Abstract
We report on an aptamer with high affinity against Salmonella typhimurium (S. typhimurium) and selected from an enriched oligonucleotide pool by a whole-cell SELEX process in a method for the fluorimetric determination of S. typhimurium using a graphene oxide platform. In the absence of target, the fluorescence was fairly weak as result of the FAM-labeled aptamer adjacent to graphene oxide. If, however, the fluorophore is released from the graphene oxide due to the formation of the target/aptamer complexes, fluorescence intensity is substantially increased. Under the optimum conditions, the assay displays a linear response to bacteria in the concentration range from 1 × 103 to 1 × 108 CFU·mL−1, with a detection limit of 100 CFU·mL−1. The method is selective in that fluorescence is not much enhanced in case of other bacteria. This aptasensor displays higher sensitivity and selectivity than others and is believed to possess a large potential with respect to the rapid detection of bacteria.

A useful fluorescence aptasensor based on a graphene oxide platform was constructed for Salmonella typhimurium detection, which has a great potential application in rapid detection of pathogen as it has high sensitivity and selectivity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Among all the food-borne pathogens that cause major public health problems worldwide, Salmonella is the most common cause of food-borne infectious disease, especially diarrheal diseases, which are caused by a range of enteropathogenic bacteria representing a major health threat in many developing countries [1]. Salmonella enterica serovar typhimurium (S. typhimurium) is one of the most frequently documented serovars associated with human infections since 1997 [2]. In 2010, its outbreak resulted in 85 infections according to the data from the Centers for Disease Control and Prevention (CDC, USA). Therefore, the detection, identification and quantification of the pathogen as well as other specific serovars from infected samples are critical for the subsequent treatment of infectious diseases. Currently, the detection methods of S. typhimurium generally focus on traditional culture methods, immunological methods and polymerase chain reaction (PCR). However, traditional culture methods require long time spans, immunological methods are time-consuming and costly, as well as producing false-positive results due to cross-reaction. Similarly, the method of PCR requires multiple touch conditions and is easily contaminated during the process of the nucleic acid sample extraction [3, 4]. In addition, several sensors for S. typhimurium detection have been reported, such as electrochemical impedance immunoassay [5], chemiluminescent immunoassay [6], colorimetric detection [7] and fluorescence detection with quantum dots [8]. However, in these cases, aggregation of metal nanoparticles might occur and result in high background signals and low sensitivity. Consequently, it is greatly desirable that a highly sensitive, selective, and cost-effective method can be constructed for detection of the bacteria.
Aptamers are special nucleic acid sequences that can bind to a wide range of targets, such as proteins, organic molecules, various cell surface receptors, and whole cells [9–12], with high affinity and specificity. They are generally selected from the pools containing randomly created sequences through a vitro systematic evolution of ligands by exponential enrichment (SELEX) [13]. Aptamers are easily synthesized and commercially available. They can be easily modified with a variety of chemical groups. Meanwhile, aptamers as newly emerging molecular recognition agents have been extensively studied in recent years, which provide an extraordinary flexibility in different assays [14]. Compared with antibody and nucleic acid probes-based biosensors, aptamer-based biosensors possess unprecedented advantages with high productivity, affinity, selectivity, and stability. Hence aptamers are an ideal method for the detection of food-borne pathogens, especially bacteria, in environment, food or clinical samples [15]. However, the number of biosensors based on aptamers for S. typhimurium detection is still scarce.
In the past 5 years, graphene-based nanomaterials have been the focus of a vast amount of attention. Graphene oxide (GO), a two-dimensional (2D)-layered precursor of graphene preparation [16], consists of sp2-bonded carbon with a hexagonal configuration. These bonds and electron configuration are the reasons for the extraordinary properties of graphene, which includes a very large surface area, a tunable band gap, room-temperature Hall effect, high mechanical strength, thermal conductivity and tunable optical properties [17–19]. It is an ideal material for the construction of biosensor-based devices with a low cost and low environmental impact [20]. Meanwhile, GO is an excellent energy acceptor in fluorescence resonance energy transfer (FRET) and makes the fluorescence detection promising application in sensing technology [21]. As fluorescence is a highly sensitive platform for biomolecular detection, GO is applied in various roles as a substrate in fluorescence quenching detection schemes. For example, the quenching principle was used for the aptamer-based detection of thrombin and ATP [22, 23]. However, little has been done to explore it in the analysis of food-borne pathogens. Therefore, it is necessary that GO based aptamer can been used as a platform for the sensitive and selective detection of bacteria.
We describe here that an aptasensor based on GO was designed, in which one end was labeled with the fluorophore 5-Carboxyfluorescein (FAM), as the energy donor and GO acting as the energy acceptor. When the FAM-aptamer binds to the GO by π-π stacking interaction, fluorescence quenching of FAM takes place because of FRET. The addition of target leads to the restoration of fluorescence due to the formation of FAM-aptamer/target complexes, which makes FAM-aptamer keep away from the GO. The detection platform not only has a lower limitation and good selectivity, but also can detect its targeted bacteria in a short time. These advantages such as simplicity, sensitivity and low background, make it a broad applied prospects in pathogen detection.
Experimental
Chemicals and materials
The sequences of the aptamer (5′-FAM-GGGAGCTCAGAATAAACGCTCAAGGGCAGGTGTTATGTGTACTGCTACAGTGTGGTTGTTCGACATGAGGCCCGGAC-3′) were synthesized by Shanghai Sangon Biotechnology Co. Ltd. (Shanghai, China, http://www.sangon.com). The sequences were dissolved in TE buffer to prepare 50 nM stock solution. The concentration of oligonucleotide was determined by using the absorbance at 260 nm. Graphite powder was purchased from Nanjing XFNANO Materials Tech Co. Ltd (Nanjing, China, http://www.xfnano.com). The clinical strains of S. typhimurium (CMCC50115), S. paratyphi A (CMCC50001), E. coli, ETEC K88 (CVCC 216), and Staphyloccocus aureus (CMCC (B) 26113) were obtained from the Basic Medicine Department of Hunan University of Traditional Chinese Medicine (Changsha, China). The classical strain of Salmonella cholerae-suis (ATCC10708) is obtained from the Changsha Food and Drug Administration (Changsha, China). The pasteurized milk was purchased from a local supermarket. All the other chemicals were of analytical reagent grade and were used as received without further purification. Milli-Q purified water was used to prepare all the solutions.
Apparatus
All fluorescence measurements and spectra were obtained with a LS55 fluorescence spectrophotometer (Perkin–Elmer, UK). The optical path length of a quartz fluorescence cell was 1.0 cm. Excitation and emission slits were all set for a 10.0 nm band-pass. The fluorescence emission spectra were collected at 517 nm with the excitation of 480 nm. Scanning electron microscope (SEM) was conducted with a NOVA NANOSEM450 microscope (FEI, USA). Atomic force microscope (AFM) characterizations were conducted on a Nanoscope III (Digital Instrument, USA). The Millipore water was produced by an Ultra-pure water system (Millipore, Boston, MA, USA).
Synthesis of GO
After being synthesized from natural graphite powder based on modified Hummers method [24, 25], GO was subjected to ultrasonication for 120 min (300 W, 20 % amplitude). Finally, a homogeneous GO aqueous dispersion (1 mg·mL−1) was obtained and stored at room temperature prior to use. The GO was then characterized with SEM and AFM.
Bacteria fluorescent detection
In fluorescence experiments, FAM-aptamer of S. typhimurium (50 nM) suspended in phosphate buffer solution (20 mM, pH 7.4) was first added to a 1.5 mL calibrated test tube. Then, 7 μL of solution with 1 mg·mL−1 GO was added, and allow to settle for 15 min. The final concentration of GO was 0.07 mg·mL−1. Subsequently, the different concentrations of tested targets were mixed with the resulting solution to obtain final volume of 100 μL. After the solution was incubated for 30 min at 37 °C, the fluorescence was measured and recorded at room temperature (RT). The experiments to optimize for sensing conditions were carried out under identical conditions. All experiments were repeated three times.
Results and discussion
Aptamer selection and identification
A panel of single-stranded DNA aptamers with high affinity and specificity against the S. typhimurium were selected from an enriched oligonucleotide pool by SELEX procedure [26]. Four other food-borne bacteria were used as counter-selection targets. After eleven rounds SELEX and four rounds counter-SELEX, aptamers were sorted, cloned, sequenced, and characterized for binding efficiency. Compared with the other three candidate aptamers, aptamer Apt6 showed relatively high binding affinity with an apparent dissociation constant (Kd value) of 30 ± 4 nM. Consequently, aptamer Apt6 was selected for further experiments.
Principle of the platform
AFM was employed to characterize the morphology and thickness of GO. As shown in Fig. S1a (Electronic Supplementary Material, ESM), a typical AFM image showed that GO was mostly single-layered with a topographic height of ca. 1.2 nm. SEM was also used to confirm the formation of GO (Fig. S2, ESM), in which its surface was very smooth, The optical image in the inset of Figure S2 showed that GO was well-dispersed in aqueous solution at the concentration of 1 mg·mL−1 and highly stable, which would be used in the following fluorescence tests. Figure S1b showed the AFM image of the aptamer/GO complexes, where the bright areas on the GO surface might be due to the presence of aptamer. Meanwhile, to verify successful coupling of aptamer to GO, a 2.5 % agarose gel was prepared. Gel electrophoresis data in the Figure S3 displayed the formation of the aptamer/GO complexes. Lane 1, 2, 3, 4 represented DNA ladder, GO alone, aptamer, and aptamer/GO complexes, respectively. As agarose gel electrophoresis is a molecular weight-dependent method, aptamer was appeared at approximately 100 bp with a bright and visible band (lane 3). When the aptamer/GO complexes were added, no migration of aptamer occurred (lane 4), which suggested that aptamer was completely retained in the loading point by GO and unable to enter the gel as a result of covalent functionalization. Therefore, aptamer was indeed attached to the GO surface.
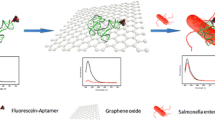
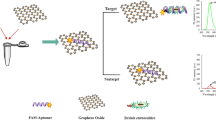
As illustrated in Fig. 1a, GO based bacteria detection platform based on turn-off/on fluorescence was constructed. In the absence of target, FAM-aptamer was adsorbed onto the GO surface by π-π stacking [22] and the fluorescence of FAM-aptamer was completely quenched by GO due to the FRET. By contrast, the aptamer would dissociate from the GO owing to the specific recognition between the FAM-aptamer and the target. It was clearly indicated that both turn-on/off fluorescence were dynamic and rapid processes, which was conducive to the achievement of rapid target detection. Figure 1b shows the fluorescence emission spectra of FAM-aptamer at different conditions. The fluorescence spectrum of FAM-aptamer without GO exhibited strong fluorescence emission (curve 1). However, in the presence of GO, more than 90 % FAM’s fluorescence was quenched (curve 2). This result reveals that GO can quench the fluorescence of FAM efficiently, which is due to the energy transfer from the FAM to GO. Meanwhile, upon addition of 1 × 109 CFU·mL−1 target, the fluorescence restored dramatically, which was nearly 80 % of the original intensity of FAM-aptamer (curve 3). Therefore, according to identify the turn-on of fluorescence, the bacteria can be quickly detected through aptamer recognition.
a Schematic illustration of graphene oxide sensing platform for detection of S. typhimurium. Based on this strategy, bacteria can be detected by the turn-on fluorescence after the recognition of its aptamer. b Fluorescence emission spectra of FAM-aptamer at different conditions: (1) FAM-aptamer; (2) FAM-aptamer + GO; (3) FAM-aptamer + GO + target (109 CFU·mL−1); (4) blank. FAM-aptamer concentration: 50 nM. GO concentration: 0.07 mg·mL−1. Excitation: 480 nm, and emission: 517 nm. Error bars indicate standard deviation (n = 3)
Optimization of assay conditions
It is reported that the ratios of nanomaterial and biomolecule was a critical factor for detection efficiency. Consequently, it is necessary to survey how many aptamer molecules could actually be loaded onto GO. Figure 2a showed the fluorescence quenching of FAM-aptamer at different concentrations of GO, the fluorescence intensity decreased substantially when the GO was added into the FAM-aptamer solution (50 nM). It can be seen that the relative fluorescence intensity F/F0-1 increased with the GO concentration over the range 0.03–0.07 mg·mL−1, indicating that excess FAM-aptamer was still free in the solution when the concentration of GO less than 0.07 mg·mL−1. Moreover, It was found that the concentration of GO reached 0.07 mg·mL−1, over 90 % fluorescence intensity of FAM-aptamer was quenched. Meanwhile, the F/F0-1 value was not increased any more when the concentration of GO was more than 0.07 mg·mL−1, which demonstrated that the GO was excess and free FAM-aptamer was not present in the solution. Hence, 0.07 mg·mL−1 GO was used in the next experiments.
a Optimization of the concentration ratio of graphene oxide to FAM-aptamer. b Kinetic curves of fluorescence show the processes for the loading/release of the FAM-aptamer on/off the GO in the present system. (1) Fluorescence quenching process of the FAM-aptamer by GO as a function of time. (2) Fluorescence restoration process of FAM-aptamer when it released from GO coursed by the recognition of target (109 CFU·mL−1) as a function of time. FAM-aptamer concentration: 50 nM. GO concentration: 0.07 mg·mL−1. Excitation: 480 nm. Error bars indicate standard deviation (n = 3)
To further understand the processes of turn-on/off fluorescence by FAM-aptamer/GO complexes with or without target, a relevant research was also completed. As shown in Fig. 2b, FAM-aptamer was adsorbed onto the surface of GO quickly (curve 1). After 15 min incubation, over 90 % fluorescence of FAM-aptamer was quenched by the addition of GO in the solution. Therefore, 15 min was chosen as the incubation time for GO and FAM-aptamer. However, when the target was added to FAM-aptamer/GO complexes, there was an increased fluorescence recovery with nearly 80 % in 30 min (curve 2). The result demonstrated that either the fluorescence quenching process of FAM-aptamer directed by GO or the fluorescence restoration process through the addition of target were both time-dependent. The reason why FAM-aptamer was no longer available to interact with GO is that aptamer has higher binding affinity to target than GO. It was clearly indicated that both turn-on/off fluorescence were dynamic and rapid processes, which was conducive to the achievement of rapid target detection.
Target detection
In Fig. 3, it depicts the fluorescence emission spectra of FAM-aptamer/GO complexes after incubation with different concentrations of target. With adding the target into the FAM-aptamer/GO solution buffer, the fluorescence were evidently restored ranging from 1 × 102 to 1 × 109 CFU·mL−1 (Fig. 3a). Then a calibration curve of fluorescence intensity was plotted as a function of concentration (Fig. 3b). The recovered fluorescence increased widely ranging from 1 × 103 to 1 × 108 CFU·mL−1. The recovered fluorescence increased widely ranging from 1 × 103 to 1 × 108 CFU·mL−1 with linear equation Y = 17.861X − 0.423, where Y is fluorescence intensity and X is the concentration of target (regression coefficient R 2 = 0.9754). More importantly, the detection limit of target is 100 CFU·mL−1 can be estimated at a signal-to-noise of 3 criterions.
a Fluorescence emission spectra of FAM-aptamer/GO complexes after incubation with different concentrations of target (blank, 102, 103, 104, 105, 106, 107, 108, 109 CFU·mL−1). b Calibration curve of fluorescence intensity as a function of concentration was plotted. Conditions: FAM-aptamer concentration: 50 nM. GO concentration: 0.07 mg·mL−1. Excitation: 480 nm, and emission: 517 nm. Error bars indicate standard deviation (n = 3)
Table S1 (ESM) summarized the analytical performances of several reported sensors for determination of S. typhimurium. The sensitivity achieved in our strategy was higher than those of electrochemical impedance immunoassay [5], chemiluminescent immunoassay [6], colorimetric detection [7] and fluorescence detection with quantum dots [8] as reported previously. The greatly improved detection limit of our aptasensor could be attributed to the prominent quenching effect of GO and the high affinity of aptamer to target. Another advantage of this method is a satisfactory response toward target over a wide range of concentrations. These features, as well as its other merits, such as low cost and fluorescence background, make it a promising candidate for the rapid detection of pathogens.
Selectivity of the aptasensor
For selectivity assay, parallel experiments were performed to evaluate the detection selectivity. In our experiments, S. typhimurium and other typical food-borne pathogens such as S. paratyphi A, S. cholerae-suis, S. aureus, E. coli k88 were investigated under the same experimental conditions (1 × 109 CFU·mL−1), respectively. As shown in Fig. 4, addition of target bacteria with concentration of 109 CFU·mL−1 induced significant fluorescence increase, whereas no obvious fluorescence restoration was observed in negative controls. Furthermore, it can be seen that the fluorescence enhancement in the presence of target was almost 5-fold than other four food-borne pathogens. The results clearly demonstrated that the detection system could detect S. typhimurium with high specificity and had no cross-reaction with other Salmonella serovars and other pathogens, which was attributed to the intrinsic properties of aptamer that can specifically recognize its target [27, 28]. Moreover, the sensing platform required neither the extraction of specific genes nor the precise probe design and synthesis [29]. Thus we can conclude that fluorescent aptasensor based on GO has a high selectivity, which is promising to use in some complicated matrixes.
Application to milk sample
In order to test the applicability of this method for real sample detection, it was challenged to determinate the bacteria in the milk sample. Figure 5 shows the fluorescence emission spectra of FAM-aptamer (50 nM) in phosphate buffer solution containing 50 % milk sample. In the absence of GO, the FAM-aptamer exhibited strong fluorescence emission (curve 1). However, the presence of GO (0.07 mg·mL−1) led to 85 % fluorescence quenching (curve 2). Upon further incubation with target (1 × 109 CFU·mL−1), nearly 64 % fluorescence recovery was observed (curve 3). The above results demonstrated the potential applicability of the present method in real samples detection.
Fluorescence emission spectra of FAM-aptamer (curve 1), the FAM-aptamer/GO complexes (curve 2) and the addition of target (109 CFU·mL−1) with FAM-aptamer/GO in phosphate buffer solution containing 50 % milk sample (curve 3). FAM-aptamer concentration: 50 nM. GO concentration: 0.07 mg·mL−1. Excitation: 480 nm, and emission: 517 nm. Error bars indicate standard deviation (n = 3)
Conclusion
In summary, we have successfully developed a simple, highly sensitive and selective fluorescent aptasensor for S. typhimurium detection by using GO as an efficient fluorescence quencher. This sensor can detect the target with a wide linear range from 1 × 103 to 1 × 108 CFU·mL−1 and a low detection limit of 100 CFU·mL−1, which is superior to other methods. Compared with other materials, GO has its intrinsic advantages such as low cost, large production scale and higher fluorescence quenching, to name a few. Meanwhile, this approach did not need labeling of fluorophore/quencher pairs, which reduced the cost and lowered the background. Furthermore, prominent fluorescence signals of this aptasensor were obtained when it was used in real samples assays. Consequently, this biosensor could be widely applied to sensitive and selective bacteria detection.
References
Plym Forshell L, Wierup M (2006) Salmonella contamination: a significant challenge to the global marketing of animal food products. Rev Sci Tech 25:541–554
Cody SH, Abbott SL, Marfin AA, Schulz B, Wagner P, Robbins K, Mohle-Boetani JC, Vugia DJ (1999) Two outbreaks of multidrug-resistant Salmonella serotype typhimurium DT104 infections linked to raw-milk cheese in Northern California. J Am Med Assoc 281:1805–1810
Prusak-Sochaczewski E, Luong JHT (1989) An improved ELISA method for the detection of Salmonella typhimurium. J Appl Microbiol 66:127–135
Widjojoatmodjo MN, Fluit AC, Torensma R, Keller BH, Verhoef J (1991) Evaluation of the magnetic immuno PCR assay for rapid detection of Salmonella. Eur J Clin Microbiol Infect Dis 10:935–938
Dong J, Zhao H, Xu M, Ma Q, Ai S (2013) A label-free electrochemical impedance immunosensor based on AuNPs/PAMAM-MWCNT-Chi nanocomposite modified glassy carbon electrode for detection of Salmonella typhimurium in milk. Food Chem 141:1980–1986
Magliulo M, Simoni P, Guardigli M, Michelini E, Luciani M, Lelli R, Roda A (2007) A Rapid multiplexed chemiluminescent immunoassay for the detection of Escherichia coli O157:H7, Yersinia enterocolitica, Salmonella typhimurium, and Listeria monocytogenes pathogen bacteria. J Agric Food Chem 55:4933–4939
Wu WH, Li M, Wang Y, Ouyang HX, Wang L, Li CX, Cao YC, Meng QH, Lu JX (2012) Aptasensors for rapid detection of Escherichia coli O157:H7 and Salmonella typhimurium. Nanoscale Res Lett 7:658
Duan N, Wu S, Yu Y, Ma X, Xia Y, Chen X, Huang Y, Wang Z (2013) A dual-color flow cytometry protocol for the simultaneous detection of Vibrio parahaemolyticus and Salmonella typhimurium using aptamer conjugated quantum dots as labels. Anal Chim Acta 804:151–158
Chen Z, Li G, Zhang L, Jiang J, Li Z, Peng Z, Deng L (2008) A new method for the detection of ATP using a quantum-dot-tagged aptamer. Anal Bioanal Chem 392:1185–1188
Joshi R, Janagama H, Dwivedi HP, Senthil Kumar TM, Jaykus LA, Schefers J, Sreevatsan S (2009) Selection, characterization, and application of DNA aptamers for the capture and detection of Salmonella enterica serovars. Mol Cell Probes 23:20–28
Dwivedi HP, Smiley RD, Jaykus LA (2010) Selection and characterization of DNA aptamers with binding selectivity to Campylobacter jejuni using whole-cell SELEX. Appl Microbiol Biotechnol 87:2323–2334
Liu GQ, Yu XF, Xue F, Chen W, Ye YJ, Lian YQ, Yan Y, Zong K (2012) Screening and preliminary application of a DNA aptamer for rapid detection of Salmonella O8. Microchim Acta 178:237–244
Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346:818–822
Sheng L, Ren J, Miao Y, Wang J, Wang E (2011) PVP-coated graphene oxide for selective determination of ochratoxin A via quenching fluorescence of free aptamer. Biosens Bioelectron 26:3494–3499
Yang M, Peng Z, Ning Y, Chen Y, Zhou Q, Deng L (2013) Highly specific and cost-efficient detection of Salmonella paratyphi A combining aptamers with single-walled carbon nanotubes. Sensors 13:6865–6881
Robinson JT, Perkins FK, Snow ES, Wei ZQ, Sheehan PE (2008) Reduced graphene oxide molecular sensors. Nano Lett 8:3137–3140
Geim AK, Novoselov KS (2007) The rise of graphene. Nat Mater 6:183–191
Bunch JS, van der Zande AM, Verbridge SS, Frank IW, Tanenbaum DM, Parpia JM, Craighead HG, McEuen PL (2007) Electromechanical resonators from graphene sheets. Science 315:490–493
Nair RR, Blake P, Grigorenko AN, Novoselov KS, Booth TJ, Stauber T, Peres NM, Geim AK (2008) Fine structure constant defines visual transparency of graphene. Science 320:1308–1311
Pérez-López B, Merkoçi A (2012) Carbon nanotubes and graphene in analytical sciences. Microchim Acta 179:1–16
He Y, Wang ZG, Tang HW, Pang DW (2011) Low background signal platform for the detection of ATP: when a molecular aptamer beacon meets graphene oxide. Biosens Bioelectron 29:76–81
Chang HX, Tang LH, Wang Y, Jiang JH, Li JH (2010) Graphene fluorescence resonance energy transfer aptasensor for the thrombin detection. Anal Chem 82:2341–2346
Wang Y, Li ZH, Hu DH, Lin CT, Li JH, Lin YH (2010) Aptamer/graphene oxide nanocomplex for in situ molecular probing in living cells. J Am Chem Soc 132:9274–9276
Xu YX, Bai H, Lu GW, Li C, Shi GQ (2008) Flexible graphene films via the filtration of water-soluble noncovalent functionalized graphene sheets. Am Chem Soc 130:5856–5857
Dong HF, Gao WC, Yan F, Ji HX, Ju HX (2010) Fluorescence resonance energy transfer between quantum dots and graphene oxide for sensing biomolecules. Anal Chem 82:5511–5517
Li H, Ding XH, Peng ZH, Deng L, Wang D, Chen H, He Q (2011) Aptamer selection for the detection of Escherichia coli K88. Can J Microbiol 57:453–459
Kawde AN, Rodriguez MC, Lee TMH, Wang J (2005) Label-free bioelectronic detection of aptamer–protein interactions. Electrochem Commun 7:537–540
Bamrungsap S, Shukoor MI, Chen T, Sefah K, Tan WH (2011) Detection of lysozyme magnetic relaxation switches based on aptamer-functionalized superparamagnetic nanoparticles. Anal Chem 83:7795–7799
Chen W, Martinez G, Mulchandani A (2000) Molecular beacons: a real-time polymerase chain reaction assay for detecting Salmonella. Anal Biochem 280:166–172
Acknowledgments
Support for this research has been provided by National Natural Science Foundation (81271660), The Cooperative Innovation Center of Engineering and New Products for Developmental Biology of Hunan, Doctoral Program of Higher Education of the Education Ministry (20114306110006) and Scientific Research Fund of Hunan Province Education Department (09K021, 12K032).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 284 kb)
Rights and permissions
About this article
Cite this article
Duan, Y.F., Ning, Y., Song, Y. et al. Fluorescent aptasensor for the determination of Salmonella typhimurium based on a graphene oxide platform. Microchim Acta 181, 647–653 (2014). https://doi.org/10.1007/s00604-014-1170-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-014-1170-4