Abstract
Non-native tree species are often used as ornamentals in urban landscapes. However, their root-associated fungal communities remain yet to be examined in detail. Here, we compared richness, diversity and community composition of ectomycorrhizosphere fungi in general and ectomycorrhizal (EcM) fungi in particular between a non-native Pinus nigra and a native Quercus macrocarpa across a growing season in urban parks using 454-pyrosequencing. Our data show that, while the ectomycorrhizosphere community richness and diversity did not differ between the two host, the EcM communities associated with the native host were often more species rich and included more exclusive members than those of the non-native hosts. In contrast, the ectomycorrhizosphere communities of the two hosts were compositionally clearly distinct in nonmetric multidimensional ordination analyses, whereas the EcM communities were only marginally so. Taken together, our data suggest EcM communities with broad host compatibilities and with a limited numbers of taxa with preference to the non-native host. Furthermore, many common fungi in the non-native Pinus were not EcM taxa, suggesting that the fungal communities of the non-native host may be enriched in non-mycorrhizal fungi at the cost of the EcM taxa. Finally, while our colonization estimates did not suggest a shortage in EcM inoculum for either host in urban parks, the differences in the fungi associated with the two hosts emphasize the importance of using native hosts in urban environments as a tool to conserve endemic fungal diversity and richness in man-made systems.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ectomycorrhizal (EcM) fungi are critical to the ecosystem function in temperate and boreal forest ecosystems (Read 1991; Smith and Read 2008). Urbanization leads to severe soil disturbance and has been proposed to compromise the mycorrhizal communities by either reducing inoculum loads (Cousins et al. 2003) and colonization (Baxter et al. 1999; Timonen and Kauppinen 2008; Bainard et al. 2011; Karpati et al. 2011) or by rearranging the fungal community composition (Baxter et al. 1999; Jumpponen et al. 2010a; Karpati et al. 2011). The reduced inoculum potential and colonization in urban environments are likely attributable to challenging urban environments — compacted and often salt or heavy metal enriched urban soils (Pouyat and McDonnell 1991; Kaye et al. 2006; Polanco et al. 2008; Pavao-Zuckerman 2008) can compromise plant performance and hinder mycorrhizal colonization. In addition to anthropogenic impacts on soil properties, urban stand management may lead to lower fungal richness and altered community composition compared to surrounding areas (Jumpponen and Jones 2010; Jumpponen et al. 2010a). Accordingly, studies that target the effects of urban management activities on fungal communities are few but necessary to identify drivers that may lead to the decline in fungal richness (Newbound et al. 2010).
Human population living in cities and suburbs is increasing (Ash et al. 2008) and ornamental urban trees are becoming increasingly important because of their high esthetic, recreational and economic value. However, urban stands may also provide means for conservation on local scales. Although studies evaluating the importance of mycorrhizal associations in urban systems are few (but see Polanco et al. 2008), it is likely that mycorrhizae are fundamental for the health and growth of urban ornamental trees. Therefore, maintaining EcM communities that may best support longevity of urban trees is crucial. Host taxon selection in the maintenance of symbiotic communities is similarly important as locally adapted symbioses may outperform introduced ones (Bonfante et al. 1998; Bahram et al. 2013). EcM communities in planted, man-constructed stands with non-native hosts often differ from native stands compositionally, even if the fungal richness may remain unaltered (O’Hanlon and Harrington 2012). Evaluating different host taxa in urban settings is important to evaluate the consequences of ornamental tree selection.
While a number of environmental and anthropogenic drivers likely interact in urban systems and can cause community rearrangement (e.g., Baxter et al. 1999; Jumpponen and Jones 2010; Jumpponen et al. 2010b), host selection is likely to be crucial to facultatively biotrophic organisms such as the EcM fungi (Kranabetter et al. 1999; Jumpponen et al. 2010a). The use of non-native species in urban ecosystems can have cascading effects. Once introduced, non-native taxa can promote further introduction of peripheral symbionts or companion taxa that may compromise the establishment, restoration, or maintenance of original native systems — resulting in loss of ecological memory (Schaefer 2011). Management practices in urban ecosystems, including selection of ornamental non-native species, use of fertilizers and pesticides, as well as inadvertent biogeochemical alterations and pollutant enrichment likely exaggerate shifts from native to non-native communities furthering the loss of ecological memory.
EcM fungi vary in host specificities and can range from narrow (limited to a level of a genus) to broad (across families and orders) compatibilities (Molina et al. 1992). The non-native EcM hosts may expediently adopt local fungal communities (Cullings et al. 2000; Bahram et al. 2013; O’Hanlon et al. 2013), or alternatively, rely on EcM fungi that are co-introduced (Vellinga et al. 2009; Dickie et al. 2010). Because of the previously observed EcM fungus selectivity for particular hosts (Jacobson and Miller 1992; Molina et al. 1992; Richard et al. 2005; Walker et al. 2005; Tedersoo et al. 2008; Morris et al. 2009), we expected that the fungal communities associated with the ectomycorrhizospheres of two non-confamilials would support distinct communities differing also in their EcM species richness (see Trocha et al. 2012). In other words, we hypothesized that the non-native host would support a limited number of broad host range fungi in our urban environment, whereas the native host would support an overlapping community of broad host range fungi but also a number of narrow host range taxa. As a result of our choice to use high throughput next generation sequencing, we acquired data that broadly characterized fungal communities in the ectomycorrhizosphere in general. Additionally, we filtered non-mycorrhizal fungi and performed additional analyses that focused on EcM taxa only. Our data suggest that both hosts support high EcM colonization and diverse communities that differ between the two hosts and are likely enriched for endemic EcM taxa in the native host.
Materials and methods
Study sites
The city of Manhattan is located in the Flint Hill region of northeastern Kansas, USA. It contains a population of approximately 50,000 residents, with an additional >20,000 transient student population. Quercus macrocarpa (Bur oak) is a native Kansas tree commonly used as an ornamental. Pinus nigra (Austrian pine) is native to Mediterranean Europe but often chosen as an ornamental in Kansas and mixed with various Quercus spp. Although exotic to North America, inoculation and common garden studies show that P. nigra is broadly compatible with cosmopolitan EcM taxa (Martin-Pinto et al. 2006) and share fungi with other Pinaceae (Lazarevic et al. 2012) as well as Fagaceae (Trocha et al. 2012). One isolated individual of each species was selected at each of the six urban sites for a total of 12 trees across the study (Table 1). The sites were sampled three times (June 13, August 8, and October 3) at 8-week intervals during the growing season of 2011 resulting in a total of 36 samples.
Sampling and evaluation of EcM colonization
At each of the three sampling occasions, three evenly distributed, 15 cm deep 5 cm (diam.) cores were sampled ~3 m from the bore of each tree resulting in a total of 36 subsamples per sampling occasion; 108 subsamples in total. The cores were stored on ice until frozen in the laboratory at −20 °C until further processing. Roots were manually separated from the soil cores and cleaned from the adhering soil. We used isolated trees to visually identify EcM from the abundant grass roots in the park soils. All EcM root tips were collected from all three cores under 10× magnification based on their visual appearance and presence of a mantel. Total number of root tips and EcM tips were enumerated in one random core to estimate colonization and volumetric densities of root tips and EcM. The EcM tips were stored in MoBio Bead Tubes (UltraClean Soil DNA, MoBio Laboratories) in the bead solution and frozen at −20 °C until DNA was extracted separately from all EcM in each core. The root colonization data were arcsine square root transformed prior to analyses to satisfy the assumptions of analysis of variance (ANOVA). The between (host species) and within (sampling time) subject effects were analyzed using a repeated-measures ANOVA. Additionally, we analyzed the differences between the two host species separately for each of the three sampling times using one-way ANOVA.
DNA extraction and qPCR to estimate inoculum loads
Total DNA was extracted with UltraClean Soil DNA kit following the manufacturer’s instructions and eluted in 50 μl of molecular grade H2O. The DNA yields were quantified with a Nanodrop ND1000 spectrometer (Thermo Scientific, Wilmington, Delaware) and adjusted to a final 2 ng/μl concentration. We selected one random sample for each experimental unit from each sampling time to estimate inoculum loads by quantitative real-time PCR (qPCR) using primers ITS1F and 5.8S as described by Fierer et al. (2005). Each qPCR reaction contained the following final concentrations or absolute amounts of reagents: 1 nM each primer, 5 ng template DNA, 12.5 μl iQ™ SYBR® Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA), and 7.5 μl molecular grade H2O for 25 μl per reaction. The qPCR reactions were performed in triplicate in an iCycler iQ™ (Bio-Rad Laboratories) RT-PCR detection system with iCycler software (v. 3.1.7050). Cycling parameters included an initial denaturing at 95 ° C for 5 min, and 55 cycles of 95 °C denaturation for 1 min, 53 °C annealing step for 30 s, and 72° extension step for 1 min. Standard curves based on a 10-fold dilution series (4×10−7 to 4×10−3 ng of the plasmid DNA per reaction) were used to estimate ITS1 copy numbers per sample (Pfaffl 2001) accounting for the target region length and assuming an average dsDNA molecular mass of 660 g mol−1. The qPCR data were log10 transformed prior to analyses to satisfy the assumptions of ANOVA. The between (host species) and within (sampling time) subject effects as well as the differences between the two host species separately for events were analyzed as described above.
PCR amplification and sequencing
The template concentration, annealing temperature, and cycle numbers were optimized for PCR amplification of the fungal large subunit (LSU) of the ribosomal RNA gene. We used a two-step PCR protocol as suggested by Berry et al. (2011) to avoid the DNA-tag related 3′-end bias during the amplification. Each primary PCR reaction contained the following final concentrations or absolute amounts of reagents: 1 nM of each primer (LR0R and LR3; Amend et al. 2010), 10 ng of the template DNA, 200 μM of each deoxynucleotide triphosphate, 2.5 mM MgCl2, 1 unit of GoTaq DNA polymerase (Promega, Madison, WI, USA), and 5 μl of PCR buffer (Promega, Madison, WI, USA). The parameters consisted of an initial denaturation at 94 °C for 4 min, then 30 cycles of denaturation at 94 °C for 1 min, annealing at 53 °C for 45 s and extension at 72 °C for 2 min, followed by a final extension step at 72 °C for 8 min. All PCR reactions were performed in 96-well PCR plates on MasterCyclers (Eppendorf, Hamburg, Germany) in three technical replicates to account for potentially heterogeneous amplification from the environmental template. In the secondary PCR, a total of 5 μl of the LSU amplicons from the primary PCR were reamplified with DNA-tagged primers that combined the 454-A adapters and the sample specific DNA tags (LR0R-A) and the 454-B adapters (LR3-B). The primers and DNA tags are available in Table S1. The reaction conditions and reagents were identical to those described above, but the number of cycles was reduced to five. Possible PCR amplification of contaminants was determined using a blank sample run through the extraction protocol simultaneously with the actual samples and a negative PCR control in which the template DNA was replaced with ddH2O. These remained free of PCR amplicons. The three technical replicates for each experimental unit were combined into one sample prior to purification with a magnetic bead clean up system (AmPure SPRI; Beckman Coulter, Inc., Brea, CA, USA). The clean amplicons were quantified with the ND1000 spectrometer and 100 ng for each experimental unit pooled for subsequent 454-sequencing. The pools representing the three seasonal samples of the 12 experimental units were aliquoted to 1,000 ng in a 100-μl volume (10 ng/μl) and sequenced in one quarter sequencing reaction on a Roche GS-FLX Titanium 454-pyrosequencer at the Integrated Genomics Facility at Kansas State University. Two samples yielded no amplicons and were omitted from further analyses.
Bioinformatics and operational taxonomic unit (OTU) designation
The sequence data were processed using mothur (v. 1.30.2; Schloss et al. 2009) following a modified standard operating protocol (Schloss et al. 2011). Briefly, we extracted the quality, flow, and sequence data from raw files (.sff). Sequences without a forward primer or sample-specific DNA-tags were omitted and remaining sequences denoised (Quince et al. 2009). Sequences with long homopolymers (maxhomop = 8) and shorter than minimum length threshold (minlength = 200) were removed. The 454-sequences were aligned against mafft-aligned (Katoh et al. 2005) LSU training dataset from the Ribosomal Database Project (RDP) compiled of high-quality public Sanger sequences spanning the first 1,400 bp of the LSU gene (Liu et al. 2012), screened for proper alignment, unique representatives pre-clustered to 99 % similar representatives to further reduce potential noise in the final data analyses, and reads with potential chimeric origin removed (uchime; Edgar et al. 2011). The data were assigned to OTUs at 97 % similarity using the average neighbor joining algorithm and global singletons omitted (see Tedersoo et al. 2010). OTUs were assigned to taxa using the naϊve Bayesian classifier (Wang et al. 2007) against the RDP’s training dataset at a 60 % bootstrap support cutoff (Liu et al. 2012). Based on the genus-level affinities, OTUs were assigned to fungi with known EcM habits (Table 2) or those with uncertain or unknown ecologies. Two datasets comprised of all fungi in the ectomycorrhizosphere and of EcM fungi only were analyzed separately. As recommended by Gihring et al. (2012), the data were randomly sub-sampled to the lowest number of OTUs per experimental unit for further analysis.
We estimated the rarefaction curves (Magurran 1988) for each experimental unit to evaluate the representativeness of our sampling effort. Observed and extrapolated OTU richness (S obs and Chao1), diversity (Simpson’s complement; 1 − D = 1 − ∑p i 2), evenness (Simpson’s equitability; E D = (1/∑p i 2)/S), and Good’s coverage (complement of the ratio between singleton OTUs and the total sequence count) were estimated after subsampling the data to 158 sequences for all ectomycorrhizosphere fungi and to 120 for EcM fungi to eliminate bias stemming from unequal sampling effort across experimental units (Gihring et al. 2012). The richness and coverage data were log10-transformed; diversity and evenness data arc sine square root transformed to satisfy the assumptions of ANOVA. Similar to the colonization and inoculum load analyses, the data were analyzed both by repeated-measures ANOVA as well as separately for each sampling time.
Community composition
To visualize and infer compositional differences in the fungal communities colonizing the two hosts, we used nonmetric multidimensional scaling (NMDS) as implemented in mothur (v. 1.30.2; Schloss et al. 2009). In brief, we derived a Bray–Curtis distance matrix and estimated NMDS axis scores for first four ordination axes in the ectomycorrhizosphere dataset and first three in the EcM dataset to obtain a stress ≤0.20. We analyzed the resultant axis scores as described above for other derived metrics. Additionally, we used analyses of molecular variance (AMOVA; PERMANOVA in Anderson 2001) to infer community-wide differences based on Bray–Curtis similarity values between the two hosts. However, this iterative permutation test does not permit repeated-measures analyses. Rather, it provides an overall test of community differences for our two hosts and is used to corroborate treatment differences inferred from ANOVA.
In addition to the ordination analyses to visualize community level differences, we also separately analyzed those OTUs that occurred in counts greater than a total of 50 sequences in the dataset. This resulted in a total of 43 focal OTUs in these analyses. The relative frequencies of the OTUs were log10 transformed and analyzed for host species separately for each of the three sampling times. Since these analyses are likely sensitive to erroneous conclusions because of multiple comparisons, the results were further evaluated after false detection rate (FDR) corrections (Q = 0.10). Few of the observed differences remained significant (those with P values ≤0.0090) after the correction suggesting limited statistical power with six replicates per hosts; we present the uncorrected tests here but advise the reader to observe the determined FDR threshold.
Results
Colonization and inoculum loads in the native and exotic hosts
Although the native Q. macrocarpa tended to have a greater density of root tips (Fig. 1a), the hosts did not differ in EcM tip densities when analyzed across the growing season (Fig. 1b; Table 2). The two hosts were heavily colonized (Q. macrocarpa: 86.1 ± 7.5 %; P. nigra: 79.6 ± 24.1 %) despite being isolated ornamentals in an urban environment (Fig. 1c). Colonization did not differ between the hosts but the nearly significant “Time” term in our repeated-measures ANOVA (Table 2) suggests seasonal dynamics, mainly driven by the small decline in Q. macrocarpa colonization during the growing season (Fig. 1c). Q. macrocarpa and P. nigra root and EcM tip densities tended to fluctuate differently during the growing season as indicated by the within-subject interaction (Time × Species; Table 2). When analyzed separately for each of the three sampling times, Q. macrocarpa total (F 1,9 = 9.89, P = 0.0118) and EcM (F 1,9 = 9.26, P = 0.0139) root tips were in higher densities than those of P. nigra in August (Fig. 1a,b). Similar differences were not visible at any other sampling events. Neither the EcM colonization (Fig. 1c) nor the qPCR-estimated ITS1 copy numbers (Fig. 1d) differed between the two hosts (Table 2).
Root and ectomycorrhizal tip densities (a, b), ectomycorrhizal colonization (c), and qPCR-estimated fungal ITS1 copy numbers (d) of the native Quercus macrocarpa (solid black bars) and exotic Pinus nigra (open grey bars) urban ornamental trees. The roots were sampled in June, August, and October to capture the root and ectomycorrhizal seasonal dynamics. Asterisks above the bars indicate differences (P < 0.05) between the two hosts at those sampling events. Note that although the root and ectomycorrhizal densities were higher in Quercus than in Pinus in August, the ITS copy numbers and ectomycorrhizal colonization were comparable between the two hosts
High-throughput sequencing of the ectomycorrhizosphere and ectomycorrhizal fungi
After quality control, we analyzed a total of 17,977 sequences that were retained from the total of 51,275 sequences after denoising. The heterogeneous yields across the experimental units necessitated subsampling of 158 sequences per sample for all ectomycorrhizosphere fungi and 120 for those representing OTUs assigned to EcM taxa. The subsampling also led to loss of three experimental units from analyses for all ectomycorrhizosphere fungi and loss of nine experimental units from the EcM analyses.
Of all sequences, a total of 52.8 % belonged to Basidiomycota, 45.9 % to Ascomycota, and less than 1 % to each of Chytridiomycota and Blastocladiomycota (Table S3). A majority of sequences (66.8 %) were assigned to EcM taxa and the remaining sequences represented a heterogeneous mix of potential pathogens and antagonists, saprobes, or soil fungi. The acquired sequences were distributed to a total of 188 non-singleton OTUs at 97 % similarity and 244 singleton OTUs, which were removed from subsequent analyses as they may represent PCR or sequencing artifacts (Tedersoo et al. 2010). Only 41 OTUs were assigned to EcM genera, whereas the large majority (147) of the non-singleton OTUs represented non-mycorrhizal fungi that occurred mainly in low frequencies. Russula dominated EcM communities with 17.3 % of all sequences and 14 OTUs, followed by Thelephoraceae (15.6 % of sequences and three OTUs), Tuber (12.3 % of sequences and 17 OTUs), Inocybe (10.7 % of sequences and 14 OTUs), and a single Hebeloma OTU (5.4 % of all sequences). The most common non-mycorrhizal OTUs were assigned to Gibberella (4.0 % of sequences and 11 OTUs) and Hypocrea (3.4 % of sequences and 12 OTUs) — likely inhabitants of dead woody tissues or common plant antagonists.
Richness and diversity
Rarefaction analyses indicated that the target sequencing depth was inadequate to saturate the species richness in the ectomycorrhizosphere, particularly so in the subsampled data set (158 reads per experimental unit; Fig. 2a–c). Good’s coverage estimators for the subsampled dataset ranged from 0.96 ± 0.02 for Q. macrocarpa and 0.95 ± 0.02 for P. nigra, suggesting an overall good sampling and providing a view different from the rarefaction analysis. In contrast to the entire ectomycorrhizosphere fungal community, the EcM richness approached saturation prior to subsampling suggesting low EcM diversity (Fig. 2d–f). Good’s coverage estimators (0.99 ± 0.01 for both Q. macrocarpa and P. nigra) for the subsampled EcM dataset corroborated the near saturation observed in the rarefaction analyses.
Rarefaction curves for the 36 samples analyzed for all fungi (a–c) and ectomycorrhizal (d–f) fungi in the ectomycorrhizosphere of the native Quercus macrocarpa (solid lines) and exotic Pinus nigra (dashed lines) urban ornamental trees. The roots were sampled in June (a,d), August (b,e), and October (c,f) to capture the seasonal dynamics in the fungal communities. The ectomycorrhizal communities were sampled more thoroughly than the ectomycorrhizosphere communities as indicated by the near plateau of many samples (d–f). Note that y-axis scales differ between the ectomycorrhizosphere (a–c) and ectomycorrhizal (d–f) communities
The overall richness (SObs; Fig. 3a), extrapolated richness (Chao1; Fig. 3b), diversity (Simpson’s 1 − D; Fig. 3c), or evenness (ED; Fig. 3d) did not differ between the native Q. macrocarpa and the exotic P. nigra ectomycorrhizospheres in repeated-measures analyses (Table 2) or at any of the three sampling times (Fig. 3; Table S4; ANOVA: P > 0.05). However, the significant or nearly significant within-subjects tests (Species × Time; Table 2) suggested differing seasonal trends in observed (Fig. 3a) and extrapolated (Fig. 3b) richness estimators between the two hosts. These differing trends were attributable to declining richness in Q. macrocarpa and stable or slightly increasing richness in P. nigra.
Observed (S Obs; a,e) and extrapolated richness (Chao1; b,f) plus diversity (Simpson’s complement, 1 − D = 1 − ∑p i 2; c,g) and evenness (Simpson’s equitability, E D = (1/∑p i 2)/S; d,h) for 36 samples analyzed for all (a–d) and ectomycorrhizal (e–h) fungi in the ectomycorrhizosphere of the native Quercus macrocarpa (solid black bars) and exotic Pinus nigra (grey open bars) urban ornamental trees. The roots were sampled in June, August, and October to capture the seasonal dynamics in the fungal communities. Asterisks above the bars indicate differences (P < 0.05) between the two hosts at those sampling events, those in parentheses indicate marginally significant differences (0.05 ≤ P < 0.10). Note that while the overall communities never differed between the hosts, the ectomycorrhizal communities differed in richness, diversity, and evenness in the early and late samplings. Note the different y-axis scales for different metrics
In contrast to the overall ectomycorrhizosphere, the EcM richness (Fig. 3e) was greater in the native Q. macrocarpa than in the exotic P. nigra in June (ANOVA: F 1,7 = 7.78, P = 0.0316), but only marginally or not significant in the subsequent samplings in August (ANOVA: F 1,7 = 2.93, P = 0.1306) and October (ANOVA: F 1,7 = 4.26, P = 0.0845). These early season differences in richness were also reflected in extrapolated richness (Chao 1; Fig. 3f) as well as diversity and evenness estimators (Fig. 3g,h). The extrapolated richness and diversity tended to be greater in the native Q. macrocarpa than in the exotic P. nigra in the early sampling in June and in the late sampling in October, but not in the August sampling (Fig. 3). In contrast, the EcM communities in the non-native P. nigra tended to be more even than those colonizing the native Q. macrocarpa (Fig. 3h), suggesting less pronounced highly dominant OTUs, such as the single dominant Thelephoraceae OTU in August and October (Fig. 4). However, none of the observed richness or diversity patterns were clear when the data were analyzed by repeated-measures ANOVA (P > 0.06; Table 3) — a result likely attributable to high variability or contrasting seasonal trends of the two hosts.
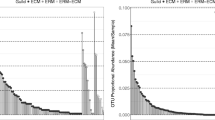
Rank orders of the 43 fungal OTUs that were represented by more than 50 sequences in June (a), August (b), October (c) in the ectomycorrhizospheres of native Quercus macrocarpa (solid black bars) and exotic Pinus nigra (open grey bars) urban ornamental trees. Asterisks above the bars indicate differences (P < 0.05) between the two hosts for that OTU, those in parentheses indicate marginally significant differences (0.05 ≤ P < 0.10). Note that only the differences in Thelephoraceae 1 frequencies remain significant after correction for multiple comparisons. The crosses identify OTUs that occur exclusively in one host. For convenience, black symbols identify OTUs associated with the native host, while grey symbols represent those with exotic host. Means and standard deviations are available in Table S6
Community composition
We analyzed the communities comprised of 188 non-singleton ectomycorrhizosphere and 41 EcM fungi by NMDS using subsampled datasets. The ectomycorrhizosphere community data were resolved on four axes, whereas the EcM community data were resolved on three axes. Our AMOVA clearly distinguished the ectomycorrhizosphere fungal communities of the two hosts indicating strong differences (F 1,30 = 4.43; P < 0.001). Similarly, ANOVA suggested that the communities differed among the two hosts on the first axis (F 1,10 = 7.48; P = 0.0230) in the June sampling (Fig. 5a) and on the fourth axis (F 1,10 = 5.16; P = 0.0491) in the August sampling, and only marginally so in the final October sampling (ANOVA: P > 0.08). In the repeated-measures analyses, these ordination analyses did not distinguish the ectomycorrhizosphere communities (Table 2). The EcM communities of the two hosts were only marginally distinguished based on AMOVA (F 1,24 = 1.89; P = 0.057). Consistently, the EcM communities were marginally distinct in NMDS analyses when analyzed separately for the three sampling events. While the EcM communities did not differ between the two hosts in August (ANOVA: P > 0.19) or October (ANOVA: P > 0.45) on any of the three NMDS axes, they differed marginally in June (ANOVA: F 1,7 = 5.33; P = 0.0603) on the third NMDS axis (Fig. 5b). Similar to the analyses focusing on all ectomycorrhizosphere fungi, the NMDS ordination did not distinguish the EcM communities based on the repeated-measures analyses (Table 3).
Nonmetric multidimensional scaling (NMDS) ordination of the ectomycorrhizosphere (a) and ectomycorrhizal (b) fungal communities in native Quercus macrocarpa (solid symbols) and exotic Pinus nigra (open symbols) urban ornamental trees sampled in three occasions (June, August, October). For clarity, sampling times that differ in ANOVA have been highlighted in black, whereas others are grey. Note that only early season (June) ectomycorrhizosphere and ectomycorrhizal samples differ
The analyses of the 43 most common OTUs identified few responsive taxa — mainly dominated by non-mycorrhizal taxa. Five EcM OTUs (OTU4 and 37 assigned to Inocybe plus OTU9, 22, and 42 assigned to Tuber) were frequent in Q. macrocarpa, but never occurred in P. nigra (Fig. 4; Table S6). Similarly, OTU29 (Inocybe) and OTU43 (Suillus) never occurred in Q. macrocarpa, while present in P. nigra. A suite of non-EcM OTUs (27, 40, 41 representing the families Vibresseaceae, Myxotrichiaceae, and Melanommataceae, respectively) was exclusive to P. nigra (Fig. 4). Only one EcM OTU (OTU1; assigned to uncultured Thelephoraceae) was more frequent in Q. macrocarpa than in P. nigra after correction for multiple comparisons (Fig. 4). This OTU was more frequent in Q. macrocarpa in the last sampling event in October (F 1,10 = 6.66; P = 0.0297), but not in samplings prior to that or when the data were analyzed by repeated-measures (F 1,3 = 0.89; P = 0.4160). However, this OTU — like many others — highlights the seasonal dynamics. While consistently low in P. nigra, this OTU peaked in August in Q. macrocarpa — a pattern visible also in the repeated-measures analyses: variability within the growing season was indicated by the within-subjects effect (Time: F 2,2 = 85.04; P = 0.0116) and the different dynamics among the two hosts by the significant between-subjects interaction term (Time × Species: F 2,2 = 103.94; P = 0.0095).
Interpretation of these results is complicated by multiple comparisons as well as the seasonally dynamic OTU frequencies. For example, OTU23 (assigned to Immersiella, Sordariales) occurred more frequently Q. macrocarpa than in P. nigra in June (F 1,11 = 10.45; P = 0.0090), but not in the subsequent samplings (P > 0.10), or when analyzed by repeated-measures ANOVA (F 1,3 = 1.14; P = 0.3638). However, some OTUs were more consistent in their responses. When data were analyzed separately for each of the three sampling events, OTU14 (assigned to Cochliobolus, Pleosporales) occurred more frequently in P. nigra than in Q. macrocarpa in August (F 1,10 = 12.77; P = 0.0060) and October (F 1,10 = 7.33; P = 0.0482). These analyses were consistent with the repeated-measures analyses, which also indicated that OTU14 was overall more frequent in P. nigra than in Q. macrocarpa (between-subjects effect: F 1,3 = 25.33; P = 0.0151), variable over the growing season as indicated by the within-subjects effect (Time: F 2,2 = 141.76; P = 0.0070), and more so in P. nigra than in Q. macrocarpa as indicated by the significant between-subjects interaction term (Time × Species: F 2,2 = 131.63; P = 0.0075). Similarly, OTU6 (assigned to Gibberella, Hypocreales), when analyzed separately for each of the three sampling events, occurred more frequently in P. nigra than in Q. macrocarpa in August (F 1,10 = 12.59; P = 0.0062) and October (F 1,10 = 6.10; P = 0.0356). Consistently, the repeated-measures analyses indicated that OTU6 was overall more frequent in P. nigra than in Q. macrocarpa (F 1,3 = 15.29; P = 0.0297), but in contrast to OTU14 did not vary over the growing season (P > 0.20).
Discussion
This research was motivated by our earlier effort that concluded that fungal communities associated with urban trees tend to be less species rich and compositionally distinct compared to adjacent non-urban trees (Jumpponen and Jones 2009, 2010; Jumpponen et al. 2010a). Accordingly, our goal was to evaluate whether or not the choice of ornamental trees in such environments would further impact the resident fungal richness, diversity, or community composition. Our data show that EcM fungi heavily colonize the selected hosts. Both ectomycorrhizosphere and EcM fungal communities show considerable overlap; the EcM communities are marginally distinct but include members that are unique to a specific host. The differences between the EcM fungi associated with the two hosts highlight the importance of thoughtful management choices in urban settings to best preserve the endemic communities that may be declining as a result of human activities (Arnolds 1991; Watling 2005; Newbound et al. 2010). Urban systems are increasing in their importance in conservation ecology (Newbound et al. 2010), as urban land use has been predicted to increase by more than 200 % by 2030 (Angel et al. 2005). Although some studies have highlighted the anthropogenic atmospheric pollution as one primary negative driver for EcM fungi (Arnolds 1991; Dighton et al. 2004; Newbound et al. 2010), pinpointing the exact drivers is difficult without experimental testing of hypotheses on specific mechanisms (Jumpponen et al. 2010b). Our current study specifically examines the effects of ornamental tree species selection in highly managed urban environment.
Although mycorrhizal fungi are present in urban environments, their inoculum loads and colonization have been proposed to be lower than in non-urban environments (Cousins et al. 2003; Ochimaru and Fukuda 2007; Bainard et al. 2011; Karpati et al. 2011). This tends to be the case if the soils have undergone recent or severe disturbances (Boerner et al. 1996; Karpati et al. 2011), although others have reported no effect of disturbance on EcM colonization in natural systems (Byrd et al. 2000; Jones et al. 2003). Other proposed mechanisms underlying the low colonization include environmental stresses that may have negative impacts on mycorrhizal associations (Gehring and Whitham 1992). The reported low colonization and its potential adverse effects on trees have led to calls for considering inoculation programs for urban trees (Stabler et al. 2001; Appleton et al. 2003; Fini et al. 2011), although commercial mycorrhizal inocula often yield inconsistent results (Gilman 2001; Ferrini and Nicese 2002; Rao et al. 2006). Our relatively high EcM colonization is in contrast with other observations (Baxter et al. 1999; Ochimaru and Fukuda 2007; Bainard et al. 2011) and suggests presence of a readily available inoculum pool. It is of note that our qPCR estimators of ITS1 copy numbers aimed to test inoculum loads, but included likely a large proportion of non-EcM taxa as indicated by the presence of many non-EcM OTUs in the ectomycorrhizospheres. Furthermore, the high colonization of the non-native pine suggests readily available and compatible inoculum, a result consistent with other studies that have shown P. nigra broad compatibility with cosmopolitan EcM fungi as well as species typically associated with members of Pinaceae or Fagaceae (Martin-Pinto et al. 2006; Lazarevic et al. 2012; Trocha et al. 2012). Our high and rather stable colonization may relate to minimal soil disturbance near our urban mature trees, the adjacency of the two hosts at each of the sites, or the relatively adjacent undisturbed non-urban inoculum sources around the small urban center. Taken together, we consider that urban inoculation programs may be unnecessary to ensure EcM colonization, if adequate inoculum sources are available and the sites have not been severely disturbed, for example, by topsoil removal or addition. Even so, if endemic inocula were to prove successful in establishing reservoirs or enriching the diversity of native fungal populations and/or communities, inoculation programs warrant attention for high-value urban trees. Furthermore, EcM taxa or strains may be locally adapted; EcM fungi and hosts naturally occurring in similar environments may show superior symbiotic performance (Bonfante et al. 1998). It is of note that a number of EcM taxa were observed only with one host.
We analyzed both general, broad fungal communities and EcM communities inhabiting roots of our hosts. The two communities show some contrasts. The overall communities did not differ in richness or diversity, but were compositionally distinguishable despite large overlaps. The EcM communities were seasonally dynamic, differed in richness and diversity in early growing season and were comprised of members that were often unique to one host. While these and earlier data (Jumpponen et al. 2010a) suggest low fungal richness and compositional shifts in urban EcM communities compared to surrounding non-urban environments, the functional consequences of these shifts remain unclear. Mycorrhizal fungi have diverse functions with a potential to facilitate nutrient uptake by accessing resources not readily available for the plants (Marschner and Dell 1994), improve drought tolerance (Lehto and Zwiazek 2011), increase disease resistance (Whipps 2004), and permit interplant transfer of nutrients through common mycelial networks (Simard and Durall 2004). EcM species differ in their functional attributes and physiological tolerances (Baxter and Dighton 2001, 2005). Thus, greater diversity of root-associated symbionts should translate to more diverse benefits to the host, or alternatively, to greater flexibility under changing conditions (Druebert et al. 2009; Pena et al. 2010). However, the importance of mycorrhiza-mediated ecosystem services and/or host benefits in intensively managed, fertilized and watered urban systems remains unclear and requires further investigation.
Habitat loss is perhaps the main threat to fungal richness and diversity, stemming from fungal requirements for specific substrates or hosts (Watling 2005; Newbound et al. 2010). To counter and minimize habitat loss and fragmentation resulting from urbanization, selection of host species that may best preserve the resident biodiversity is of particular importance. Our data highlight the differences in the ectomycorrhizosphere and EcM fungal communities associated with two host species in an urban setting. While the former was minimally influenced by the host taxon, the EcM communities of the non-native host were less specious and lacked community constituents when compared to the native host. A common garden study evaluating pairs of native and non-native con-generics (species of Pinus and Quercus) found that, while the native Quercus hosted a more diverse EcM community than the non-native one, the opposite was true for the two Pinus taxa compared (Trocha et al. 2012). Other studies focusing on non-native species have reported that introduced or invasive non-natives expediently recruit native EcM symbionts (Cullings et al. 2000; Gebhardt et al. 2007; Tedersoo et al. 2007; Kohout et al. 2011; Bahram et al. 2013), although the host establishment may be facilitated by co-introduction with compatible root symbionts (Tedersoo et al. 2007; Vellinga et al. 2009; Dickie et al. 2010). Additionally, while native forests and adjacent non-native plantations can host rather similar EcM fungus richness (O’Hanlon and Harrington 2012; Bahram et al. 2013), the community constituents may differ (O’Hanlon and Harrington 2012), particularly so in stands isolated from inoculum sources (Bahram et al. 2013). Similar to O’Hanlon and Harrington (2012), our data suggest host-taxon driven EcM community rearrangement and re-ordering in urban environment in absence of a dramatic loss of biodiversity. Our study is among first to so comprehensively dissect the EcM and ectomycorrhizosphere fungal communities in native and non-native hosts in an urban environment and our results are in line with an expedient recruitment of compatible EcM fungi by non-native hosts in a fragmented urban environment.
The fungal communities in our study included a large proportion of non-EcM fungi. Among the 43 most abundant OTUs were putative plant antagonists (Gibberella, Bionectria), saprobes (Chaetomium, Gerronema), and common soil fungi (Eupenicillium, Aspergillus). It is notable that while the ectomycorrhizosphere communities of the native host included a large number of common EcM taxa (Russulaceae, Thelephoraceae, Tuberaceae), many top-ranked taxa that do not form mycorrhizae (e.g., Cochliobolus, Gerronema, Gibberella, Vibrissea) were exclusively or more commonly associated with the non-native host. It is open to speculation whether this observation suggests a lesser abundance of compatible EcM partners for the non-native host, or a compromised health of the host plant resulting in greater susceptibility to colonization by non-EcM fungi. Curiously, our observations on the fungal communities colonizing the native and non-native hosts seem to suggest patterns analogous to those in response to nutrient enrichment: EcM richness often declines, whereas the lignicolous and saprobic taxa either remain unchanged or increase (Peter et al. 2001; Högberg et al. 2003; Trudell and Edmonds 2004).
To evaluate the differences of native and non-native hosts in an urban setting, we sampled co-occurring isolated ornamental trees. Characteristically to urban stands, the sampled trees occurred in small landscape fragments. This may limit the dispersal, viability, and genetic variability of fungi (Edman et al. 2004; Peay et al. 2007). While it is possible that fungi associated with non-native hosts suffer from the isolation and dispersal limitation more than those associated with native hosts, the observed differences among the two hosts are more likely attributable to incompatibility between non-native host and indigenous fungal communities (see Jacobson and Miller 1992; Walker et al. 2005; Tedersoo et al. 2008). This is supported by our observation that EcM taxa consistently and exclusively occurred in the native host (common EcM in Q. macrocarpa: three Inocybe OTUs and two Tuber OTUs). However, the non-native host also included exclusive EcM taxa (Inocybe and Suillus) as well as many shared EcM taxa (Inocybe, Russula, and Tuber). The former is in line with other studies that observed a small number of unique EcM taxa in a non-native host (Bahram et al. 2013), whereas the latter is in line with the broad compatibility between non-native hosts and native EcM fungi (O’Hanlon and Harrington 2012; O’Hanlon et al. 2013). It is unclear whether the observed unique EcM taxa indicate differing preferences among hosts or introduction of novel fungal taxa with the exotic host as suggested for Amanita phalloides and European oaks in Australia (Nicholson and Korman 1997) and for Rhizopogon vinicolor and Pseudotsuga menziesii in New Zealand (Chou-Chou and Grace 1983). Alternatively, our finding may suggest differing environmental preferences among the fungal taxa when associated with different host species. Our studies fail to clearly pinpoint the mechanisms that drive EcM community rearrangements. Yet, our study directly compares two common ornamental hosts and identifies differences in a fixed environmental setting afforded by the coexistence of the two hosts in close proximity in the selected six stands.
Our studies described here aimed to evaluate the importance of host selection in urban settings to maintain fungal biodiversity. Our data show that while the richness and diversity of host associated fungal communities did not perhaps consistently differ between a native and non-native host, the communities included a strong signal of community rearrangement compositionally and functionally: the fungal communities associated with the native host were enriched or included exclusive EcM taxa, whereas the non-native host communities were enriched with non-EcM fungi. We speculate that this observation signals either absence of compatible EcM symbionts or poor health of the exotic trees outside their non-native range. Our observations emphasize the need for caution in selection of ornamental trees as native trees may serve as a superior reservoir for native fungal symbionts and the non-natives may bear a higher load of putative antagonists.
References
Amend AS, Seifert KA, Samson R, Bruns TD (2010) Indoor fungal composition is geographically patterned and more diverse in temperate zones than in the tropics. Proceedings of the National Academy of Sciences of the United States of America 107:13748–13753
Angel S, Sheppard SC, Civco DL (2005) The dynamics of global urban expansion. The World Bank, Washington, DC
Appleton B, Koci J, French S, Lestyan M, Harris R (2003) Mycorrhizal fungal inoculation of established street trees. J Arboric 29:107–110
Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Aust Ecol 26:32–46
Arnolds E (1991) Decline of ectomycorrhizal fungi in Europe. Agric Ecosyst Environ 35:209–244
Ash C, Jasny BR, Roberts L, Stone R, Sugden A (2008) Reimagining cities—introduction. Science 319:739–739
Bahram M, Köljalg U, Kohout P, Mirshahvaladi S, Tedersoo L (2013) Ectomycorrhizal fungi of exotic pine plantations in relation to native host trees in Iran: evidence of host range expansion by local symbionts to distantly related host taxa. Mycorrhiza 23:11–19
Baxter JW, Dighton J (2001) Ectomycorrhizal diversity alters growth and nutrient acquisition of grey birch (Betula populifolia) seedlings in host-symbiont culture conditions. New Phytol 152:139–159
Baxter JW, Dighton J (2005) Diversity–functioning relationships in ectomycorrhizal fungal communities. In: Dighton J, White JF, Oudemans P (eds) The fungal community—its role and organization in the ecosystem, 3rd edn. CRC Press, Boca Raton, pp 383–398
Baxter JW, Pickett STA, Carreiro MM, Dighton J (1999) Ectomycorrhizal diversity and community structure in oak forest stands exposed to contrasting anthropogenic impacts. Can J Bot 77:771–782
Bainard LD, Klironomos JN, Gordon AM (2011) The mycorrhizal status and colonization of 26 tree species growing in urban and rural environments. Mycorrhiza 21:92–96
Berry D, Mahfoudh KB, Wagner M, Loy A (2011) Barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl Environ Microbiol 77:7846–7849
Boerner REJ, DeMars BG, Leicht PN (1996) Spatial patterns of mycorrhizal infectiveness of soils long a successional chronosequence. Mycorrhiza 6:79–90
Bonfante P, Balestrini R, Martino E, Perotto S, Plassard C, Mousain D (1998) Morphological analysis of early contacts between pine roots and two ectomycorrhizal Suillus strains. Mycorrhiza 8:1–10
Byrd KB, Parker VT, Vogler DR, Cullings KW (2000) The influence of clear-cutting on ectomycorrhizal fungus diversity in a lodgepole pine (Pinus contorta) stand, Yellowstone National Park, Wyoming, and Gallatin National Forest, Montana. Can J Bot 78:149–156
Chou-Chou M, Grace LJ (1983) Characterization and identification of mycorrhizas of Douglas-fir in New Zealand. Eur J For Pathol 13:251–260
Cousins JR, Hope D, Gries C, Stutz JC (2003) Preliminary assessment of arbuscular mycorrhizal fungal diversity and community structure in an urban ecosystem. Mycorrhiza 13:319–326
Cullings KW, Vogler DR, Parker VT, Finley SK (2000) Ectomycorrhizal specificity in a mixed Pinus contorta and Picea engelmannii forest in Yellowstone National park. Appl Environ Microbiol 66:4988–4991
Dickie IA, Bolstridge N, Cooper JA, Peltzer DA (2010) Co-invasion by Pinus and its mycorrhizal fungi. New Phytol 187:475–484
Dighton J, Tuininga AR, Gray DM, Huskins RE, Belton T (2004) Impacts of atmospheric deposition on New Jersey pine barren forest soils and communities of ectomycorrhizae. For Ecol Manag 201:133–144
Druebert C, Lang C, Valtanen K, Polle A (2009) Beech carbon productivity as driver of ectomycorrhizal abundance and diversity. Plant Cell Environ 32:992–1003
Edman M, Gustafsson M, Stenlid J, Ericson L (2004) Abundance and viability of fungal spores along a forestry gradient—responses to habitat loss and isolation? Oikos 104:35–42
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Ferrini F, Nicese FP (2002) Response of English oak (Quercus robur L.) trees to biostimulants application in the urban environment. J Arboric 28:70–75
Fierer N, Jackson JA, Vilgalys R, Jackson RB (2005) Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl Environ Microbiol 71:4117–4120
Fini A, Frangi P, Amoroso G, Piatti R, Faoro M, Bellasio C, Ferrini F (2011) Effect of controlled inoculation with specific mycorrhizal fungi from the urban environment on growth and physiology of containerized shade tree species growing under different water regimes. Mycorrhiza 21:703–719
Gebhardt S, Neubert K, Wöllecke J, Münzenburger B, Hütl RF (2007) Ectomycorrhiza communities of red oak (Quercus rubra L.) of different age in the Lusatian lignite mining district, East Germany. Mycorrhiza 17:279–290
Gehring CA, Whitham TG (1992) Reduced mycorrhizae on Juniperus monosperma with mistletoe: the influence of environmental stress and tree gender on a plant parasite and a plant–fungal mutualism. Oecologia 89:298–303
Gihring TM, Green SJ, Schadt CW (2012) Massively parallel rRNA gene sequencing exacerbates the potential for biased community diversity comparisons due to variable library sizes. Environ Microbiol 14:285–290
Gilman EF (2001) Effect of nursery production method, irrigation, and inoculation with mycorrhizae-forming fungi on establishment of Quercus virginiana. J Arboric 27:30–39
Högberg MN, Bååth E, Nordgren A, Arnebrant K, Högberg P (2003) Contrasting effects of nitrogen availability on plant carbon supply to mycorrhizal fungi and saprotrophs—a hypothesis based on field observations in boreal forest. New Phytol 160:225–238
Jacobson KM, Miller OK (1992) Physiological variation between tree associated populations of Suillus granulatus as determined by in vitro mycorrhizal synthesis experiments. Can J Bot 70:26–31
Jones MD, Durall DM, Cairney JWG (2003) Ectomycorrhizal fungal communities in young forest stands regenerating after clearcut logging. New Phytol 157:399–422
Jumpponen A, Jones KL (2009) Massively parallel 454 sequencing indicates hyperdiverse fungal communities in temperate Quercus macrocarpa phyllosphere. New Phytol 184:438–448
Jumpponen A, Jones KL (2010) Seasonally dynamic fungal communities in Quercus macrocarpa phyllosphere differ among urban and rural environments. New Phytol 186:496–513
Jumpponen A, Jones KL, Mattox JD, Yeage C (2010a) Massively parallel 454-sequencing of fungal communities in Quercus spp. ectomycorrhizas indicates seasonal dynamics in urban and rural sites. Mol Ecol 19(Suppl. 1): 41–53
Jumpponen A, Keating K, Gadbury GL, Jones KL, Mattox JD (2010b) Multi-element fingerprinting and high throughput sequencing identify multiple elements that affect fungal communities in Quercus macrocarpa foliage. Plant Signal Behav 5:1157–1161
Karpati AS, Handel SN, Dighton J, Horton TR (2011) Quercus rubra-associated ectomycorrhizal fungal communities of disturbed urban sites and mature forests. Mycorrhiza 21:537–547
Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518
Kohout P, Sýkorová Z, Bahram M, Hadincová V, Albrechtová J, Tedersoo L, Vohník M (2011) Ericaceous dwarf shrubs affect ectomycorrhizal fungal community of the invasive Pinus strobus and native Pinus sylvestris in a pot experiment. Mycorrhiza 21:403–412
Kaye JP, Groffman PM, Grimm NB, Baker LA, Pouyat RV (2006) A distinct urban biogeochemistry? Tree 21:192–199
Kranabetter JM, Hayden S, Wright EF (1999) A comparison of ectomycorrhiza communities from three conifer species planted on forest gap edges. Can J Bot 77:1193–1198
Lazarevic J, Keca N, Martinovic A (2012) Mycorrhization of containerized Pinus nigra seedlings with Suillus granulatus under open field conditions. For Syst 21:498–507
Lehto T, Zwiazek JJ (2011) Ectomycorrhizas and water relations of trees: a review. Mycorrhiza 21:71–91
Liu K-L, Kuske CR, Porras-Alfaro A, Eichorst S, Xie G (2012) Accurate, rapid taxonomic classification of fungal large subunit rRNA genes. Appl Environ Microbiol 78:1523–1533
Magurran AE (1988) Ecological diversity and its measurement. Princeton University Press, Princeton
Marschner H, Dell B (1994) Nutrient-uptake in mycorrhizal symbiosis. Plant Soil 159:89–102
Martin-Pinto P, Pajares J, Diez J (2006) In vitro effects of four ectomycorrhizal fungi, Boletus edulis, Rhizopogon roseolus, Laccaria laccata and Lactarius deliciosus on Fusarium damping off in Pinus nigra seedlings. New For 32:323–334
Molina R, Massicotte H, Trappe JM (1992) Specificity phenomena in mycorrhizal symbiosis: community-ecological consequences and practical implications. In: Allen MF (ed) Mycorrhizal functioning. An integrative plant–fungal process. Chapman and Hall, New York, pp 357–423
Morris MH, Perez-Perez MA, Smith ME, Bledsoe CS (2009) Influence of host species on ectomycorrhizal communities associated with two co-occurring oaks (Quercus spp.) in a tropical cloud forest. FEMS Microbiol Ecol 69:274–287
Newbound M, Mccarthy MA, Lebel T (2010) Fungi and the urban environment: a review. Landsc Urban Plan 96:138–145
Nicholson FB, Korman MG (1997) Death from Amanita poisoning. Aust NZ J Med 27:448–449
O’Hanlon R, Harrington TJ (2012) Similar taxonomic richness but different communities in native forests and non-native plantation forests. Mycorrhiza 22:371–382
O’Hanlon R, Harrington TJ, Berch SM, Outerbridge RA (2013) Comparisons of macrofungi in plantations of Sitka spruce (Picea sitchensis) in its native range (British Columbia, Canada) versus non-native range (Ireland and Britain) show similar richness but different species composition. Can J For Res 43:450–458
Ochimaru T, Fukuda K (2007) Changes in fungal communities in evergreen broad-leaved forests across a gradient of urban to rural areas in Japan. Can J For Res 37:247–258
Pavao-Zuckerman MA (2008) The nature of urban soils and their role in ecological restoration in cities. Restor Ecol 16:642–649
Peay KG, Bruns TD, Kennedy PG, Bergemann SE, Garbelotto M (2007) A strong species–area relationship for eukaryotic soil microbes: island size matters for ectomycorrhizal fungi. Ecol Lett 10:470–480
Pena R, Offermann C, Simon J, Naumann PS, Geßler A, Holst J, Mayer H, Kögel-Knabner I, Rennenberg H, Polle A (2010) Girdling affects ectomycorrhizal diversity and reveals functional differences of EM community composition in a mature beech forest (Fagus sylvatica). Appl Environ Microbiol 76:1831–1841
Peter M, Ayer F, Egli S (2001) Nitrogen addition in a Norway spruce stand altered macromycete sporocarp production and below-ground ectomycorrhizal species composition. New Phytol 149:311–325
Pfaffl M (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:2002–2007
Polanco MC, Zwiazek JJ, Voicu MC (2008) Responses of ectomycorrhizal American elm (Ulmus americana) seedlings to salinity and soil compaction. Plant Soil 308:189–200
Pouyat RV, McDonnell MJ (1991) Heavy metal accumulations in forest soils along an urban–rural gradient in southeastern New York, USA. Water Air Soil Pollut 57–8:797–807
Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT (2009) Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods 6:639
Rao B, Marx DH, Jeffers B (2006) Response of oaks and elm to soil inoculations with mycorrhizal fungi and rhizobacteria in a nursery. Arboric Urban For 32:62–66
Read DJ (1991) Mycorrhizas in ecosystems. Experentia 47:376–390
Richard F, Millot S, Gardes M, Selosse MA (2005) Diversity and specificity of ectomycorrhizal fungi retrieved from an old-growth Mediterranean forest dominated by Quercus ilex. New Phytol 166:1011–1023
Schaefer VH (2011) Remembering our roots: a possible connection between loss of ecological memory, alien invasions and ecological restoration. Urban Ecosyst 14:35–44
Schloss PD, Gevers D, Westcott SL (2011) Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE 6:e27310
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesneiwski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Van Thallinger GC, Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Simard SW, Durall DM (2004) Mycorrhizal networks: a review of their extent, function, and importance. Can J Bot 82:1140–1165
Smith SE, Read DJ (2008) Mycorrhizal symbiosis. Elsevier, Amsterdam
Stabler LB, Martin CA, Stutz JC (2001) Effect of urban expansion on arbuscular mycorrhizal fungal mediation of landscape tree growth. J Arboric 27:193–202
Tedersoo L, Suvi T, Beaver K, Kõljalg U (2007) Ectomycorrhizal fungi of the Seychelles: diversity patterns and host shifts from the native Vateriopsis seychellarum (Dipterocarpaceae) and Intsia bijuga (Caesalpiniaceae) to the introduced Eucalyptus robusta (Myrtaceae), but not Pinus caribea (Pinaceae). New Phytol 175:321–333
Tedersoo L, Jairus T, Horton BM, Abarenkov K, Suvi T, Saar I, Köljalg U (2008) Strong host preference of ectomycorrhizal fungi in a Tasmanian wet sclerophyll forest as revealed by DNA barcoding and taxon-specific primers. New Phytol 180:479–490
Tedersoo L, Nilsson RH, Abarenkov K, Jairus T, Saar I, Bahram M, Bechem E, Chuyong G, Kõljalg U (2010) 454 pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New Phytol 188:291–301
Timonen S, Kauppinen P (2008) Mycorrhizal colonisation patterns of Tilia trees in street, nursery and forest habitats in southern Finland. Urban For Urban Green 7:265–276
Trocha L, Kalucha I, Stasinska M, Nowak W, Dabert M, Leski T, Rudawska M, Oleksyn J (2012) Ectomycorrhizal fungal communities of native and non-native Pinus and Quercus species in a common garden of 35-year-old trees. Mycorrhiza 22:121–134
Trudell SA, Edmonds RL (2004) Macrofungus communities correlate with moisture and nitrogen abundance in two old-growth conifer forests, Olympic National Park, Washington, USA. Can J Bot 82:781–800
Vellinga EC, Wolfe BE, Pringle A (2009) Global patterns of ectomycorrhizal introductions. New Phytol 181:960–973
Walker JF, Miller OK, Horton JL (2005) Hyperdiversity of ectomycorrhizal fungus assemblages on oak seedlings in mixed forests in the southern Appalachian Mountains. Mol Ecol 14:829–838
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian Classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Watling R (2005) Fungal conservation: some impressions—a personal view. In: Dighton J, White JF, Oudemans P (eds) The fungal community—its organisation and role in the ecosystem, 3rd edn. CRC Press, Boca Raton
Whipps JM (2004) Prospects and limitations for mycorrhizas in biocontrol of root pathogens. Can J Bot 82:1198–1227
Acknowledgments
We are grateful to the city of Manhattan for permitted sampling within the city limits and Kansas State University for allowing sampling on campus. Kale Lothamer was supported by Undergraduate Research Mentoring (URM) in Ecological Genomics program (NSF Grant # 1041199). Shawn Brown was supported by GAANN (Graduate Awards in Areas of National Need) from Department of Education and GK-12 (NSF DGE-0841414, PI Ferguson) awards. This is Kansas Agricultural Experimental Station publication number 14-162-J.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Table S1
Primer and dna-tag sequences used for sample specific PCR and 454-sequencing. (TXT 1 kb)
Supplemental Table S2
Putative assignments to ecological roles of the 378 genera detected in the 36 ectomycorrhosphere samples. AM – Arbuscular Mycorrhiza; ECM – EctoMycorrhizal; ENT – ENTomopahtogen; PAR – PARastitic, PAT – PAThogenic; SAP – SAProbic; UNK – UNKnown. (TXT 6 kb)
Supplemental Table S3
Taxon assignments of 188 OTUs that occurred more than once in the entire dataset. K_Sup, P_Sup, C_Sup, O_Sup, F_Sup, G_Sup refer to Ribosomal Database Project’s naïve Bayesian classifier bootstrap support for Kingdom, Phylum, Class, Order, Family and Genus, respectively. Ecology refers to assignments using list in Supplemental Table S2. Last two columns list total counts of sequences assigned to each OTU for Quercus macrocarpa and Pinus nigra. (TXT 21 kb)
Supplemental Table S4
Mean (± 1 standard deviation) root and ectomycorrhizal tip densities, EcM colonization, qPCR derived inoculum load (ITS1 copy number), Good’s coverage, observed richness (SObs), Simpson’s diversity complement (1-D), Shannon’s diversity, extrapolated richness estimate (Chao I), and evenness as estimated by Simpson’s equitability (ED) for all ectomycorrhizosphere inhabiting fungi. Note that only the root and ectomycorrhizal tip densities differ in the second sampling in August, other comparisons of the two hosts at each sampling time (one way ANOVA) are non-significant (see Figs 1-2). Compare these ectomycorrhizosphere analyses to those for ectomycorrhizal fungi only (Supplemental Table S5). (TXT 1 kb)
Supplemental Table S5
Mean (± 1 standard deviation) Good’s coverage, observed richness (SObs), Simpson’s diversity complement (1-D), Shannon’s diversity, extrapolated richness estimate (Chao I), and evenness as estimated by Simpson’s equitability (ED) for all ectomycorrhizal fungi. Note that the two hosts did not differ in repeated measures ANOVA. In contrast, many richness and diversity estimators differed in early sampling in June and marginally so in the late sampling in October (see Fig. 3). (TXT 0 kb)
Supplemental Table S6
Frequencies (mean ± standard deviation) of the 43 most abundant OTUs analyzed for OTU-level responses in June, August, and October 2011. (TXT 5 kb)
Rights and permissions
About this article
Cite this article
Lothamer, K., Brown, S.P., Mattox, J.D. et al. Comparison of root-associated communities of native and non-native ectomycorrhizal hosts in an urban landscape. Mycorrhiza 24, 267–280 (2014). https://doi.org/10.1007/s00572-013-0539-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-013-0539-2