Abstract
Diversity of arbuscular mycorrhizal fungi (AMF) was assessed in two 9.2 × 9.2-m plots planted with landscape trees and shrubs at an experimental site in Phoenix, AZ, USA. Twenty-five soil samples were collected in a regular grid pattern from each plot, and AMF species were identified using trap cultures. A total of 12 species were detected, with 7 species detected in one plot and 11 in the other. We found that sampling effort had a major impact on assessing species richness and composition in this local community. Fifteen samples would be necessary to detect 70–80% of species present in each plot. A limited number of additional undetected species are likely to be present in both plots, based on the sampling effort curves and jackknife estimates. Only two species, Glomus eburneum and Glomus microaggregatum, were detected in over 50% of the samples from both plots, and rank–frequency plots revealed a lognormal species distribution. Despite the patchiness of plants in the plots, the number of species detected per point exhibited spatial structuring only at the smallest sampling scale in a single plot, and only a single species in each plot was not randomly distributed. These results indicate that sampling effort and strategy can affect perceptions of AMF community structure.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Arbuscular mycorrhizal fungi (AMF) form associations with the majority of terrestrial plant species and have been shown to improve the growth and nutrition of individual plants (Smith and Read 1997). AMF have recently been shown to have a pivotal role in plant community ecology by altering plant productivity and diversity (Klironomos et al. 2000), changing the course of succession (Gange et al. 1990), and affecting plant competition (Hartnett and Wilson 1999). Until recently, AMF species were generally assumed to be functionally similar, so there was little focus on AMF diversity in natural habitats. However, recent work has shown that plant diversity (van der Heijden et al. 1998) and ecosystem variability and productivity (Hart and Klironomos 2002) are directly influenced by AMF diversity, making an accurate assessment of species richness and community composition crucial to understanding the role of AMF in ecosystem functioning.
Unfortunately, assessing AMF diversity is complicated by their clonal, cryptic nature (Bever et al. 2001). Several inherent problems complicate the measurement of species richness (the number of species in a community) and community evenness (the abundance and distribution of each species in the community) (Magurran 1988). One problem is the detection and identification of AMF species. AMF are entirely clonal and their vegetative structures are nearly indistinguishable, making it impossible to identify individuals, and even difficult to distinguish between species based on vegetative characteristics. A second problem is in attempting to measure AMF abundance and species dominance. Many researchers have relied on counting the number of spores in a soil sample as a measurement of abundance, but this technique underestimates the abundance of species that are not sporulating at the time of sampling (Cousins et al. 2003). Another major confounding factor in assessing AMF diversity is the high spatial heterogeneity of the soil environment (Ettema and Wardle 2002). AMF propagules and spore densities are known to exhibit strong spatial structuring at small scales. Boerner et al. (1996) quantified variation in arbuscular mycorrhizal (AM) infectivity within two disturbed sites, a pseudo-stripmine and a soybean field, and found that spatial structure accounted for 44–50% of the variance among samples in the 5 × 5-m plots. In an undisturbed forest, however, spatial variability accounted for only 18% of variance in infectivity. Using nested quadrats to sample AM spores in a prairie, Anderson et al. (1983) found that spores were distributed randomly within 30 × 30-cm quadrats, but were highly aggregated at a slightly larger scale (0.5 × 0.5 m), and their presence was positively correlated with plant cover. Similarly, Carvalho et al. (2003) utilized nested grid sampling to quantify spore numbers in a salt marsh and a shrubland and discovered that the four AMF genera in their 5 × 5-m plots all exhibited spatial structuring at various scales. Hart and Klironomos (2002) documented the patchiness of spores of four AMF species in a 50 × 50-m plot in a Canadian old field, and Klironomos et al. (1999) also found that spores of Acaulospora, Glomus, and Scutellospora species were very patchily distributed within a 3 × 10-m plot in a Southern California burned chaparral site. Pringle and Bever (2002) found spatial structuring of spores of both frequently and infrequently detected AMF species in a North Carolina grassland site. Given the spatially heterogeneous nature of AMF spores and biomass, it is reasonable to predict that the number and composition of AMF species in a local community is also patchy at small spatial scales. However, the spatial patterning of AMF species at the plot scale has received little attention.
Spatial patterning of AMF species in a local community could have major implications when considering sampling effort. Klironomos et al. (1999) note that the combination of clustered spatial distribution of soil biota and the low number of samples (5–10) typically collected in soil ecology field experiments often result in low statistical power and nonsignificant treatment results. Because of this problem, the use of taxon accumulation curves (Gotelli and Colwell 2001) may be especially important to determine if adequate sampling has been used to capture AMF diversity. This technique allows for valid comparison between species richness counts when accumulation curves reach an asymptote. When species area curves were used to analyze species richness of local communities of ectomycorrhizal fungi, more than 30 samples were found to be necessary before accumulation curves would reach an asymptote (Horton and Bruns 2001).
Details about spatial patterning of AMF species could also inform sampling strategies. When organisms are patchily distributed within a plot, it is unlikely that individuals sampled “randomly” are collected in a truly random fashion due to spatial autocorrelation (Ettema and Wardle 2002). Spatial autocorrelation is a measure of the degree to which phenomena are correlated with themselves in space. Samples exhibiting positive autocorrelation tend to have values that are more similar to neighboring samples than to more distant samples. Conversely, negatively autocorrelated samples tend to be dissimilar to their neighbors. Autocorrelation leads to a violation of the assumption of sample independence, upon which parametric statistics are dependent. AMF are generally assumed to be highly aggregated around host plants, especially in arid and disturbed environments (Allen and MacMahon 1985), so that AMF species diversity values are likely to be autocorrelated at scales similar to those of plant patches. Although testing of these assumptions is rare, most sampling strategies for assessing AMF species diversity appear to be based on these assumptions or employ random sampling.
Due to these problems, sampling effort and sampling strategies could have a major influence on how AMF community structure is perceived. This study was undertaken to better understand how AMF communities are structured on a small scale in an urban environment. Woody plants in urban areas often have very patchy distributions, especially in landscapes designed for water conservation, where plants are often planted together in groups and surrounded by open areas covered with organic or inorganic mulch. Although there is some evidence that urban development directly affects AMF communities (Cousins et al. 2003; Stutz and Martin 1998), the effects of urban planting designs on the distribution of AMF have not yet been studied. The hypotheses tested in this study were (1) the sampling effort needed to detect the number of AMF species in this local community is greater than those typically collected in soil ecology field studies, and (2) AMF species number in urban landscapes exhibits spatial autocorrelation with a greater number of AM fungi at sampling points in proximity to landscape plants and lower numbers in the interspaces between plants.
Materials and methods
Site description
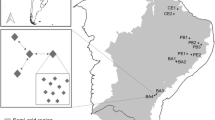
The study was carried out at an urban experimental site associated with Central Arizona - Phoenix Long-Term Ecological Research (CAP LTER) located at the Desert Botanical Garden in Phoenix, Arizona (111°57′ W longitude, 33°28′ N latitude). Before establishment of the experimental plots in 1999, the land was used to grow Prosopis velutina for horticultural use for 5 years. The area surrounding the plot is part of a Sonoran desert remnant creosotebush-white bursage community (Turner and Brown 1982) in the Phoenix metropolitan area. The soil is a Rillito gravelly loam with a pH of 8.1 and an electrical conductivity of 1.25 dS/m. Rainfall in Phoenix is bimodal, occurring in July–August and December–January, with a mean annual precipitation of 180 mm. The minimum mean monthly temperature of 5.1°C occurs in January; the maximum mean monthly temperature of 41.0°C occurs in July.
Experimental design and sampling
Experimental landscape plots (9.2 × 9.2 m) were established in April 1999 at the site after trees were removed and ruderal vegetation disced. Each plot was planted with six Leucophyllum frutescens “Green Cloud™” and six Nerium oleander “Sister Agnes” shrubs, and one each of Quercus virginiana, Eucalyptus microtheca “Blue Ghost,” Rosmarinus officinalis, and Opuntia violacea var. santa-rita plants (V and P Nursery, Chandler, AZ, USA). Woody plant species were selected because they are representative landscape plants in the Phoenix metropolitan area. Plants had been propagated using stem cuttings and grown in containers in a mixture of native soil, bark, sand and Turflite™ (Sierra Cascade, Chemult, OR, USA).
After planting, water was applied by drip irrigation at the base of each plant at an average rate of 2,401 l plant−1 year−1. Plants were fertilized during April of each year with 200 g of controlled release 20N-4.4P-4K plus micronutrients fertilizer (Best Paks™, J. R. Simplot, Boise, ID, USA). Weeds were managed with winter and summer broadcast applications of preemergent herbicide (XL 2G, Dow Elanco Specialty Products, Indianapolis, IN, USA) and regular handweeding.
Soil samples were collected at two plots (designated plot A and plot B) located 4 m apart in late fall 2000 (plot A) or winter 2001 (plot B). Although the plots were sampled in two different seasons, this difference was not considered a treatment and no between-plot comparison were made. Each plot was divided into 25 equally sized quadrats and a soil core was collected from a sampling point at the center of each quadrat. The soil was removed with a sanitized hand shovel to a depth of 10 cm and stored in sealed plastic bags at 4°C.
Roots were collected from plants and stained in trypan blue (Koske and Gemma 1989) to verify fungal colonization. Colonization by AM fungi was detected in roots of all plant species using a light microscope. In addition, ectomycorrhizal root tips were detected on Q. virginiana and E. microtheca.
Analysis of mycorrhizal fungi diversity
AMF diversity has been studied by counting asexual spores isolated from field soils (Morton et al. 1995). However, this method may not adequately capture species richness at a site when spore numbers are low and cryptic AMF species are present that do not readily sporulate in field soils (Bever et al. 2001; McGee 1989; Stutz and Morton 1996). Soil samples from the sites were examined for the presence of AM fungal spores and found to have very low spore densities (<10 spores/100 cm3 soil), typical of arid urban environments (Cousins et al. 2003). Although the use of molecular methods has begun to revolutionize studies of AMF diversity (Clapp et al. 2002; Redecker 2002), we decided not to employ these methods because of the lack of a single primer to amplify AMF DNA and the difficulty in matching sequence groups to morphospecies (Redecker et al. 2003). We decided to use the successive trap cultures technique (Stutz and Morton 1996), in which host plants are grown in diluted field soils in a greenhouse for several months to induce sporulation of recalcitrant fungi (Bever et al. 1996; Brundrett et al. 1999; Stutz and Morton 1996), enabling the detection of fungi that may be present in roots and soil but not sporulating at the time of sampling. Trap cultures were established by mixing a 250-cm3 portion of each soil sample with 250 cm3 of a 1:1 mixture of autoclaved #12 and #20 silica sand and placing the mixture in a D40 Deepot™ (Stuewe and Sons, Corvallis, OR, USA). The pots were planted with 50–60 seeds of Sorghum bicolor, and the plants were grown in a greenhouse for 4 months, then the grass was clipped, and the pots were reseeded. After an additional 4 months of growth, the soil was air-dried, removed from the pots, and stored in sealed plastic bags at 4°C until further processing.
AMF spores were isolated from a 100-cm3 soil subsample of each trap culture using the sucrose density gradient centrifugation method of Daniels and Skipper (1982). Isolated spores were examined using a dissecting microscope and spores of each morphotype were mounted in polyvinyl-lacto-glycerol (PVLG) and 1:1 PVLG/Melzer’s reagent for identification (Koske and Tessier 1983). Species identification was based on examination of spore morphological and subcellular characteristics using stereo and light microscopes and compared to voucher specimens and species descriptions on the International Culture Collection of Arbuscular and Vesicular-Arbuscular Mycorrhizal Fungi web site (Morton et al. 1993).
Data analysis
Average sampling effort curves for each plot were generated with the EstimateS 5.0 program (Colwell 1997). First-order jackknife estimates were calculated to estimate the expected number of species in each plot (Smith and van Belle 1984).
The spatial structure of the number of AMF species detected in each quadrat was visualized using ArcView 3.3 (ESRI). The number of species detected at each of the 25 sampling points in the plots was mapped and locations of plants were overlain onto these maps.
The number of AMF species detected at each sampling point was used to determine the spatial structure of the AMF species in each plot using Moran’s I spatial autocorrelation coefficient, implemented with the ROOKCASE program (Sawada 1999). Moran’s I is a global measure of spatial autocorrelation, determining the extent to which a variable is surrounded by variables with similar values. Moran’s I coefficients were calculated at a series of lag distances, indicating the distance from one sample location to another. Moran’s I values normally range between −1 and +1. A positive value at a given lag indicates that samples at that distance are more similar to each other than to the overall mean. A negative value at a given lag indicates that samples at that distance are less similar to each other than they are to the overall mean. Regular distribution of AMF species should produce corresponding peaks and troughs in the Moran’s I correlogram (Roberts and Jones 2000).
Spore abundance in trap cultures was not used as a measure of species dominance because it is probably more indicative of the culture environment rather than having any ecological significance. In an effort to overcome this problem, we used relative frequency as an indication of dominance. This method often underestimates the occurrence of the most common species (Magurran 1988) and could give a different perception of community evenness than the use of abundance (Horton and Bruns 2001). However, the use of a large number of sampling points has been found to overcome some of these problems (Magurran 1988). Relative frequency was calculated as the number of times a species was detected per total number of samples in each plot.
AMF community structure was depicted by plotting rank frequency of species within each experimental plot. The join-count statistic, implemented in the ROOKCASE program (Sawada 1999), was used to determine the amount of clustering among the four most frequently encountered species in each plot. This method codes each sampling point as present or absent for a given species, then determines the type of join (present/present, present/absent, or absent/absent) for each pair of neighboring points. If present/present joins are higher than expected, that species is clustered, and if absent/absent joins are higher than expected, that species is overdispersed. If both types of joins are near the expected values, the species is randomly distributed across the plot. Significance was tested by calculating the Z score for each type of join (Sawada 1999).
Results
AMF community and sampling effort
A total of 12 AMF species were detected in pot cultures established with soil from both experimental plots. Seven species were detected in plot A and 11 species in plot B. AMF spores were recovered from trap cultures in 96% of samples in plot A and 100% of samples in plot B. In both plots, there was a maximum of five species detected per sampling point. In plot A, the mean number of species detected per sampling point was 2.4 (SEM = 0.20), and in plot B a mean of 3.6 species (SEM = 0.21) was detected per sampling point.
Sampling effort curves continue to rise in both plots, indicating that there were still species to be found (Fig. 1). The logarithmic curve for plot A approached a horizontal asymptote, with more than 90% of all species detected in the first 20 samples. The logarithmic curve for plot B did not approach an asymptote after 25 samples. Nonlinear regressions of number of species against number of samples yielded the equation y = 2.89x 0.28 (P < 0.001) for plot A and y = 3.56x 0.34 (P < 0.001) for plot B. Based on first-order jackknife estimates, the expected number of species in plot A was 9 and the expected number in plot B was 16.
Analysis of spatial patterns
Maps of the number of AMF species detected per quadrat in each plot are depicted in Fig. 2. All but one quadrat contained at least one AMF species, even in areas that were bare of plants. Visual inspection appears to reveal a patchier distribution of the number detected per point in plot B, but spatial statistical analysis does not support this observation.
The mean nearest neighbor distance of 1.84 m was used as the lag in calculating Moran’s I, with six lags in each plot. In plot A, values were negatively autocorrelated (Moran’s I = −0.556, z = −3.03) at the first lag, indicating that number of species for samples within 1.84 m of each other are less similar to each other than they are to the overall mean number of species per sample for the plot. Moran’s I values were not significant at any other distance in this plot. Plot B demonstrated a similar pattern, but Moran’s I was not significant at any distance (Fig. 3). These results indicate that at the resolution of the sampling design, AMF species are fairly randomly distributed across both plots.
Individual taxon-level analysis
Six species (Glomus eburneum, Glomus intraradices, Glomus microaggregatum, Glomus mosseae, and Glomus sp. AZ 123) were detected in both experimental plots, and the remaining six species were unique to a single plot. G. eburneum and G. microaggregatum were detected in more than 50% of sampling points in both plots. G. intraradices was also detected in more than 50% of points in plot B but at ∼30% of the points in plot A. Glomus spurcum was detected at more than 50% of points in plot B but was not detected in plot A. G. mosseae was detected in ∼40% of points in both plots, and the remaining species were detected in less than 30% (Fig. 4).
Plots of species rank in frequency vs relative frequency of the species imply that there is a lognormal distribution in both plots, with a few common species and a longer tail of rarer species (Fig. 5).
The spatial distribution of the four most frequently detected species in each plot is depicted in Fig. 6. Based on visual observation, G. mosseae and G. eburneum appear to have patchy distributions in plot A, while G. microaggregatum appears to be evenly distributed. Likewise, G. microaggregatum, G. mosseae, and G. intraradices all appear to be nonrandomly distributed in plot B. According to the join-count statistic, however, only a single species in each plot was not randomly distributed (Table 1). The sampling points at which G. eburneum was detected in plot A were negatively autocorrelated (absent/absent joins are higher than expected), indicating an overdispersed pattern. In contrast, points at which G. microaggregatum was detected in plot B were positively autocorrelated (present/present joins are higher than expected), suggesting a clustered distribution.
Discussion
We found that sampling effort had a major impact on assessing species richness in this local community. A limited number of additional undetected species are likely to be present in both plots based on the estimation procedures such as sampling effort curves and first-order jackknife estimate calculations. Based on sampling effort curves, 15 samples would have been adequate to detect 70–80% of species present in each plot. These results confirm the findings of Tews and Koske (1986), who estimated that 30 samples would be adequate to enumerate AMF species richness in a barrier dune system. The species richness detected in this experimental urban landscaped site was similar to the number found in similarly sized sites in other semiarid and mesic ecosystems, including Brazilian sand dunes (Stürmer and Bellei 1994), an Iowa poplar plantation (Walker et al. 1982), and Sonoran and Chihuahuan desert scrub (Stutz and Morton 1996). However, when Bever et al. (2001) used intensive sequential sampling over a longer period and a variety of culture techniques, they detected a total of 37 species in a single 1-ha old field, an area about 50 times larger than our study area.
Our results illustrate the importance of using taxon sampling curves when comparing AMF species richness between sites or treatments. If a small number of samples is collected in a study, it would be difficult to ascertain if differences in detected species richness between sites and/or treatments are due to underlying differences in species richness or reflect sampling effort (Gotelli and Colwell 2001; Taylor 2002). Sampling effort curves in this study did not reach an asymptote in the five to ten samples that Klironomos et al. (1999) report are typically collected per treatment in soil ecology field experiments. Gotelli and Colwell (2001) suggest that species richness counts can only be compared when taxon-sampling curves reach a clear asymptote.
Klironomos et al. (1999) suggested that the location of plants could be used to predict below-ground “hotspots” because the spatial organization of soil microorganisms and chemical cycles is linked to primary producers. Because of the patchy distribution of plants in the experimental plots, we expected that the number of AMF would be higher at sampling points in proximity to landscape plants and lower in the interspaces between plants. Our results indicated that the density of AMF species (number/sample point) was fairly randomly distributed across both plots with no detectable hotspots. The relatively recent establishment of our experimental plots could be one explanation for the lack of detectable hotspots associated with landscape plants. AM fungi detected in the interspaces between landscape plants could be a legacy effect of the previous land use (Schlesinger et al. 1966) as a tree farm and creosote desert flat. AMF propagules may have survived from previous association with plants in the tree farm or the original desert community, remaining dormant in the bare areas between landscape plants. Ettema and Wardle (2002) report that dormancy contributes to the intrinsic heterogeneity of soil organisms, with dormant propagules being analogous to soil seed banks. AMF propagules may have also migrated to the sampling points lacking plants from the surrounding areas. In arid environments, wind serves as an effective dispersal agent for AMF propagules (Allen et al. 1989; Warner et al. 1987). Wind-blown propagules of AMF species that were deposited at sampling points would have been part of the original soil sample and these species would be detected in the resulting trap culture.
Like Pringle and Bever (2002), we found that AMF communities were highly variable on a local scale, with the species richness and species composition of one site quite different from that of a second nearby site. Six species were detected at both plots, but the remaining six species were unique to a single plot. Some species, such as Glomus intraradices and G. spurcum, were detected at more than 50% of the sampling points in one plot but were rare or not detected in the other plot. The distribution of AMF species was clustered to the extent that half the species detected occurred in less than 10% of the soil samples taken. Although there were differences in species composition and richness between the two plots, these differences may be due to seasonal differences in growth of the AMF species, as plot A was sampled during the dry autumn season and plot B was sampled 4 months later during the winter wet season. Camargo-Ricalde and Esperon-Rodriguez (2005) observed greater numbers of AMF spores in semiarid soils during the wet season than during the dry season in Tehuacan-Cuicatlan, Mexico. Due to the difference in sampling times, the two plots were not considered true replicates for the purposes of this study.
In conclusion, this study illustrates some of the pitfalls in assessing AMF diversity and community structure. Sampling effort had a major influence on detected AMF species richness, showing similarity to results reported using molecular techniques with ectomycorrhizal fungi (Horton and Bruns 2001; Taylor 2002). Our results also indicated that the number of AMF species detected at sample points was not autocorrelated at scales similar to those of plant patches and that “plant-stratified” approach may not be best for sampling in an arid urban ecosystem or possibly in other recently disturbed environments. While these results are specific to an arid urban site and cannot be extrapolated to other locations, they do illustrate the difficulties in quantifying AMF diversity and the complexity of AMF communities.
References
Allen MF, MacMahon JA (1985) Impact of disturbance on cold desert fungi: comparative microscale dispersion patterns. Pedobiologia 28:215–224
Allen MF, Hipps LE, Wooldridge GL (1989) Wind dispersal and subsequent establishment of VA mycorrhizal fungi across a successional arid landscape. Landsc Ecol 2:165–171
Anderson RC, Liberta AE, Dickman LA, Katz AJ (1983) Spatial variation in vesicular-arbuscular mycorrhiza spore density. Bull Torrey Bot Club 110:519–525
Bever JD, Morton JB, Antonovics J, Schultz PA (1996) Host-dependent sporulation and species diversity of arbuscular mycorrhizal fungi in a mown grassland. J Ecol 84:71–82
Bever JD, Schultz PA, Pringle A, Morton JB (2001) Arbuscular mycorrhizal fungi: more diverse than meets the eye, and the ecological tale of why. BioScience 51:923–932
Boerner REJ, DeMars BG, Leicht PN (1996) Spatial patterns of mycorrhizal infectiveness of soils long a successional chronosequence. Mycorrhiza 6:79–90
Brundrett MC, Abbott LK, Jasper DA (1999) Glomalean mycorrhizal fungi from tropical Australia I. Comparison of the effectiveness and specificity of different isolation procedures. Mycorrhiza 8:305–314
Camargo-Ricalde SL, Esperon-Rodriguez M (2005) Effect of the spatial and seasonal soil heterogeneity over arbuscular mycorrhizal fungal spore abundance in the semi-arid valley of Tehuacan-Cuicatlan, Mexico. Rev Biol Trop 53:339–352
Carvalho LM, Correia PM, Ryel RJ, Martins-Louçầo MA (2003) Spatial variability of arbuscular mycorrhizal fungal spores in two natural plant communities. Plant Soil 251:227–236
Clapp JP, Helgason T, Daniell TJ, Young JPW (2002) Genetic studies of the structure and diversity of arbuscular mycorrhizal fungal communities. In: van der Heijden MGA, Sanders IR (eds) Mycorrhizal ecology. Springer, Berlin Heidelberg New York, pp 201–224
Colwell RK (1997) EstimateS: statistical estimation of species richness and shared species from samples, version 5. Available at http://viceroy.eeb.uconn.edu/estimates. Accessed 8 Sep 2002
Cousins JR, Hope D, Gries C, Stutz JC (2003) Preliminary assessment of arbuscular mycorrhizal fungal diversity and community structure in an urban ecosystem. Mycorrhiza 13:319–326
Daniels BA, Skipper HD (1982) Methods for the recovery and quantitative estimation of propagules from soil. In: Schenck NC (ed) Methods and principles of mycorrhizal research. American Phytopathological Society Press, St. Paul, pp 29–35
Ettema CH, Wardle DA (2002) Spatial soil ecology. Trends Ecol Evol 17:177–183
Gange AC, Brown VK, Farmer LM (1990) A test of mycorrhizal benefit in an early successional plant community. New Phytol 115:85–91
Gotelli NJ, Colwell RK (2001) Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecol Lett 4:379–391
Hart MM, Klironomos JN (2002) Diversity of arbuscular mycorrhizal fungi and ecosystem functioning. In: van der Heijden MGA, Sanders IR (eds) Mycorrhizal ecology. Springer, Berlin Heidelberg New York, pp 225–242
Hartnett DC, Wilson GWT (1999) Mycorrhizae influence plant community structure and diversity in tallgrass prairie. Ecology 80:1187–1195
Horton TR, Bruns TD (2001) The molecular revolution in ectomycorrhizal ecology: peeking into the black-box. Mol Ecol 10:1855–1871
Klironomos JN, Rillig MC, Allen MF (1999) Designing belowground field experiments with the help of semi-variance and power analyses. Appl Soil Ecol 12:227–238
Klironomos JN, McCune J, Hart M, Neville J (2000) The influence of arbuscular mycorrhizae on the relationship between plant diversity and productivity. Ecol Lett 3:137–141
Koske RE, Gemma JN (1989) A modified procedure for staining roots to detect VA mycorrhizas. Mycol Res 92:486–486
Koske RE, Tessier B (1983) A convenient, permanent slide mounting medium. Nwsl Mycol Soc Am 34:59
Magurran AE (1988) Ecological diversity and its measurement. Princeton University Press, Princeton
McGee PA (1989) Variation in propagule numbers of vesicular-arbuscular mycorrhizal fungi in a semi-arid soil. Mycol Res 92:28–33
Morton JB, Bentivenga SP, Wheeler WW (1993) Germ plasm in the International Collection of Arbuscular and Vesicular-Arbuscular Mycorrhizal Fungi (INVAM) and procedures for culture development, documentation and storage. Mycotaxon 48:491–528
Morton JB, Bentivenga SP, Bever JD (1995) Discovery, measurement and interpretation of diversity in symbiotic endomycorrhizal fungi (Glomales, Zygomycetes). Can J Bot 73:S25–S32
Pringle A, Bever JD (2002) Divergent phenologies may facilitate the coexistence of arbuscular mycorrhizal fungi in a North Carolina grassland. Am J Bot 89:1439–1446
Redecker D (2002) Molecular identification and phylogeny of arbuscular mycorrhizal fungi. Plant Soil 244:67–73
Redecker D, Hijri I, Wiemken A (2003) Molecular identification of arbuscular mycorrhizal fungi in roots: perspectives and problems. Folia Geobot 38:113–124
Roberts C, Jones JA (2000) Soil patchiness in juniper–sagebrush–grass communities of central Oregon. Plant Soil 223:45–61
Sawada M (1999) ROOKCASE: an Excel 97/2000 visual basic (VB) add-in for exploring global and local spatial autocorrelation. Bull Ecol Soc Am 80:231–234
Schlesinger WH, Raikes JA, Hartley AE, Cross AF (1966) On the spatial pattern of soil nutrients in desert ecosystems. Ecology 77:364–374
Smith SE, Read DJ (1997) Mycorrhizal symbiosis, 2nd edn. Academic, London
Smith EP, van Belle G (1984) Nonparametric estimation of species richness. Biometrics 40:119–129
Stürmer SL, Bellei MM (1994) Composition and seasonal variation of spore populations of arbuscular mycorrhizal fungi in dune soils on the island of Santa Catarina, Brazil. Can J Bot 72:359–363
Stutz JC, Martin CA (1998) Arbuscular mycorrhizal fungal diversity associated with ash trees in urban landscapes in Arizona. Phytopathology 88:586
Stutz JC, Morton JB (1996) Successive pot cultures reveal high species richness of arbuscular mycorrhizal fungi in arid ecosystems. Can J Bot 74:1883–1889
Taylor AFS (2002) Fungal diversity in ectomycorrhizal communities: sampling effort and species detection. Plant Soil 244:19–28
Tews LL, Koske RE (1986) Toward a sampling strategy for vesicular-arbuscular mycorrhizas. Trans Br Mycol Soc 87:353–358
Turner RM, Brown DE (1982) Sonoran desertscrub. Desert Plants 4:181–221
van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis P, Strietwolf-Engel R, Boller T, Wiemken A, Sanders IR (1998) Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396:69–72
Walker C, Mize CW, McNabb HS (1982) Populations of endogonaceous fungi at two locations in central Iowa. Can J Bot 60:2518–2529
Warner NJ, Allen MF, MacMahon JA (1987) Dispersal agents of vesicular-arbuscular mycorrhizal fungi in a disturbed arid ecosystem. Mycologia 79:721–730
Acknowlegements
We would like to thank Chris Martin and Brooke Stabler for setting up and maintaining the study site, and Elizabeth Wentz for help with the spatial statistics. This material is based upon work supported by the National Science Foundation (NSF) under grant no. DEB 9714833, CAP LTER. Any opinions, findings and conclusions, or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the NSF.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Whitcomb, S., Stutz, J.C. Assessing diversity of arbuscular mycorrhizal fungi in a local community: role of sampling effort and spatial heterogeneity. Mycorrhiza 17, 429–437 (2007). https://doi.org/10.1007/s00572-007-0118-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-007-0118-5