H at 3459 cm−1, C–H at 2927 cm−1 and 2904 cm−1, C–C at 1637 cm−1, C-O stretching at 1724 cm−1 1132 cm−1 and 1078 cm−1 and, finally C-N stretching vibrations at 1388 cm−1 (Fig. 2). Therefore, the FTIR results confirmed that the QDs and CNPs were water-soluble with a large number of hydroxyl, carbonyl, or carboxylic acid groups on their surfaces. More specifically, this large number of hydroxyl, carbonyl, or carboxylic acid groups on the surface of the QDs indicated that the QDs were successfully bound to the carboxylic acid functional groups of the TGA. In addition, the presence of hydroxyl, carbonyl, or carboxylic acid groups on the surface of the CNPs was indicative of the oxidizing effect of HNO3 on some of the surface carbon atoms converting them into carboxylic groups during the HNO3 treatment step [28].
It is worth quoting that the ability of carbon nano tubes (CNTs) [29], graphene [17], GO, carbon nanofibers (CNFs) and graphite [30] in quenching QDs has been investigated previously, however, the quenching mechanism of QDs by CNPs was first taken into account by the present investigation. Recently, Ma et al. (2015) also employed carbon dots (CDs) as a fluorophore to detect mucin 1 protein (MUC1). More specifically, they labeled CDs with specific antibodies and aptamers against MUC1. In the presence of MUC1, the immune-reaction between CD-labeled antibody and of the CD-labeled aptamer to MUC1, led to the formation of a sandwich structure that was accompanied by aggregation of CDs and quenching of fluorescence [31].
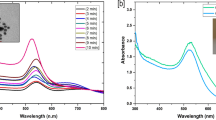
The immobilization of the antibodies onto the surface of the QDs and the conjugation of the antigens with the CNPs were verified by the spectrophotometric analysis. The observed decrease in the optical density (OD) of the QD-Ab compared with the pure QDs confirmed successful covalent coupling of the antibody and the QDs. Moreover, the maximum peaks for the bare and conjugated QDs were recorded at 548 nm and 551nm, respectively (Fig. 3). The antibodies were in fact attached onto the surface of the QDs through covalent interactions between the carboxyl groups of the TGA-capped QDs and the amino groups of the antibody.
As for the CNPs, a reduction in OD was observed for the CNP-CP in comparison with the pure CNPs and the spectral position of the CNPs extinction peak (λmax) was also changed from 295 nm to 314 nm (Fig. 4). In fact, the existence of the free amine groups (arginine or lysine) in the antigens led to an efficient labeling of the antigen with the CNPs via interactions between the carboxyl groups of the CNPs and the amine groups of the CP.
The quenching efficiency (E) was estimated by using E = 1- (F/F0) where F and F0 are the fluorescence emission intensity of the donor in the presence and absence of the acceptor, respectively. The E is directly dependent on the QD-Ab/CNP-CP ratio, and hence, in order to find the optimum ratio, varying concentrations of the CNP-CP (0.2 to 0.9 nM) were added in sequence to a constant concentration of the QD-Ab (6 nM). The maximum quenching efficiency of the nanobiosensor was observed at the QD-Ab/CNP-CP ratio of 1:10, and the quenching efficiency decreased by using lower or higher ratios (Fig. 5). Moreover, the CNPs led to a significant quenching of the fluorescence at about 63%. More specifically, at ratios higher than the optimum, surplus QD-Ab did not participate in the formation of the immuno-complex and, therefore, the excess QD-Ab reacted with free CP in the samples. This would jeopardize the sensing process when low concentrations of CP exist in a sample and subsequently no considerable shifts in the fluorescence signals were recorded. On the other hand, at lower molar ratios, due to the presence of excess CNP-CP, higher amounts of CP would be required to win the competition over the binding sites of the antibodies immobilized on the QDs` surfaces. This would result in low accuracy and sensitivity of the nanobiosensor in detecting low values of CP in samples.
The cut-off value of the nanobiosensor, i.e., 0.418 ± 0.00567 was obtained using the Eq. 9 and was employed to differentiate the healthy and infected samples. Based on this cut-off value, 14 samples out of the 15 positive (infected) samples and 15 samples out of the 17 healthy samples were successfully detected by the nanobiosensor (Table 1). Moreover, all four WBDL infected samples were found negative. Therefore, the sensitivity and specificity of the nanobiosensor were calculated at 93% and 88%, respectively without cross reactivity to WBDL (Table 1).
Concerning the mechanism of the nanobiosensor, the QD-Ab and CNP-CP formed an immuno-complex based on the antibody–antigen interaction phenomenon. The vicinity of the moieties then led to the quenching phenomena to occur. As a result, the emission spectrum of the QD-Ab decreased from 0.96 a.u. to 0.30 a.u. in the presence of the CNP-CP. But when an infected sample was added to the solution, the fluorescence intensity of the reaction mixture was recovered into the original value (Fig. S1 in the electronic supplementary material). In better words, since the antibody-antigen attachment is not of robust kind; thus, when the free CP was added to the solution containing both the QD-Ab and CNP-CP, the CNP-CP was displaced by the free CP. This caused an upward shift in the fluorescence intensity in comparison with the fluorescence intensity recorded formerly for the solution containing both the QD-Ab and CNP-CP. Hence, in the case of fluorometric-based nanobiosensors, QDs and CNPs may be considered as an appropriate donor and acceptor pair.
The limit of detection (LOD) for the designed system and ELISA were investigated by using the following equation:
where S0 is the standard deviation of blank measurements (n = 6) and K is the slope of the calibration curve.
Accordingly, the LOD of the nanobiosensor was estimated at 0.22 ± 0.0012 μgmL−1 (the standard curve was linear with a correlation coefficient, R2 of 0.94), while the LOD for the ELISA was estimated at 2.85 ± 0.031 μgmL−1 (R2 = 0.91). The detection limit of the designed system at 220 ngmL−1 was partially in agreement with the results obtained by Safarpour et al. (2012). According to the results obtained herein, it was possible to detect even very low concentrations of CP in field samples by comparing the obtained fluorescence signals.
The estimated LOD was mostly higher than the LOD values reported in the previous studies where CNPs were used as fluorophore molecule (Table 2) [21, 32, 33]. This is attributed to the fact that in the previous studies, artificial samples were investigated; where background signal would be decreased dramatically. While in the present study, field samples were used to investigate the sensitivity of the developed sensor. Biological field samples mostly contain different kinds of proteins, ions, and nucleic acids. Therefore, the background signal is at the highest value resulting in higher levels of LOD. On the other hand, the higher LOD values observed can also be ascribed to the fact that in the former studies the CNPs were used as donor while, the CNPs herein were used as acceptor. So, further research is needed to confirm the capability of CNPs in quenching QDs in the fluorometric-based systems.
The dynamic quenching mechanism is typically attributed to the collision and is demonstrated by the linear Stern–Volmer equation [19]. In order to obtain the Stern–Volmer plot, the experimentally-recorded values of I0 and I were used. Literally, the Stern–Volmer quenching curve explained the I0/I values of the QDs as a function of CNPs concentrations [Q].
As shown in Fig. 6, the Stern–Volmer plots were nonlinear with a positive deviation from linearity (Fig. 6).
Based on Eq. 3, the dynamic quenching constant, Ksv, determined from the lower portion of the Stern–Volmer plot was found to be approximately 3.31 M−1 (Table S1 in the electronic supplementary material). The obtained positive deviation from linearity in Stern–Volmer curve suggested that the attributed quenching mechanism was not simply dynamic, but the mechanism rather involved dynamic and static quenching simultaneously [22]. Such positive deviation from linearity was also reported by other studies [34, 35]. While some studies demonstrated that the quenching mechanism of CNPs by ions was mostly of the static type [21]. The differences in the quenching mechanism of CNPs in the present study and the previous studies can be attributed to the different quencher molecules used.
As observed in Fig. S2 in the electronic supplementary material, the plot of [(I0/I) - 1]/[Q] against [Q] showed a positive deviation with a correlation coefficient of about 87% with the intercept (KSV + Kg) and slope (KSVKg) (Table S1 in the electronic supplementary material). The observed positive deviation in the plot of [(I0/I) - 1]/[Q] against [Q] proved that the quenching mechanism was not completely dynamic and that dynamic and static quenching were present simultaneously (Fig. S2 in the electronic supplementary material).
According to the data presented in Table S1 in the electronic supplementary material, the Kg value was much smaller than the KSV value. In addition, the KSV value estimated using Eq. 4 was different from the value calculated using Eq. 3. Hence, in the current case, the positive deviation due to the ground-state complex formation was rejected [36]. As a result, the data were investigated by applying the sphere of action static quenching model to calculate the quantity of effective quenching radius.
The effective quenching radius involves the capacity and strength of a quencher in quenching a fluorophore molecule. As a result, a nanoparticle with a smaller radius will exhibit a lower quenching capacity but a higher accuracy in fluorometric-based platforms compared with a nanoparticle possessing a larger radius. According to the data presented in Table S1 in the electronic supplementary material, the calculated large effective quenching radius of 100.23 Å was a suitable theoretical explanation for the observed false results when detecting CTV.
Conclusion
In summary, by taking advantage of the CNPs, a fluorometric-based nanobiosensor was designed to detect CTV. To the best of our knowledge, CNPs have not been employed before as a QDs quencher in fluorometric-based sensors to detect any analytes. Therefore, it was demonstrated that CNPs can be applied as efficient quencher in a protein sensing platform. The findings of the present study are significantly important as CNPs application was further extended to protein detection. Nevertheless, due to the high level of LOD obtained herein, further studies are required to improve the accuracy of the fluorometric-based systems when CNPs are applied as acceptor. Moreover, due to the large effective quenching radius estimated for the applied CNPs, further investigation is needed to fabricate CNPs with lower quenching radius in order to advance the efficiency and analytical performance of protein platforms.
References
Moreno P, Ambrós S, Albiach-Martí MR, et al. (2008) Citrus tristeza virus: a pathogen that changed the course of the citrus industry. Mol Plant Pathol 9:251–268. doi:10.1111/J.1364-3703.2007.00455.X
Bertolini E, Moreno A, Capote N, et al. (2008) Quantitative detection of citrus tristeza virus in plant tissues and single aphids by real-time RT-PCR. Eur J Plant Pathol 120:177–188. doi:10.1007/s10658-007-9206-9
Sankaran S, Mishra A, Ehsani R, Davis C (2010) A review of advanced techniques for detecting plant diseases. Comput Electron Agric 72:1–13. doi:10.1016/j.compag.2010.02.007
Loconsole G, Saponari M, Savino V (2010) Development of real-time PCR based assays for simultaneous and improved detection of citrus viruses. Eur J Plant Pathol 128:251–259. doi:10.1007/s10658-010-9653-6
Barbarossa L, Savino V (2006) Sensitive and specific digoxigenin-labelled RNA probes for routine detection of citrus tristeza virus by dot-blot hybridization. J Phytopathol 154:329–335. doi:10.1111/j.1439-0434.2006.01102.x
Garibotti AV, Pérez-Rentero S, Eritja R (2011) Functionalization and self-assembly of DNA bidimensional arrays. Int J Mol Sci 12:5641–5651. doi:10.3390/ijms12095641
Cerf A, Alava T, Barton RA, Craighead HG (2011) Transfer-printing of single DNA molecule arrays on graphene for high-resolution electron imaging and analysis. Nano Lett 11:4232–4238. doi:10.1021/nl202219w
Liu W, Zhong H, Wang R, Seeman NC (2011) Crystalline two-dimensional DNA-origami arrays. Angew Chem Int Ed Eng 50:264–267. doi:10.1002/anie.201005911
Hongyang S, Yalei Z, Chunmin Z, et al. (2011) Cultivation of Chlorella pyrenoidosa in soybean processing wastewater. Bioresour Technol 102:9884–9890. doi:10.1016/j.biortech.2011.08.016
Wu P, Li Y, Yan X-P (2009) CdTe quantum dots (QDs) based kinetic discrimination of Fe 2+ and Fe 3+, and CdTe QDs-Fenton hybrid system for sensitive photoluminescent detection of Fe 2 +. Anal Chem 81:6252–6257. doi:10.1021/ac900788w
Pan J, Feng S-S (2009) Targeting and imaging cancer cells by folate-decorated, quantum dots (QDs)- loaded nanoparticles of biodegradable polymers. Biomaterials 30:1176–1183. doi:10.1016/j.biomaterials.2008.10.039
Fei X, Gu Y, Wang J, et al. (2011) Preparation and fluorescent properties of a complex probe based on inorganic QDs and organic dye. J Lumin 131:291–296. doi:10.1016/j.jlumin.2010.10.015
Zuo P, Lu X, Sun Z, et al. (2015) A review on syntheses, properties, characterization and bioanalytical applications of fluorescent carbon dots. Microchim Acta 183:519–542. doi:10.1007/s00604-015-1705-3
Safarpour H, Safarnejad MR, Tabatabaei M, et al. (2012) Development of a quantum dots FRET-based biosensor for efficient detection of Polymyxa betae. Can J Plant Pathol 34:507–515. doi:10.1080/07060661.2012.709885
Zekavati R, Safi S, Hashemi SJ, et al. (2013) Highly sensitive FRET-based fluorescence immunoassay for aflatoxin B1 using cadmium telluride quantum dots. Microchim Acta 180:1217–1223. doi:10.1007/s00604-013-1047-y
Roodbar Shojaei T, Mohd Salleh MA, Tabatabaei M, et al. (2014) Development of sandwich-form biosensor to detect mycobacterium tuberculosis complex in clinical sputum specimens. Braz J Infect Dis 18:600–608. doi:10.1016/j.bjid.2014.05.015
Liu M, Zhao H, Quan X, et al. (2010) Distance-independent quenching of quantum dots by nanoscale-graphene in self-assembled sandwich immunoassay. Chem Commun 46:7909–7911. doi:10.1039/c0cc02085k
Cui D, Pan B, Zhang H, et al. (2008) Self-assembly of quantum dots and carbon nanotubes for ultrasensitive DNA and antigen detection. Anal Chem 80:7996–8001. doi:10.1021/ac800992m
Lakowicz JR (2006) Principles of fluorescence spectroscopy, 3rd ed. doi:10.1007/978–0–387-46312-4
Chao M-R, Hu C-W, Chen J-L (2014) Comparative syntheses of tetracycline-imprinted polymeric silicate and acrylate on CdTe quantum dots as fluorescent sensors. Biosens Bioelectron 61:471–477. doi:10.1016/j.bios.2014.05.058
Mohd Yazid SNA, Chin SF, Pang SC, Ng SM (2013) Detection of Sn(II) ions via quenching of the fluorescence of carbon nanodots. Microchim Acta 180:137–143. doi:10.1007/s00604-012-0908-0
Al-Kady AS, Gaber M, Hussein MM, Ebeid E-ZM (2011) Structural and fluorescence quenching characterization of hematite nanoparticles. Spectrochim Acta A Mol Biomol Spectrosc 83:398–405. doi:10.1016/j.saa.2011.08.052
Nemzek TL (1975) Kinetics of diffusion-controlled reactions: transient effects in fluorescence quenching. J Chem Phys 62:477–489. doi:10.1063/1.430501
Bhunia SK, Saha A, Maity AR, et al. (2013) Carbon nanoparticle-based fluorescent bioimaging probes. Sci Rep 3:1473–1479. doi:10.1038/srep01473
Zhu A, Qu Q, Shao X, et al. (2012) Carbon-dot-based dual-emission nanohybrid produces a ratiometric fluorescent sensor for in vivo imaging of cellular copper ions. Angew Chem Int Ed Eng 51:7185–7189. doi:10.1002/anie.201109089
Adkar-Purushothama CR, Maheshwar PK, Sano T, Janardhana GR (2011) A sensitive and reliable RT-nested PCR assay for detection of citrus tristeza virus from naturally infected citrus plants. Curr Microbiol 62:1455–1459. doi:10.1007/s00284-011-9883-7
Mathews DM, Riley K, Dodds JA (1997) Comparison of detection methods for citrus tristeza virus in field trees during months of nonoptimal titer. Plant Dis 81:525–529. doi:10.1094/PDIS.1997.81.5.525
Zhang S, He Q, Li R, et al. (2011) Study on the fluorescence carbon nanoparticles. Mater Lett 65:2371–2373. doi:10.1016/j.matlet.2011.05.025
Yang R, Tang Z, Yan J, et al. (2008) Noncovalent assembly of carbon nanotubes and single-stranded DNA: an effective sensing platform for probing biomolecular interactions. Anal Chem 80:7408–7413. doi:10.1021/ac801118p
Morales-Narváez E, Pérez-López B, Pires LB, Merkoçi A (2012) Simple förster resonance energy transfer evidence for the ultrahigh quantum dot quenching efficiency by graphene oxide compared to other carbon structures. Carbon N Y 50:2987–2993
Ma N, Jiang W, Li T, et al. (2015) Fluorescence aggregation assay for the protein biomarker mucin 1 using carbon dot-labeled antibodies and aptamers. Microchim Acta 182:443–447. doi:10.1007/s00604-014-1386-3
Du F, Zeng F, Ming Y, Wu S (2013) Carbon dots-based fluorescent probes for sensitive and selective detection of iodide. Microchim Acta 180:453–460. doi:10.1007/s00604-013-0954-2
Wang Y, Bao L, Liu Z, Pang D-W (2011) Aptamer biosensor based on fluorescence resonance energy transfer from upconverting phosphors to carbon nanoparticles for thrombin detection in human plasma. Anal Chem 83:8130–8137. doi:10.1021/ac201631b
Evale BG, Hanagodimath SM (2009) Fluorescence quenching of newly synthesized biologically active coumarin derivative by aniline in binary solvent mixtures. J Lumin 129:1174–1180. doi:10.1016/j.jlumin.2009.05.017
Melavanki RM, Kusanur RA, Kadadevaramath JS, Kulakarni MV (2009) Quenching mechanisms of 5BAMC by aniline in different solvents using stern–Volmer plots. J Lumin 129:1298–1303. doi:10.1016/j.jlumin.2009.06.011
Biradar DS, Thipperudrappa J, Hanagodimath SM (2007) Fluorescence quenching of 2,2″ dimethyl-p-terphenyl by carbon tetrachloride in different solvents and temperatures. J Lumin 126:339–346. doi:10.1016/j.jlumin.2006.08.066
Author information
Authors and Affiliations
Corresponding authors
Electronic Supplementary Material
ESM 1
(DOC 95 kb)
Rights and permissions
About this article
Cite this article
Shojaei, T.R., Salleh, M.A.M., Sijam, K. et al. Fluorometric immunoassay for detecting the plant virus Citrus tristeza using carbon nanoparticles acting as quenchers and antibodies labeled with CdTe quantum dots. Microchim Acta 183, 2277–2287 (2016). https://doi.org/10.1007/s00604-016-1867-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-016-1867-7