Abstract
Arbuscular mycorrhizal fungi are able to improve plant establishment in polluted soils but little is known about the genes involved in the plant protection against pollutant toxicity by mycorrhization, in particular in the presence of polycyclic aromatic hydrocarbons (PAH). The present work aims at studying in both symbiotic partners, Medicago truncatula and Rhizophagus irregularis: (i) expression of genes putatively involved in PAH tolerance (MtSOD, MtPOX, MtAPX, MtGST, MtTFIIS, and MtTdp1α), (ii) activities of antioxidant (SOD, POX) and detoxification (GST) enzymes, and (iii) H2O2 and the heavy PAH, benzo[a]pyrene (B[a]P) accumulation. In the presence of B[a]P, whereas induction of the enzymatic activities was detected in R. irregularis and non-mycorrhizal roots as well as upregulation of the gene expressions in the non-mycorrhizal roots, downregulation of the gene expressions and decrease of enzyme activities were observed in mycorrhizal roots. Moreover, B[a]P increased H2O2 production in non-mycorrhizal roots and in R. irregularis but not in mycorrhizal roots. In addition, a lower B[a]P bioaccumulation in mycorrhizal roots was measured in comparison with non-mycorrhizal roots. Being less affected by pollutant toxicity, mycorrhizal roots did not activate any defense mechanism either at the gene expression regulation level or at the enzymatic level.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Polycyclic aromatic hydrocarbons (PAH) are ubiquitous pollutants formed by incomplete combustion or pyrolysis of organic matter (Shen et al. 2006). Owing to their great persistence in soils, their natural dissipation is limited (Wild et al. 1991). Moreover, because of their carcinogenic and/or mutagenic properties and their bioaccumulation in the food chain, PAH are harmful (Khan et al. 2008). Numerous physical and chemical methods, such as excavation, soil washing, or oxidoreduction, have been developed to remove PAH from contaminated soils (Soleimani et al. 2011). However, these methods are expensive, often only partially effective, and mainly destroy soil life leading to an inert material (Pimda and Bunnag 2012). So far, many studies have indicated that phytoremediation is an attractive alternative with environmental-friendly properties and low cost compared to traditional approaches to clean contaminated soils (Chigbo and Batty 2013). This “green technology” uses plants and their associated microorganisms to degrade, stabilize, reduce, and/or remove pollutants from the environment (Pilon-Smits 2005).
Possible mechanisms by which the most effective plants enhance removal of PAH have been proposed to involve the stimulation of PAH degrading rhizosphere microorganisms or, alternatively, uptake by the plant with subsequent accumulation in the plant tissues, enzymatic degradation, or volatilization (Martin et al. 2014). Indeed, some plants are able to metabolize organic pollutants, after absorption, through two major steps: phase I involving oxidation of lipophilic xenobiotics and phase II consisting of conjugation of the metabolite product of phase I to endogenous hydrophilic molecules such as glutathione using glutathione-S-transferases (GST) (Dietz and Schnoor 2001). But, in the main cases, organic pollutant degradation results from the stimulation of degrading rhizosphere microorganism activities (bacteria and saprotrophic fungi) thanks to root exudates released by the plants.
Among the microorganisms that affect rhizosphere processes, arbuscular mycorrhizal fungi (AMF) induce a series of changes in plant physiology, nutrient availability, and microbial composition that may determine the outcome of a phytoremediation attempt. Many studies have reported that AMF are not only able to protect plants directly against PAH (Lenoir et al. 2016a; Rajtor and Piotrowska-Seget 2016) but also enhance bioremediation processes by stimulating soil microbial activity and improving soil structure (Joner and Leyval 2003; Gao et al. 2011). Whereas for lignolytic fungi, a correlation was observed between PAH degradation and the presence of extracellular lignolytic enzymes, such as peroxidases and laccases (Verdin et al. 2004; Dodor et al. 2004; Baborová et al. 2006), no evidence of direct PAH catabolism by AMF has been reported yet. No genes involved in lignin decomposition, such as class II peroxidases, were found in the AMF Rhizophagus irregularis (Tisserant et al. 2013). Nevertheless, a positive contribution of the arbuscular mycorrhizal symbiosis in anthracene dissipation was demonstrated using monoxenic cultures of chicory roots colonized by R. irregularis in the absence of soil microorganisms (Verdin et al. 2006).
A common consequence of most abiotic environmental stresses is an imbalance between production and detoxification of reactive oxygen species (ROS), such as superoxide anion radical (O2 .-), hydrogen peroxide (H2O2), hydroxyl radicals (.OH), and singlet oxygen (O1 .-). Sudden and dramatic increases in cellular ROS production can lead to protein and lipid oxidation as well as DNA breakage, upsetting cell homeostasis (Miller et al. 2008). Fortunately, plants have the capacity to cope with these ROS by using several antioxidant enzymes and metabolites located in different plant cell compartments (Dat et al. 2000). For example, to fight against DNA damage, plants can implement DNA repair systems. In Medicago truncatula, two genes were described as being involved in DNA repair: MtTdp1 and MtTFIIS encode the tyrosyl-DNA phosphodiesterase enzyme and the transcription elongation factor II-S, respectively (Yang et al. 1996; Pommier 2003). Plants also possess ROS scavenging systems including antioxidant enzymes such as superoxide dismutases (SOD), peroxidases (POX), ascorbate peroxidases (APX), catalases, and non-enzymatic defense like ascorbic acid and glutathione (Niyogi 1999).
Arbuscular mycorrhizal fungi are able to improve plant growth and health under PAH pollution through alleviation of the oxidative stress caused by the pollutant (Debiane et al. 2008; Debiane et al. 2009). Indeed, several investigations have addressed mycorrhiza-induced reduction of oxidative stress. It was suggested that the tolerance of PAH phytotoxicity by mycorrhizal plants resulted from the induction of enzymatic antioxidant systems such as SOD (Debiane et al. 2008; Debiane et al. 2009; Li et al. 2011) and POX (Criquet et al. 2000; Rabie 2005; Li et al. 2011; Yu et al. 2011). But to date, no genes implicated in mycorrhizal plant protection in the presence of PAH have been described. Concerning AMF, only a few genes encoding proteins putatively involved in ROS homeostasis have been identified and characterized: GmarCuZn-SOD1 (Lanfranco et al. 2005) and GintSOD1 (González-Guerrero 2005) encoding superoxide dismutases; GintGRX1 encoding a glutaredoxin, a multifunctional protein with oxidoreductase, peroxidase, and glutathione-S-transferase activity (González-Guerrero 2005); and GintPDX1 encoding a pyridoxine 5′-phosphate synthase, a protein involved in vitamin B6 biosynthesis which acts as antioxidant (González-Guerrero 2005).
At present, the role played by mycorrhization in plant tolerance of organic pollutants, in particular PAH, is less documented than for other abiotic stresses (i.e., metal pollution, salinity, drought, acidity). Therefore, our work delves into this context and aims to study it in both symbiotic partners, M. truncatula and R. irregularis, as well as in their symbiosis, including (i) the expression of genes putatively involved in PAH tolerance, (ii) the activities of antioxidant (SOD, POX) and detoxification (GST) enzymes, and (iii) H2O2 and B[a]P accumulation. M. truncatula and R. irregularis have been recognized for their phytoremediation capacity (Chen et al. 2005, Lenoir et al. 2016a, 2016b, 2016c). Their genomes have been sequenced completely, and the selected genes of which expressions were studied in this work already have been described as involved in various stress tolerance responses (Lenoir et al. 2016a). Our experiments were carried out using monoxenic cultures of non-mycorrhizal and mycorrhizal roots grown in the absence or in the presence of B[a]P, a high molecular weight PAH usually detected in polluted soils. These in vitro cultures are appropriate tools to monitor the PAH impact on arbuscular mycorrhizal symbiosis because they allow non-destructive observations of AMF and facilitate obtaining a large quantity of biological material free of contaminant microorganisms.
Materials and methods
Root and fungal growth conditions
All the experiments were conducted in in vitro with Ri T-DNA-transformed alfalfa roots (M. truncatula (L.) Gaertn. var. Jemalong A17) colonized or not by the AMF R. irregularis, DAOM 197198 (Schüßler and Walker 2010). Cultures were established in bi-compartmental Petri dishes (9 cm). One compartment was filled with 25 ml of solidified M medium (Becard and Fortin 1988) [solidified with 0.25% (w/v) gellan gum (Phytagel, Sigma, St. Louis, USA)] without B[a]P (control) where a piece of 1 cm2 of mycorrhizal or non-mycorrhizal roots was added on medium. After 2 weeks, the second compartment was filled with 20 ml of liquid M medium without vitamins, sucrose, and gellan gum in the absence of the pollutant to allow root development for 4 weeks at 27 °C in the dark. In this experiment, we allowed root growth into the second compartment in order to facilitate the passage of the AMF to the liquid compartment, and subsequently to expose the fungus and the roots to the pollutant as in polluted soil. After 4 weeks, the liquid medium was removed and 12 ml of fresh liquid M medium was added supplemented or not with B[a]P (1400 μM) (Sigma-Aldrich, St. Louis, USA). This sub-lethal concentration was chosen to resemble PAH contamination levels generally found in soils worldwide, and after checking its action on the expression of the studied genes. Seven days after B[a]P addition, roots and AMF extraradical structures were harvested under a low-power microscope at ×10–40 magnification using forceps.
Determination of root and fungal development
After 7 days of exposure or not to B[a]P, root and hyphal lengths and the number of spores were measured in the second compartment using a gridline-intersect technique (Newman 1966). The number of branched absorbing structures (BAS) was counted under a low-power microscope at ×10–40 magnification (Newman 1966). Roots and AMF structures (extraradical hyphae and spores) were lyophilized over 48 h and then weighed. Root biomass was calculated from five Petri dishes (one Petri dish per replicate) and fungal biomass (spores and extraradical hyphae) were calculated from 50 Petri dishes (ten Petri dishes pooled per replicate).
Determination of arbuscular mycorrhizal colonization rate
After exposure or not to B[a]P, roots collected from each replicate were cleared in KOH (10%) and stained with Trypan blue (Phillips and Hayman 1970) to determine mycorrhizal root colonization rate (McGonigle et al. 1990). For each replicate, three slides each containing 15 stained root fragments (randomly selected) and three sections per root fragment were observed under a microscope at ×50 magnification.
Detection of H2O2 accumulation
The detection of H2O2 was carried out using the DAB (3,3′-diaminobenzidine) staining method (Fester and Hause 2005). After exposure or not to B[a]P, roots and spores were collected from each replicate and stained. For each replicate, 100 spores and three slides each containing 15 stained root fragments (randomly selected) and three sections per root fragment were observed under a microscope at ×50 magnification. Every section or spore showing detectable brown DAB staining was regarded as synthesizing H2O2.
Preparation of crude cell-free extracts
For RNA extraction and dosage of enzyme activities, roots and AMF structures were sampled, liquid nitrogen-frozen, and stored at −80 °C until using. One hundred milligrams of frozen tissues from roots (from 1 plate per replicate) and AMF structures (from 30 pooled plates per replicate) were ground for RNA extraction using the grinder Precellys 24 (Bertin Technologies, Montigny-Le-Bretonneux, France). To assess enzyme activities, 100 mg of frozen roots (from 1 plate per replicate) and only 50 mg of frozen AMF structures (from 15 pooled plates per replicate) were ground similarly.
RNA extraction and real-time RT-PCR
Total RNA was extracted from ground AMF extraradical structures (100 mg) and roots (200 mg) with the Qiagen RNeasy® Plant Mini Kit (Qiagen, Courtaboeuf, France) followed by DNase digestion (RNase-free DNase Set; Qiagen) according to the manufacturer’s protocol. The RNA integrity was assessed by visualization of ribosomal RNA bands on 1.2% agarose gels, and total RNA was quantified spectrophotometrically by measuring the absorbance at 260 nm (UVIKON 942 UV/visible, Kontron Instruments, Milan, Italy). One microgram of total RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Villebon-sur-Yvette, France) according to the manufacturer’s protocol. Expression of six targeted genes of M. trunculata and four targeted genes from R. irregularis was tracked by quantitative Reverse-Transcripts Polymerase Chain Reaction (qRT-PCR), including the 18S ribosomal RNA genes (Mt18S and Gint18S) which were used as the internal standard to normalize the starting template of cDNA. This analysis was performed using the gene-specific primers presented in Table 1. We measured the expression of targeted genes MtPOX, MtAPOX, MtSOD, MtTFIIS, MtTdp1α, GintGRX1, and GintPDX1, selected from previous studies performed on the stress tolerance of M. trunculata or R. irregularis (Puckette et al. 2008; Benabdellah et al. 2009a, 2009b; Balestrazzi et al. 2011). Primers of the MtGST, Mt18S, GintGST, GintSOD, and Gint18S genes were designed by using Primer 3.0 to amplify fragments from 55 to 60 °C Tm of 100 bp in order to obtain an efficient amplification (Bustin 2000), and were tested for secondary structure using the NetPrimer software. The GintGST sequence was obtained from the R. irregularis Database (http://genome.jgi-psf.org/Gloin1), whereas other sequences were obtained from the GenBank Database. The amplicon sequences obtained were tested under NCBI Blast nucleotide to check their theoretical specific character for the targeted gene and not to a gene family. The amplification specificity of each qRT-PCR was confirmed by the presence of a single peak in the melt curve analysis, and no primer dimers were detected using agarose electrophoresis. Reactions were performed with a 7300 Real-Time PCR system (Applied Biosystems, Life Technologies, Carlsbad, CA, USA) using the SYBR green master mix (Applied Biosystems, Villebon-sur-Yvette, France) and the following thermal profile: 15 s at 95 °C (denaturation), 30 s at annealing temperatures (55 °C for M. trunculata and 60 °C for R. irregularis), and 30 s at 72 °C (extension) for 40 cycles. The efficiencies of the primer sets were estimated by performing real-time PCR on several dilutions. The results were normalized with the Mt18S and Gin18S genes and expressed relative to the respective control, corresponding to a fixed value of 1. The analyses were performed using the relative expression software tool REST® (V.2.0.13, 2009, QIAGEN GmbH) as described by Pfaffl et al. (2002). The analyzed genes were considered to be significantly up- or downregulated in the presence of B[a]P when changes of their expressions were >2× or <0.5×, respectively, versus the controls.
Determination of SOD, POX, and GST activities
Ground samples were suspended in 1 ml of phosphate buffer (10 mM) and centrifuged 3 min at 10,000g. SOD, POX, and GST activities were measured by colorimetric assay kits. POX activity was determined according to the method of Mitchell et al. (1994). SOD activity was measured using the SOD determination kit (Sigma-Aldrich, Saint Louis, MO, USA). GST activity dosages were performed as described by Riechers et al. (1996) with slight modifications according to El Chartouni et al. (2012). Protein concentrations were determined using the Total Protein Kit, Micro Lowry, Peterson’s Modification (Sigma-Aldrich, Saint Louis, Missouri, USA).
Determination of B[a]P accumulation
Washed roots were lyophilized, dry weight was determined, and the B[a]P accumulated in the dried roots (200 mg) was extracted by soxhlet (120 ml dichloromethane during 16 h at 70 °C, 1 recycling per hour). B[a]P quantification was performed by GCMS (QP2010, Shimadzu, Marne la Vallée, France). Samples were injected at 250 °C by a split injector with a ratio of 80. The separation was performed with a Zebron Phenomenex column (50% phenyl and 50% dimethylpolysiloxane, length 10 m, diameter 0.10 mm, width of film 0.1 μm). Samples were run on a programmed temperature profile: initial temperature of 70 °C for 15 s, 60 °C/min until 150 °C, 30 °C/min until 310 °C, and 3 min at 310 °C. The detection was realized by mass spectrometry by electronic impact (70 eV, 280 °C). B[a]P was quantified using B[a]P (Sigma-Aldrich, St. Louis, USA) as standard with a retention time of 6.24 min.
Statistical analysis
Means were calculated from five replicates except relative quantification of gene expression calculated from four replicates. Except for length and dry weight of M. truncatula roots, H2O2 accumulation, SOD, POX, and GST activities in roots, for which two-way ANOVAs were conducted to compare mycorrhizal status and B[a]P addition using SPSS 17.01 (IBM, Inc., Chicago, USA), all other data were subjected to the Mann-Whitney U test (P < 0.05) using Statgraphics release 5.1 (Manugistic, Inc., Rockville, MD, USA). Relative quantification of gene expression and its statistical analysis were performed using the REST software using the Pair Wise Fixed Reallocation Randomisation test (Pfaffl et al. 2002).
Results
B[a]P impacts on M. truncatula root growth and AMF development
The effect of B[a]P on root development was evaluated by measuring root length and dry weight after 1 week of exposure to B[a]P (Table 2). Although B[a]P reduced root lengths by about 15% both in mycorrhizal and non-mycorrhizal roots, the inoculation increased significantly root length whether or not in the presence of B[a]P. The dry weights obtained in the absence of B[a]P averaged almost 70 mg/Petri dish both in mycorrhizal and non-mycorrhizal roots. B[a]P exposure had no-detectable effect on dry weight of mycorrhizal and non-mycorrhizal roots.
B[a]P toxicity on R. irregularis was determined by measuring mycorrhizal rate, sporulation, extraradical hyphae development, BAS formation, and biomass (Table 3). After 1 week of exposure, microscopic observations of stained roots showed intraradical hyphae in roots grown on both B[a]P-supplemented and non-supplemented media. Nevertheless, root colonization was reduced by 21% in the presence of B[a]P. The spore production, the extraradical hyphae development, and BAS formation were significantly lower in the presence of B[a]P. The relative decreases were about 16, 18, and 26% for spores, hyphae, and BAS, respectively. No significant difference was observed in the AMF dry weight between control and polluted medium.
Hydrogen peroxide accumulation in roots and in the extraradical AMF
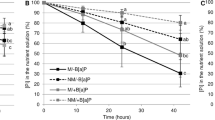
The H2O2 accumulation in mycorrhizal and non-mycorrhizal roots as well as in AMF extraradical structures in the presence or not of B[a]P was evaluated. In the absence of B[a]P, the proportions of stained roots were about 14 and 10% in non-mycorrhizal and mycorrhizal roots, respectively (Table 2). Whereas the presence of B[a]P induced an increase of H2O2 accumulation by 49% in non-mycorrhizal roots, no change in H2O2 accumulation was detected in mycorrhizal roots. Concerning the AMF, while 7% of spores accumulated H2O2 in the absence of B[a]P, 52% accumulated this ROS in the presence of B[a]P (Fig. 1).
Accumulation of H2O2 into R. irregularis spores. Production of H2O2 was visualized by microscopy (×50 magnification) using the DAB staining. Spore without H2O2 (a), spore accumulating H2O2 (b), percentage of spores containing H2O2 in the absence (control) or in the presence of benzo[a]pyrene (polluted) (c). Data are presented as means ± SD. Bars topped by different letters indicate significant differences between spores produced in the control and in the polluted medium according to the Mann-Whitney U test (P < 0.05)
B[a]P disturbs M. truncatula root and R. irregularis gene expression
Gene expression in non-mycorrhizal and mycorrhizal roots was studied after 7 days of exposure to B[a]P (Fig. 2). Genes involved in antioxidant system, MtSOD, MtPOX, and MtAPX, were found to be upregulated by 14-, 11-, and 3-fold, respectively, in non-mycorrhizal roots exposed to B[a]P in comparison with the non-polluted control. In contrast, the expressions of these three genes were decreased in mycorrhizal roots exposed to B[a]P in comparison also with the non-polluted control. In the presence of B[a]P, the levels of MtSOD, MtPOX, and MtAPX gene expressions were 8, 33, and 4 times (relative expression of 0.13, 0.03, and 0.23) less important in mycorrhizal roots than in non-mycorrhizal roots, respectively. For the MtGST gene, the transcript analysis suggested an upregulation (18-fold) in the non-mycorrhizal roots exposed to B[a]P and a downregulation (17-fold, relative expression of 0.06) in the mycorrhizal roots exposed to B[a]P. A similar pattern was observed for DNA repair genes, with an accumulation of the transcripts in non-mycorrhizal roots exposed to B[a]P (between 3- and 5-fold) and a decrease in mycorrhizal roots exposed to B[a]P (between 33- and 50-fold, relative expressions of 0.03 and 0.02) in comparison to the non-polluted controls.
Effect of benzo[a]pyrene on the relative expression (fold change) of genes involving in antioxidant system (MtSOD, MtPOX, and MtAPX), detoxification (MtGST) and DNA repair (MtTFIIS and MtTdp1α) determined by qRT-PCR in non-mycorrhizal (white bar) and mycorrhizal roots (grey bar). Gene expression was considered as significantly up- or downregulated related to the 1× controls (dashed line), when changes in relative expression were >2× or <0.5×, respectively
In contrast, no differences were observed for all gene expressions tested (GintSOD, GintGRX1, GintPDX1, and GintGST) in AMF extraradical structures after 7 days of exposure to B[a]P in comparison with the control (Fig. 3).
Effect of benzo[a]pyrene on the relative expression (fold change) of genes involving in antioxidant system (GintSOD, GintGRX1, and GintPDX1) and detoxification (GintGST) determined by qRT-PCR in extraradical structures of R. irregularis. Gene expression was considered as significantly upregulated related to the 1× controls (non-exposed roots), when changes in relative expression were >2×
B[a]P impacts M. truncatula and R. irregularis enzyme activities
As shown in Table 2, mycorrhizal inoculation and pollution affect individually SOD, POX, and GST activities. Moreover, the interaction between inoculation and pollution showed also significant impact on these three enzyme activities. Whereas inoculation was found to reduce SOD, POD, and GST activities in polluted conditions by 2, 1.4, and 2 folds respectively, they were increased by 1.1, 1.6, and 2 folds in the control (non-polluted condition).
In the AMF extraradical mycelium exposed to B[a]P, SOD, POX, and GST activities were induced by 1.6-, 2.5-, and 1.2-fold, respectively, by comparison with the control (Table 4).
B[a]P bioaccumulation in roots
B[a]P accumulation was evaluated in mycorrhizal and non-mycorrhizal roots (Fig. 4). It was 2.6-fold higher in non-mycorrhizal roots by comparison with mycorrhizal ones.
Discussion
Arbuscular mycorrhizal fungi are able to improve plant establishment and growth in extreme environments such as polluted soils, but little is known about the genes involved in plant protection by mycorrhization against pollutant toxicity, in particular the presence of B[a]P, a high molecular weight PAH frequently detected in contaminated soils.
First, the responses of non-mycorrhizal roots to B[a]P exposure were investigated. Our findings showed that the expression of the genes MtSOD, MtPOX, and MtAPX, encoding antioxidant enzymes, were induced after B[a]P exposure. These results are in accordance with the literature where it was reported that expression of several M. truncatula antioxidant genes encoding SOD, POX, APX, catalases, and glutathione reductases were upregulated in the presence of different abiotic stresses (Aloui et al. 2009; Marino et al. 2013; Rahoui et al. 2014). Although, it is well known that these antioxidant enzymes play a crucial role in ROS elimination and therefore in plant protection against pollutants like PAH (Liu et al. 2009; Martí et al. 2009; DongXue et al. 2011; Song et al. 2011) or metal trace elements (López-Millán et al. 2005; Marino et al. 2013), this is the first time that a correlation between the upregulation of the antioxidant MtSOD, MtPOX gene expression and the induction of SOD and POX activities correlated with H2O2 production in non-mycorrhizal roots after B[a]P exposure has been shown.
Furthermore, our results also showed that expression of two DNA repair genes (MtTFIIS and MtTdp1α) was upregulated in non-mycorrhizal roots under B[a]P exposure in comparison with the non-polluted condition. These upregulations suggest that B[a]P provoked DNA alteration in non-mycorrhizal roots. Indeed, a previous study highlighted the formation of 8-hydroxy-2-deoxyguanosine DNA adduct, a biomarker of oxidative DNA damage, in non-mycorrhizal chicory roots exposed to B[a]P pollution (Debiane et al. 2009). Our results, also corroborate previous data that demonstrated a good correlation between high levels of DNA oxidative damage and MtTFIIS and MtTdp1α upregulation in response to copper and osmotic stress in M. truncatula (Macovei et al. 2010; Macovei et al. 2011; Balestrazzi et al. 2011).
In addition, to implement protection mechanisms, plants can perform pollutant detoxification using enzymes such as GST. In fact, in the present study, concomitant increases in GST gene expression and in GST enzymatic activity were observed in non-mycorrhizal roots cultivated in the presence of B[a]P. Glutathione-S-transferases are heterogenous group of cell detoxifying enzymes, which catalyze the conjugation of tripeptide glutathione (GSH) to electrophilic sites on a wide range of phytotoxic substrates (Kampranis et al. 2000).
Second, H2O2 accumulation was shown in the AMF mycelium after B[a]P exposure. Although no changes in expression of the genes that we examined (GintSOD, GintGRX1, GintPDX1, and GintGST) were found, increases of the antioxidant enzyme activities SOD and POX were observed in R. irregularis grown in the presence of B[a]P. This result suggests that the AMF enhances its ROS scavenging enzymatic systems under stressful conditions. In addition, our results showed an increase of GST activity, involved in xenobiotic detoxification in the AMF during B[a]P exposure. Although the ability of GST to detoxify xenobiotics has been reported in several saprotrophic (McGoldrick et al. 2005; Morel et al. 2009) and pathogenic fungi (Prins 2001), it has not been described previously in AMF.
Finally, the responses to B[a]P exposure were investigated in the symbiosis M. truncatula root/R. irregularis. Unlike non-mycorrhizal roots, concomitant downregulation of genes encoding antioxidant enzymes (MtSOD, MtPOX, and MtAPX) and decreases of antioxidant enzyme activities (SOD and POX) were observed in mycorrhizal roots cultivated in the presence of B[a]P. These results are in agreement with those of Aloui et al. (2009) which revealed decreases of SOD transcripts and SOD proteins in M. truncatula mycorrhizal roots in the presence of cadmium. A similar behavior was described to in mycorrhizal tomato roots after metal trace element exposure. Whereas downregulation of two genes, Lemt2 and LeNrmp1, encoding metallothionein and metal trace element transporter, respectively, was observed in mycorrhizal roots, upregulation was observed in non-mycorrhizal roots (Ouziad et al. 2005). Moreover, our data also show, concomitant GST gene downregulation together with decreased GST enzymatic activity in mycorrhizal roots. In addition, B[a]P led to the downregulation of the two DNA repair genes (MtTFIIS and MtTdp1α) in mycorrhizal roots. These data support the study of Debiane et al. (2009) which reported less DNA damage in mycorrhizal chicory roots in comparison to non-mycorrhizal roots under B[a]P exposure.
All of these results suggest that mycorrhizal roots will be less sensitive to B[a]P toxicity than will non-colonized roots. According to all of our results, it seems that the upregulation of the genes MtSOD, MtPOX, MtAPX, MtGST, MtTFIIS, MtTdp1α, and GintPDX1 and the increase of activities of the enzymes SOD, POX, and GST detected under B[a]P pollution in both of the symbiosis partners, the non-mycorrhizal roots and the AMF, could be defense reactions against the oxidative stress induced by the PAH. In contrast, the downregulation of the genes MtSOD, MtPOX, MtAPX, MtGST, MtTFIIS, and MtTdp1α and the decrease of activities of SOD, POX, and GST observed in mycorrhizal roots in the presence of B[a]P could be a consequence of a protective effect against the pollutant toxicity provided by the AMF to the plant. In fact, our results showed that B[a]P exposure increased H2O2 production in non-mycorrhizal roots and in the AMF mycelium but not in mycorrhizal roots. These observations are in accordance with studies reporting that pollutants such as metal trace elements induced ROS accumulation by Medicago cells (Rahoui et al. 2014) and by the AMF (Benabdellah et al. 2009b). Our data suggest an alleviation of oxidative stress in the presence of the AMF. This assumption corroborates previous results that revealed a lower malondialdehyde (a lipid peroxidation biomarker) and 8-hydroxy-2′-deoxyguanosine (a DNA damage biomarker) content in mycorrhizal chicory roots under B[a]P exposure (Debiane et al. 2009).
It can be hypothesized that the fungus brought a protective effect to the mycorrhizal plant by accumulating the pollutant, making it less available to the plant. Indeed, our results showed a lower bioaccumulation of B[a]P in mycorrhizal roots than in non-mycorrhizal roots, suggesting that the extraradical structures could reduce the pollutant availability to the plant. AMF develop an extended mycelial network, which is several orders of magnitude longer than plant roots (Khan et al. 2000). Such a lower concentration of pollutant in mycorrhizal than in non-colonized roots also was observed in the presence of different PAH (Verdin et al. 2006; Zhou et al. 2013) and in the presence of metal trace elements (Li and Christie 2001; Vivas et al. 2006; Rahmaty and Khara 2011; Abdel Latef 2011). This phenomenon is probably a consequence of the ability of AMF to store PAH in their hyphal lipid bodies as described by Verdin et al. (2006). Thus, colonization of roots might have lowered the concentration of B[a]P in plant cells below the level needed to induce gene expression and enzymatic activities involved in PAH tolerance and detoxification.
We cannot exclude, however, that the AMF directly affects the host plant metabolism. Our findings showed that, in the absence of B[a]P, some tolerance and detoxification enzyme activities are induced by mycorrhization possibly leading to plant protection. Regulation of plant gene expression by AMF has been described, in particular, during the establishment of the symbiosis. Indeed, it is known that during root colonization, AMF penetration is accompanied by a transient increase in SOD and POX enzyme activities suggesting the existence of feedback defense mechanisms in plants (García-Garrido and Ocampo 2002; Güimil et al. 2005; Zamioudis and Pieterse 2012). A protein “effector” SP7 secreted by R. irregularis and emitted into the plant cell by the AMF interacts with the transcription factor ERF19 to modulate the plant defense reactions (Kloppholz et al. 2011). It can be expected that a similar regulation mechanism could be involved in the mycorrhizal roots exposed to B[a]P, resulting in the downregulation observed for some genes studied in the present work. Moreover, it is likely that other genes, not investigated in the present study, are activated by the AMF to fight against oxidative stress induced by the presence of B[a]P. In addition, as the fungal gene expression was carried out on extraradical AMF structures in the present study, it would be interesting to investigate the fungal response to the pollutant in its intraradical structures by using micro-dissection.
In conclusion, our findings demonstrated a beneficial contribution of the AMF R. irregularis inoculation in the protection of M. truncatula roots against B[a]P toxicity. Mycorrhizal inoculation decreased B[a]P bioaccumulation in roots and thereby alleviated oxidative stress. Being less affected by pollutant toxicity than non-colonized roots, mycorrhizal roots did not activate any defense mechanism either at the gene expression regulation level or at the enzymatic level. This work further emphasizes the utility of in vitro cultures to investigate the mechanisms behind the impact of pollutants on the plant-beneficial soil microorganisms such AMF.
References
Abdel Latef AAH (2011) Influence of arbuscular mycorrhizal fungi and copper on growth, accumulation of osmolyte, mineral nutrition and antioxidant enzyme activity of pepper (Capsicum annuum L.). Mycorrhiza 21:495–503
Aloui A, Recorbet G, Gollotte A et al (2009) On the mechanisms of cadmium stress alleviation in Medicago truncatula by arbuscular mycorrhizal symbiosis: a root proteomic study. Proteomics 9:420–433
Baborová P, Möder M, Baldrian P et al (2006) Purification of a new manganese peroxidase of the white-rot fungus Irpex lacteus, and degradation of polycyclic aromatic hydrocarbons by the enzyme. Res Microbiol 157:248–253
Balestrazzi A, Confalonieri M, Macovei A, Carbonera D (2011) Seed imbibition in Medicago truncatula Gaertn.: expression profiles of DNA repair genes in relation to PEG-mediated stress. J Plant Physiol 168:706–713
Becard G, Fortin JA (1988) Early events of vesicular-arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytol 108:211–218
Benabdellah K, Azcón-aguilar C, Valderas A et al (2009a) GintPDX1 encodes a protein involved in vitamin B6 biosynthesis that is up-regulated by oxidative stress in the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol 184:682–693
Benabdellah K, Merlos M-A, Azcón-Aguilar C, Ferrol N (2009b) GintGRX1, the first characterized glomeromycotan glutaredoxin, is a multifunctional enzyme that responds to oxidative stress. Fungal Genet Biol 46:94–103
Bustin S (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25:169–193
Chen, B., Jakobsen, I., Roos, P., & Zhu, Y. G. (2005). Effects of the mycorrhizal fungus Glomus intraradices on uranium uptake and accumulation by Medicago truncatula L. from uranium-contaminated soil. Plant and soil 275:349–359
Chigbo C, Batty L (2013) Phytoremediation potential of Brassica juncea in Cu-pyrene co-contaminated soil: comparing freshly spiked soil with aged soil. J Environ Manag 129:18–24
Criquet S, Joner E, Leglize P, Leyval C (2000) Anthracene and mycorrhiza affect the activity of oxidoreductases in the roots and the rhizosphere of lucerne (Medicago sativa L.). Biotechnol Lett 22:1733–1737
Dat J, Vandenabeele S, Vranová E et al (2000) Dual action of the active oxygen species during plant stress responses. Cellular and Molecular Life Sciences CMLS 57:779–795
Debiane D, Garçon G, Verdin A et al (2008) In vitro evaluation of the oxidative stress and genotoxic potentials of anthracene on mycorrhizal chicory roots. Environ Exp Bot 64:120–127
Debiane D, Garçon G, Verdin A et al (2009) Mycorrhization alleviates benzo[a]pyrene-induced oxidative stress in an in vitro chicory root model. Phytochemistry 70:1421–1427
Dietz AC, Schnoor JL (2001) Advances in phytoremediation. Environ Health Perspect 1:163–168
Dodor DE, Hwang H-M, Ekunwe SI (2004) Oxidation of anthracene and benzo[a]pyrene by immobilized laccase from Trametes versicolor. Enzym Microb Technol 35:210–217
DongXue Y, YuHong S, Min Q (2011) Bio-toxicity effect of polycyclic aromatic hydrocarbons pyrene on Arabidopsis thaliana. J Anhui Agric Sci 12:818–822
El Chartouni L, Randoux B, Duyme F et al (2012) Correlation of cytological and biochemical parameters with resistance and tolerance to Mycosphaerella graminicola in wheat. Plant Biol 14:11–21
Fester T, Hause G (2005) Accumulation of reactive oxygen species in arbuscular mycorrhizal roots. Mycorrhiza 15:373–379
Gao Y, Li Q, Ling W, Zhu X (2011) Arbuscular mycorrhizal phytoremediation of soils contaminated with phenanthrene and pyrene. J Hazard Mater 185:703–709
García-Garrido JM, Ocampo JA (2002) Regulation of the plant defence response in arbuscular mycorrhizal symbiosis. J Exp Bot 53:1377–1386
González-Guerrero M (2005) Estudio de los mecanismos implicados en la homeostasis de metales pesados en el hongo formador de micorrizas arbusculares Glomus intraradices. University of Granada, Granada
Güimil S, Chang HS et al (2005) Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc Natl Acad Sci 102:8066–8070
Joner EJ, Leyval C (2003) Rhizosphere gradients of polycyclic aromatic hydrocarbon ( PAH ) dissipation in two industrial soils and the impact of arbuscular mycorrhiza. Environ Sci Technol 37:2371–2375
Kampranis SC, Damianova R, Atallah M et al (2000) A novel plant glutathione S-transferase/peroxidase suppresses Bax lethality in yeast. J Biol Chem 275:29207–29216
Khan A, Kuek C, Chaudhry T et al (2000) Role of plants, mycorrhizae and phytochelators in heavy metal contaminated land remediation. Chemosphere 41:197–207
Khan S, Aijun L, Zhang S et al (2008) Accumulation of polycyclic aromatic hydrocarbons and heavy metals in lettuce grown in the soils contaminated with long-term wastewater irrigation. J Hazard Mater 152:506–515
Kloppholz S, Kuhn H, Requena N (2011) A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr Biol 21:1204–1209
Lanfranco L, Novero M, Bonfante P, Torino S (2005) The mycorrhizal fungus Gigaspora margarita possesses a CuZn superoxide dismutase that is up-regulated during symbiosis with legume hosts 1. Plant Physiol 137:1319–1330
Lenoir I, Fontaine J, Lounès-Hadj Sahraoui A (2016b) Arbuscular mycorrhizal fungal responses to abiotic stresses: a review. Phytochemistry 123:4–15
Lenoir I, Lounès-Hadj Sahraoui A, Fontaine J (2016c) Arbuscular mycorrhizal fungal-assisted phytoremediation of soil contaminated with persistent organic pollutants: a review. Eur J Soil Sci 67:624–640
Lenoir I, Lounès-Hadj Sahraoui A, Frédéric L et al (2016a) Arbuscular mycorrhizal wheat inoculation promotes alkane and polycyclic aromatic hydrocarbon biodegradation: microcosm experiment on aged-contaminated soil. Env Pol 213:549–560
Li X, Christie P (2001) Changes in soil solution Zn and pH and uptake of Zn by arbuscular mycorrhizal red clover in Zn-contaminated soil. Chemosphere 42:201–207
Li JH, Yu XZ, Wu SC et al (2011) Responses of bioaugmented ryegrass to PAH soil contamination. Int J Phytoremediation 13:441–455
Liu H, Weisman D, Ye Y et al (2009) An oxidative stress response to polycyclic aromatic hydrocarbon exposure is rapid and complex in Arabidopsis thaliana. Plant Sci 176:375–382
López-Millán AF, Ellis DR, Grusak MA (2005) Effect of zinc and manganese supply on the activities of superoxide dismutase and carbonic anhydrase in Medicago truncatula wild type and raz mutant plants. Plant Sci 168:1015–1022
Macovei A, Balestrazzi A, Confalonieri M, Carbonera D (2010) The tyrosyl-DNA phosphodiesterase gene family in Medicago truncatula Gaertn.: bioinformatic investigation and expression profiles in response to copper- and PEG-mediated stress. Planta 232:393–407
Macovei A, Balestrazzi A, Confalonieri M et al (2011) The TFIIS and TFIIS-like genes from Medicago truncatula are involved in oxidative stress response. Gene 470:20–30
Marino D, Damiani I, Gucciardo S et al (2013) Inhibition of nitrogen fixation in symbiotic Medicago truncatula upon Cd exposure is a local process involving leghaemoglobin. J Exp Bot 64:5651–5660
Martí MC, Camejo D, Fernández-García N et al (2009) Effect of oil refinery sludges on the growth and antioxidant system of alfalfa plants. J Hazard Mater 171:879–885
Martin BC, George SJ, Price CA et al (2014) The role of root exuded low molecular weight organic anions in facilitating petroleum hydrocarbon degradation: current knowledge and future directions. Sci Total Environ 472:642–653
McGoldrick S, O’Sullivan SM, Sheehan D (2005) Glutathione transferase-like proteins encoded in genomes of yeasts and fungi: insights into evolution of a multifunctional protein superfamily. FEMS Microbiol Lett 242:1–12
McGonigle TP, Miller MH, Evans DG et al (1990) A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol 115:495–501
Miller G, Shulae V, Mittler R (2008) Reactive oxygen signaling and abiotic stress. Physiol Plant 133:481–489
Mitchell H, Hall J, Barber M (1994) Elicitor-induced cinnamyl alcohol dehydrogenase activity in lignifying wheat (Triticum aestivum 1.) leaves. Plant Physiol 104:551–556
Morel M, Ngadin AA, Droux M et al (2009) The fungal glutathione S-transferase system. Evidence of new classes in the wood-degrading basidiomycete Phanerochaete chrysosporium. Cell Mol Life Sci 66:3711–3725
Newman EI (1966) A method of estimating the total length of root in a sample. J Appl Ecol 3:139–145
Niyogi KK (1999) Photoprotection revisited: genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol 50:333–359
Ouziad F, Hildebrandt U, Schmelzer E, Bothe H (2005) Differential gene expressions in arbuscular mycorrhizal-colonized tomato grown under heavy metal stress. J Plant Physiol 162:634–649
Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool ( REST © ) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:1–10
Phillips JM, Hayman DS (1970) Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. Trans Br Mycol Soc 55:158–IN18
Pilon-Smits E (2005) Phytoremediation. Annu Rev Plant Biol 56:15–39
Pimda W, Bunnag S (2012) Biodegradation of used motor oil by single and mixed cultures of cyanobacteria. African J Biotechnol 11:9074–9078
Pommier Y (2003) Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat Res Mol Mech Mutagen 532:173–203
Prins T (2001) Identification and functional analysis of Botrytis cinerea genes induced during infection of tomato. Wageningen University, Wageningen
Puckette MC, Tang Y, Mahalingam R (2008) Transcriptomic changes induced by acute ozone in resistant and sensitive Medicago truncatula accessions. BMC Plant Biol 8:1–15
Rabie GH (2005) Role of arbuscular mycorrhizal fungi in phytoremediation of soil rhizosphere spiked with poly aromatic hydrocarbons. Mycobiology 33:41–50
Rahmaty R, Khara J (2011) Effects of vesicular arbuscular mycorrhiza Glomus intraradices on photosynthetic pigments, antioxidant enzymes, lipid peroxidation, and chromium accumulation in maize plants. Turkish J Biol 35:51–58
Rahoui S, Ben C, Chaoui A et al (2014) Oxidative injury and antioxidant genes regulation in cadmium-exposed radicles of six contrasted Medicago truncatula genotypes. Environ Sci Pollut Res 21:8070–8083
Rajtor M, Piotrowska-Seget Z (2016) Prospects for arbuscular mycorrhizal fungi (AMF) to assist in phytoremediation of soil hydrocarbon contaminants. Chemosphere 162:105–116
Riechers DE, Yang K, Irzyk GP et al (1996) Variability of glutathioneS-transferase levels and dimethenamid tolerance in safener-treated wheat and wheat relatives. Pestic Biochem Physiol 56:88–101
Schüßler, A., Walker C (2010) The Glomeromycota. A species list with new families and new genera. Libraries at The Royal Botanic Garden Edinburgh, The Royal Botanic Garden Kew, Botanische Staatssammlung Munich, and Oregon State University
Shen G, Lu Y, Hong J (2006) Combined effect of heavy metals and polycyclic aromatic hydrocarbons on urease activity in soil. Ecotoxicol Environ Saf 63:474–480
Soleimani M, Akbar S, Hajabbasi MA (2011) Enhancing phytoremediation efficiency in response to environmental pollution stress, Plants and. InTech
Song H, Wang Y-S, Sun C-C et al (2011) No title. Oceanol Hydrobiol Stud 40:9–18
Tisserant E, Malbreil M, Kuo A et al (2013) Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc Natl Acad Sci U S A 110:20117–20122
Verdin A, Lounès-Hadj Sahraoui A, Durand R (2004) Degradation of benzo[a]pyrene by mitosporic fungi and extracellular oxidative enzymes. Int Biodeterior Biodegradation 53:65–70
Verdin A, Lounès-Hadj Sahraoui A, Fontaine J et al (2006) Effects of anthracene on development of an arbuscular mycorrhizal fungus and contribution of the symbiotic association to pollutant dissipation. Mycorrhiza 16:397–405
Vivas A, Biró B, Németh T et al (2006) Nickel-tolerant Brevibacillus brevis and arbuscular mycorrhizal fungus can reduce metal acquisition and nickel toxicity effects in plant growing in nickel supplemented soil. Soil Biol Biochem 38:2694–2704
Wild SR, Berrow ML, Jones KC (1991) The persistence of polynuclear aromatic hydrocarbons (PAHs) in sewage sludge amended agricultural soils. Environ Pollut 72:141–157
Yang S, Burgin AB, Huizenga BN et al (1996) A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Biochemistry 93:11534–11539
Yu XZ, Wu SC, Wu FY, Wong MH (2011) Enhanced dissipation of PAHs from soil using mycorrhizal ryegrass and PAH-degrading bacteria. J Hazard Mater 186:1206–1217
Zamioudis C, Pieterse CM (2012) Modulation of host immunity by beneficial microbes. Mol Plant Microbe In 25:139–150
Zhou X, Zhou J, Xiang X (2013) Impact of four plant species and arbuscular mycorrhizal (AM) fungi on polycyclic aromatic hydrocarbon (PAH) dissipation in spiked soil. Polish J Environ Stud 22:1239–1245
Acknowledgements
The authors wish to thank the Région Nord-Pas-de-Calais and the Pôle Métropolitain de la Côte d’Opale for providing financial supports for I. Lenoir’s Ph.D thesis. The laboratory participates in the Institut de Recherche en Environnement Industriel which is financed by the Communauté Urbaine de Dunkerque, the Région Nord-Pas-de-Calais, the Ministère de l’Enseignement Supérieur et de la Recherche, the CNRS and European Regional Development Fund. We thank Mrs. Natacha Bourdon and M. Frédéric Laruelle for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lenoir, I., Fontaine, J., Tisserant, B. et al. Beneficial contribution of the arbuscular mycorrhizal fungus, Rhizophagus irregularis, in the protection of Medicago truncatula roots against benzo[a]pyrene toxicity. Mycorrhiza 27, 465–476 (2017). https://doi.org/10.1007/s00572-017-0764-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-017-0764-1