Abstract
Computer-aided medical image segmentation has been applied widely in diagnosis and treatment to obtain clinically useful information of shapes and volumes of target organs and tissues. In the past several years, convolutional neural network (CNN)-based methods (e.g., U-Net) have dominated this area, but still suffered from inadequate long-range information capturing. Hence, recent work presented computer vision Transformer variants for medical image segmentation tasks and obtained promising performances. Such Transformers modeled long-range dependency by computing pair-wise patch relations. However, they incurred prohibitive computational costs, especially on 3D medical images (e.g., CT and MRI). In this paper, we propose a new method called Dilated Transformer, which conducts self-attention alternately in local and global scopes for pair-wise patch relations capturing. Inspired by dilated convolution kernels, we conduct the global self-attention in a dilated manner, enlarging receptive fields without increasing the patches involved and thus reducing computational costs. Based on this design of Dilated Transformer, we construct a U-shaped encoder–decoder hierarchical architecture called D-Former for 3D medical image segmentation. Experiments on the Synapse and ACDC datasets show that our D-Former model, trained from scratch, outperforms various competitive CNN-based or Transformer-based segmentation models at a low computational cost without time-consuming per-training process.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Medical image segmentation, as one of the critical computer-aided medical image analysis problems, aims to capture precisely the shapes and volumes of target organs and tissues by pixel-wise classification, obtaining clinically useful information for diagnosis, treatment, and intervention. With the recent development of deep learning methods and computer vision algorithms, medical image segmentation has been revolutionized and remarkable progresses have been achieved (e.g., automatic liver and tumor lesion segmentation [1], brain tumor segmentation [2], and multiple sclerosis (MS) lesion segmentation [3]).
Fully convolutional network (FCN) [4] was first proved effective for general image segmentation tasks, which became a predominant technique for medical image segmentation [5,6,7,8,9]. However, it was observed that vital details can be missing with the decrease of feature map sizes when FCN models went deeper. To this end, a family of U-shaped networks [10,11,12,13,14,15,16,17] was proposed to extend the sequential FCN frameworks to encoder–decoder-type architectures, alleviating the spatial information loss using skip connections. In DeepLab models [18,19,20,21], atrous convolutions instead of pool layers were applied to expand the receptive field and fully connected conditional random field (CRF) was introduced to maintain fine details. Although these CNN-based methods have achieved great performances on medical image segmentation tasks, they still suffered from limited receptive fields and were unable to capture long-range dependencies, leading to sub-optimal accuracy and failing to meet the needs of various medical image segmentation scenarios.
Inspired by the success of Transformer with its self-attention mechanism in natural language processing (NLP) tasks [22, 23], researchers tried to adapt Transformers [24,25,26,27] to computer vision (CV) in order to compensate the locality of CNNs. The self-attention mechanism in Transformers enabled to compute pair-wise relations between patches globally, consequently achieving feature interactions across a long range. The self-attention mechanism was first adopted by non-local neural networks [28] to complement CNNs for modeling pixel-level long-range dependency for visual recognition tasks. Then, a pure Transformer framework was proposed by the Vision Transformer (ViT) [24] for vision tasks, treating an image as a collection of spatial patches. Recently, Transformers have achieved excellent outcomes on a variety of vision tasks [29,30,31,32,33,34], including image recognition [29,30,31, 35, 36], semantic segmentation [32], and object detection [33, 34]. On semantic medical image segmentation, Transformer-combined architectures can be divided into two categories: The main one adopted self-attention like operations to complement CNNs [37,38,39,40] and the other used pure Transformers to constitute encoder–decoder architectures so as to capture deep representations and predict the class of each image pixel [32, 41,42,43].
Although the above medical image segmentation methods were promising and yielded good performance to some extent, they still suffered considerable drawbacks. (1) The majority of these Transformer segmentation models were designed for 2D images [37, 39, 41,42,43]. For 3D medical images (e.g., 3D MRI scans), they divided the input images into 2D slices and processed the individual slices with the 2D models, which could lose useful 3D contextual information. (2) Compared with common 2D natural scene images, processing 3D medical images inevitably incurred larger model sizes and computational costs, especially when computing global feature interactions with self-attention in vanilla Transformer [22] (see more details in Sect. 3.3). Although some adaptations were proposed to reduce the operation scopes of self-attention [29,30,31, 44,45,46,47] (e.g., progressive scaling pyramids were used in the Pyramid Vision Transformer [30] to reduce the computation costs of large feature maps), insufficient global information fusion incurred. (3) The self-attention operation in Transformers was shown to be permutation-equivalent [22], which omitted the order of patches in an input sequence. However, the permutation equivalence nature can be detrimental to medical image segmentation since segmentation results are often highly position-correlated. In prior works, absolute position encoding (APE) [22] and relative position encoding (RPE) [29, 48] were utilized to supplement position information. But, APE required a pre-given and fixed patch amount and thus failed to generalize to different image sizes, while RPE ignored the absolute position information that could be a vital cue in medical images (e.g., the positions of bones are often relatively stable).
To address the above drawbacks, we propose a new efficient model called Dilated Transformer (D-Former) to directly process 3D medical images (instead of dealing with 2D slices of 3D images independently) and predict volumetric segmentation masks. Our proposed D-Former is a 3D U-shaped architecture with hierarchical layers, and employs skip connections from encoder to decoder following [10,11,12,13,14,15,16,17]. This model’s stem is constructed with eight D-Former blocks, each of which consists of several local scope modules (LSMs) and global scope modules (GSMs). The LSM conducts self-attention locally, focusing on fine information capturing. The GSM performs global self-attention on uniformly sampled patches, aiming to explore rough and long-range-dependent information at low cost. The LSMs and GSMs are arranged in an alternate manner to achieve local and global information interaction. For drawback (3), we manage to incorporate position information among patches in a more dynamic manner. Inspired by [44, 49], we utilize depth-wise convolutions [50] to learn position information, which can help provide useful position cues in medical image segmentation.
Benefiting from these designs, our proposed D-Former model could be more suitable for medical image segmentation tasks and yield better segmentation accuracy. The main contributions of this work are as follows:
-
(1)
We construct a 3D Transformer-based architecture which allows to process volumetric medical images as a whole and thus spatial information along the depth dimension of 3D medical images can be fully captured.
-
(2)
We design local scope modules (LSMs) and global scope modules (GSMs) to increase the scopes of information interactions without increasing the patches involved in computing self-attention, which helps reduce computational costs.
-
(3)
To further incorporate relative and absolute position information among patches, we apply a dynamic position encoding method to learn it from the input directly. As a result, an inherent problem of common Transformers, permutation equivalence [22], could be considerably alleviated.
-
(4)
Extensive experimental evaluations show that our model outperforms state-of-the-art segmentation methods in different domains (e.g., CT and MRI), with smaller model sizes and less FLOPs than the known methods.
2 Related work
2.1 CNN-based segmentation networks
Since the advent of the seminal U-Net model [10], many CNN-based networks have been developed [17, 51,52,53,54]. As for the design of skip connections, U-Net++ [55] and U-Net3+ [56] were proposed to attain dense connections between encoder and decoder. In addition, regarding the locality of CNNs, different kinds of mechanisms were designed to enlarge the receptive field, such as larger kernel [57], dilated convolution module [58, 59], pyramid pooling module [60, 61], and deformable convolution module [62, 63]. In particular, dilated convolution was an ingenious design in which the convolution kernel was expanded by inserting holes between its consecutive elements. This design has been adopted by various segmentation models, achieving good performance compared with the original convolution-based methods. Our Dilated Transformer also obtains a key idea from this design and aims to conduct self-attention in a patch skipping manner (see Sect. 3.3 for details).
2.2 Visual Transformer variants
Transformer and its self-attention mechanism were first designed for sequence modeling and transduction tasks in the domain of natural language processing (NLP), achieving state-of-the-art performance [22, 23]. Inspired by tremendous success of Transformers in NLP, Transformers were adapted for computer vision tasks. The first attempt was vision Transformer (ViT) [24] which needed huge pre-training datasets. To overcome this weakness, a wide range of training strategies with knowledge distillation was proposed by DeiT [25], which contributed to better performances of vanilla Transformer. There were different kinds of adaptations for vanilla Transformer, such as Swin Transformer [29], pyramid vision Transformer [30], Transformer in Transformer [64], and aggregating nested Transformers [31]. In particular, Swin Transformer showed great success in various computer vision tasks with its elegant shift window mechanism and hierarchical architecture. Our proposed D-Former is inspired by Swin Transformer’s local–global combining scopes of information interactions.
2.3 Transformers for segmentation tasks
As mentioned above, Transformers used in medical image segmentation methods can be divided into two categories. In the main category, Transformer and its self-attention mechanism were utilized as a supplement for the convolution-based stem. SETR [65] was proposed to apply Transformer as encoder to extract features for segmentation tasks. In medical images, many models with Transformers focused on segmentation tasks. In TransUNet [37], convolutional layer was used as a feature extractor to obtain detailed information from raw images, and the generated feature maps were then put into Transformer layer to obtain global information. UNETR [38] proposed a 3D Transformer-combining architecture for medical images, which treated Transformer layer as encoder to extract features and convolutional layer as decoder. A great amount of such work focused on taking advantage of both Transformer’s long-range dependency and CNN’s inductive bias. In the other category, Transformer was regarded as the main stem for building the whole architecture [32, 41,42,43]. In MedT [66], the gated axial attention mechanism and Gated Axial Transformer layer were proposed to build the architecture. Swin-Unet [41] was constructed with the basic units of Swin Transformer blocks, which was further extended in DS-TransUNet [42] by adding another encoder pathway for input of different sizes. Compared with these previous methods, our proposed D-Former model has several advantages: (1) Our method focuses on 3D medical image segmentation, which is a topic with little previous exploration in the context of Transformer; (2) our D-Former avoids cumbersome design for fusing CNN and Transformer specifically, constructing the architecture stem based on Transformer only; and (3) by designing LSMs and GSMs (see Sect. 3.3), our model complexity is significantly lower than the compared methods.
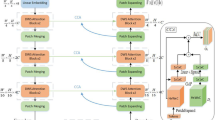
Overall architecture of our D-Former model. Each D-Former block is constructed with one dynamic position encoding block (DPE) and several local scope modules (LSMs) and global scope modules (GSMs). The input size of the D-Former block i is reported sideward, and the output sizes are the same as the corresponding input sizes. The values in round brackets denote the numbers of patches, which are regarded as one dimension when computed in Transformers (i.e., \((\frac{W}{4} \times \frac{H}{4} \times \frac{D}{2})\), \((\frac{W}{8} \times \frac{H}{8} \times \frac{D}{4})\), \((\frac{W}{16} \times \frac{H}{16} \times \frac{D}{8})\), \((\frac{W}{32} \times \frac{H}{32} \times \frac{D}{16})\))
3 Method
3.1 The overall architecture
Our proposed D-Former model is outlined in Fig. 1, which is a hierarchical encoder–decoder architecture. The encoder pathway consists of one patch embedding layer for transforming 3D images into sequences and four proposed D-Former blocks for feature extraction with three down-sampling layers in between them. The first, second, and fourth D-Former blocks each consist of one local scope module (LSM) and one global scope module (GSM), respectively, while the third D-Former block has three LSMs and three GSMs, in which the LSMs and GSMs are arranged in an alternate manner. The decoder pathway is symmetric to the encoder pathway, which also has four D-Former blocks, three up-sampling layers, and one patch expanding layer. In addition, skip connections are used to transfer information from the encoder to the decoder at the corresponding levels. The feature maps from the encoder are concatenated with the corresponding feature maps along the channel dimension, which may compensate for the loss of fine-grained information as the model goes deep.
In this section, we will present the components of D-Former one by one, including the patch embedding and patch expanding layers (Sect. 3.2), the D-Former block and its major modules, the local scope module and global scope module (Sect. 3.3), the down-sampling and up-sampling operations (Sect. 3.4), and the dynamic position encoding block (Sect. 3.5).
3.2 Patch embedding and patch expanding
Similar to common Transformers in computer vision, after data augmentation, an input 3D medical image \(x \in {\mathbb {R}}^{W \times H \times D}\) is first processed by a patch embedding layer and is divided into a series of patches of size \(4\times 4\times 2\) each, and then is projected into C channel dimensions by linear projection to yield a feature map (denoted by \(x_1\)) of size \((\frac{W}{4} \times \frac{H}{4} \times \frac{D}{2}) \times C\), where \((\frac{W}{4} \times \frac{H}{4} \times \frac{D}{2})\) denotes the number of patches and C is the number of the channel dimensions. Hence, the input 3D image is reorganized as a sequence (of length \((\frac{W}{4} \times \frac{H}{4} \times \frac{D}{2})\)) and can be directly fed to a Transformer architecture. The final patch expanding layer is used to restore the feature map to the original input size, and a segmentation head (like 3D UNet [67]) is utilized to attain pixel-wise segmentation masks.
3.3 D-former blocks
After patch embedding, \(x_{1}\) is directly fed to D-Former block 1. In the processing by Transformer block 1, \(x_1\) is first processed by a new dynamic position encoding block that embeds position information into feature maps (see details in Sect. 3.5), and then it is operated by the Local Scope Module (LSM) and Global Scope Module (GSM) alternatively to extract higher-level features. The other D-Former Blocks process the corresponding input features similarly, and the feature map sizes are provided in Fig. 1.
3.3.1 Local scope module and global scope module
The local scope module (LSM) and global scope module (GSM) are designed to capture local and global features, respectively, for which two different self-attention operations are employed, called local scope multi-head self-attention (LS-MSA) and global scope multi-head self-attention (GS-MSA). As shown in Fig. 2, an LSM is composed of a LayerNorm layer [68], a proposed LS-MSA, another LayerNorm layer, and a multilayer perceptron (MLP), in sequence, with two residual connections to prevent gradient vanishing [22]. In a GSM, the LS-MSA is replaced by a proposed GS-MSA, and the other components are kept the same as the LSM. To allow local features and global features to be captured and fused well, LSM and GSM are arranged alternatively in each D-Former block. With these components, their operations are formally defined as:
where \({\hat{z}}^{l}\) and \(z^{l}\) denote the outputs of LS-MSA and the corresponding MLP, respectively, and \({\hat{z}}^{l+1}\) and \(z^{l+1}\) denote the outputs of GS-MSA and the corresponding MLP, respectively.
a Local scope multi-head self-attention: The self-attention is conducted in a local unit (colored in blue) where the patches are adjacent. b Gobal scope multi-head self-attention: The self-attention is conducted in a global unit (colored in blue) where patches are picked every gth patch across the feature map. A small cube represents one patch. The feature map size is set as \(6\times 6\times 6\) and the unit size is \(3\times 3\times 3\) as an example. We color only the patches of one unit in blue for illustration; the other gray patches are also utilized to construct seven other units in both LS-MSA and GS-MSA
3.3.2 Local scope multi-head self-attention (LS-MSA)
Self-attention is conducted in the vanilla Transformer in a global scope in order to capture pair-wise relationships between patches, leading to quadratic complexity with respect to the number of patches. However, due to the fact that 3D medical images would increase computation inevitably, this original self-attention would not be suitable for 3D medical image related tasks, especially for semantic segmentation with dense prediction targets. Under such circumstances, as illustrated in Fig. 3a, a whole feature map is first divided evenly into non-overlapping units (the number of patches in each unit is denoted by \(u_{d} \times u_{h} \times u_{w}\), where \(u_d\) denotes the number of patches in one unit along the depth dimension D, \(u_h\) along the height dimension H, and \(u_w\) along the width dimension W), and self-attention is conducted within each unit. In this way, the computational complexity will be reduced to linear in terms of the number of patches in the whole feature map. The computational complexity (\(\Omega\)) of these two different self-attention mechanisms is computed as:
where \(u_{d} u_{h} u_{w}\) denotes the number of patches in one unit and dhw denotes the number of patches in the whole feature map. (d, h, and w denote the depth, height, and width of the feature map, respectively.) In most cases, \(u_{d} u_{h} u_{w}\ll dhw\). The Softmax operation is omitted when computing the computational complexity.
3.3.3 Global scope multi-head self-attention (GS-MSA)
The LS-MSA performs self-attention only within each local unit, which lacks global information interaction and long-range dependency. To address this issue, we design a global scope multi-head self-attention mechanism to attain information interaction across different units in a dilated manner. As illustrated in Fig. 3b, for a whole feature map, we pick one patch every g distance along each dimension and form a unit with all the patches thus picked, on which self-attention would then be conducted. Likewise, we pick the other patches to form new units, until all the patches are utilized. Hence, the receptive field in computing self-attention will be enlarged but the number of patches involved will not be increased, which means that it would not increase the computational cost while getting access to long-range information interaction. To keep consistency between LSM and GSM, we set \(d=g_{d} \times u_{d}\), \(h=g_{h} \times u_{h}\), and \(w=g_{w} \times u_{w}\), which ensures that the numbers of units in LSM and GSM are kept the same. Here, \(d \times h \times w\) denotes the number of patches in the whole feature map, \(u_{d} \times u_{h} \times u_{w}\) denotes the number of patches in one unit, and \(g_{d}\), \(g_{h}\), and \(g_{w}\) denote the distance between two nearest patches picked along the depth dimension D, height dimension H, and width dimension W, respectively.
3.4 Down-sampling and up-sampling
Between every two adjacent D-Former blocks of the encoder, a down-sampling layer is utilized to merge patches for further feature fusion. Specifically, a down-sampling layer concatenates the feature maps of \(2\times 2\times 2\) neighboring patches (2 neighboring patches along the width, height, and depth dimensions, respectively), reducing the number of patches by 8 times. Then, a fully connected layer is utilized to reduce the feature channel size by 4 times to ensure that the channel size can be doubled after each down-sampling layer. Thus, the output feature maps of each down-sampling layer will be \(x_{2} \in {\mathbb {R}}^{(\frac{W}{8} \times \frac{H}{8} \times \frac{D}{4}) \times 2C}\), \(x_{3} \in {\mathbb {R}}^{(\frac{W}{16} \times \frac{H}{16} \times \frac{D}{8}) \times 4C}\), and \(x_{4} \in {\mathbb {R}}^{(\frac{W}{32} \times \frac{H}{32} \times \frac{D}{16}) \times 8C}\), respectively. In reverse to the down-sampling layers, four up-sampling layers of the decoder are used to enlarge the low-resolution feature maps and reduce the number C of the channel dimensions. In this way, our model will be able to extract features in a multi-scale manner and yield better segmentation accuracy.
3.5 The dynamic position encoding block
The depth-wise convolution (DW-Conv) is a type of convolution that applies a single convolutional filter for each input channel instead of for all channels as in a common convolution, which can decrease the computational cost. We apply 3D depth-wise convolution [50] to the input feature maps (or images) once in every D-Former block to learn position information. Then the learned position information will be added to the original input \(x_i\) as:
where \(x_i\) denotes the input feature maps of the ith D-Former block and \(x_i^\prime\) denotes the output feature maps embedded with position information. Resize is used to adjust the dimensions of feature maps \(x_i\) to cater the input need of DW Convolution.
In this way, position information among patches can be extracted by a DW-Convolution. Given the fact that position information could be dynamically learned based on the input x itself, a drawback in the previous work that requires a fixed number of patches can be avoided. In addition, the convolution’s inherent nature of translation invariance can be utilized to increase the stability and generalization performance [69].
4 Experiments
4.1 Datasets
The Synapse multi-organ segmentation (Synapse) dataset includes 30 axial contrast-enhanced abdominal CT scans. Following the training–test split in [37], 18 of the 30 scans are used for training and the remaining ones are for testing. The average dice similarity coefficient (DSC) [17] is used as the measure for evaluating the segmentation performances of the eight target organs, including aorta, gallbladder, kidney (L), kidney (R), liver, pancreas, spleen, and stomach.
The Automated Cardiac Diagnosis Challenge (ACDC) dataset contains 150 magnetic resonance imaging (MRI) 3D cases collected from different patients, and each case covers a heart organ from the base to the apex of the left ventricle. Following the setting in [37], only 100 well-annotated cases are used in the experiments, and the training, validation, and test data are partitioned with the ratio of 7: 1: 2. For fair comparison, the average DSC is employed to evaluate the segmentation performances following the previous work [37], and three key parts of the heart are chosen as targets, including the right ventricle (RV), myocardium (Myo), and left ventricle (LV).
4.2 Implementation setup
Pre-training. Our D-Former model is trained from scratch, which means that we initialize the model’s weights randomly. Note that in common practice, pre-training is important to Transformer-based models. This is because the pre-training process provides generalized representations and prior knowledge for downstream tasks. For example, in vision Transformer (VIT) [24], it considered that the model performance depends heavily on pre-training, and its experiments verified this view. Besides, lots of known medical image segmentation methods used pre-trained weights to initialize their models [32, 37, 39, 70, 71]. However, the pre-training process of Transformer-based models brings up two issues. First, the pre-training process usually incurs high computational complexity in terms of time or computation consumed. Second, for medical images, there are few complete and acknowledged sizable datasets for pre-training (in comparison, ImageNet [72] is available for natural scene images), and the domain gap between natural images and medical images makes it hard for medical image segmentation models to use existing large natural image datasets directly. For these reasons, we choose to train our D-Former model from scratch, which nevertheless yields promising performance that surpasses state-of-the-art methods with pre-training.
Implementation details. Our proposed D-Former is implemented on PyTorch 1.8.0, and all the experiments are trained on an NVIDIA GeForce RTX 3090 GPU with 24 GB memory. The batch size during training is 2 and during inference in 1. The SGD optimizer [73] with momentum 0.99 is used. The initial learning rate is 0.01 with weight decay of 3e–5. The polylearning rate strategy [74] is utilized with the maximum training epochs of 3000 for the Synapse dataset and 1500 for the ACDC dataset. The training takes about 8 h for the Synapse dataset and about 6.5 h for the ACDC dataset, and the test time of one sample takes about 1.3 s for the Synapse dataset and about 1.2 s for the ACDC dataset.
Loss function. The cross-entropy loss and Dice loss are both widely used for general segmentation tasks. However, since the cross-entropy loss is apt to perform well for uniform class distribution while Dice loss is more suitable for target objects of large sizes [75], each of them alone may not be effective for medical image segmentation tasks that involve imbalanced classes and target objects of small sizes. Thus, our loss function combines the binary cross-entropy loss Y [76] and Dice loss \({\hat{Y}}\) [17] together, which is defined as:
where \(Y_{n}\) and \({\hat{Y}}_{n}\) denote the ground truth and predicted probabilities of the \(n^{th}\) image, respectively, and N is the batch size.
4.3 Quantitative results
We evaluate the performance of our proposed D-Former model on the Synapse and ACDC datasets, and compare with various state-of-the-art models, including V-Net [17], DARR [77], R50 U-Net [10], R50 Att-UNet [78], U-Net [10], Att-UNet [78], VIT [24], R50 VIT [24], TransUNet [37], Swin-UNet [41], LeVit-Unet-384 [71], nnFormer [32], and MISSFormer [43].
Quantitative results on the Synapse dataset are reported in Table 1, which show that our method outperforms the previous work by a clear margin. It is notable that the concurrent Transformer-based methods nnFormer and MISSFormer achieve some performance gains compared to the CNN-based methods, while our method still brings further improvement in the average DSC by 0.0143 compared to nnFormer and by 0.0687 compared to MISSFormer. Besides, our D-Former obtains accuracy improvement on almost every organ class, except for the pancreas and stomach, which verifies that our D-Former is a promising and robust framework.
Quantitative results on the ACDC dataset are reported in Table 2, and a similar conclusion can be drawn. D-Former achieves the best average DSC of 0.9229 without pre-training. Compared with the other methods, our method brings improvements in the average DSC by 0.0474 over R50 U-Net and by 0.0554 over R50 Att-UNet. Compared to the concurrent Transformer-based methods, our method still achieves 0.0051 performance gain over nnFormer and 0.0439 over MISSFormer in the average DSC. Specifically, among all the key parts of the heart, including the right ventricle (RV), myocardium (Myo), and left ventricle (LV), our D-Former obtains the best segmentation accuracy compared to the other methods in the average DSC.
The results in Tables 1 and 2 show that our D-Former attains excellent generalization on both CT data and MRI data, outperforming the previous methods. Notably, different from most of the known Transformer-based frameworks that require a pre-training process, D-Former is initialized randomly and is trained from scratch, yet still obtains competitive performances. This implies that our model could be more suitable for medical imaging tasks when general large size medical image pre-training datasets (such as ImageNet [72] for natural scene images) are lacking.
To verify the statistical difference between our proposed method and other compared methods, Sign test [79] and Paired T-test [80] are conducted. (1) For the Synapse dataset, the Sign test is conducted between our method and other compared methods one by one, where the inputs are the organ’s segmentation accuracy (i.e., average DSC) of two paired groups (i.e., one is our method and the other is one compared method). In comparing our proposed method with nnFormer, the output p-value is 0.09, and p-values are 0.02 between our proposed method and other methods. (2) For the ACDC dataset, the Paired T-test is utilized, where the inputs are the organ’s segmentation accuracy of two paired groups (i.e., one is our method and the other is one compared method). The detailed p values are shown in Table 2. One can see that our proposed method slightly outperforms nnFormer and TransUNet, while significantly outperforms other methods. Meanwhile, our method still achieves a lower computational cost compared to nnFormer and other methods (see Sect. 4.4).
4.4 Comparison of model complexity
In Table 3, we compare the numbers of parameters and floating point operations (FLOPs) of our proposed D-Former with those of different 3D medical image segmentation models, including UNETR [38], CoTr [40], TransBTS [70], and nnFormer [32]. The number of FLOPs is calculated based on the input image size of 64\(\times\)128\(\times\)128 for fair comparison. We should note that we omit the part of the complexity brought by activation functions and normalization layers. Table 3 shows that our D-Former has 44.26M parameters and 54.46G FLOPs, which has a lower computational cost compared to nnFormer (157.88G FLOPs), TransBTS (171.30G FLOPs), CoTr (377.48G FLOPs), UNETR (86.02G FLOPs), and 3D U-Net (947.69G FLOPs). The CNN-based model of 3D U-Net has less parameters, but it is burdened with a high model complexity of 947.69G FLOPs, which is much bigger than our D-Former method. Moreover, compared with the other Transformer-based models, our model still shows comparable model complexity while outperforming these models by a large margin.
To further explore the effectiveness of our model in reducing model complexity, we remove two key designs, respectively, and compute the corresponding numbers of parameters and floating point operations (FLOPs). As shown in Table 4, one can see that our patch embedding layer, and local scope multi-head self-attention (LS-MSA) and global scope multi-head self-attention (GS-MSA) contribute to decreasing the model complexity considerably. Specifically, without the patch embedding layer, an input image is directly projected into C channel dimensions and fed to the subsequent Transformer architecture, leading to 1189.47G FLOPs. Besides, the introduction of LS-MSA and GS-MSA helps decrease the FLOPs from 477.19G to 54.46G, and this is consistent with the theoretical analysis in Sect. 3.3.2.
4.5 Qualitative visualizations
To intuitively demonstrate the performances of our D-Former model, we compare some qualitative results of our model with several other methods (including Swin-Unet, TransUnet, and UNet) on the Synapse dataset, and some hard samples are shown in Fig. 4. One can see that the predicted organ masks of our model are much more similar to the ground truth in general. As for specific organs, our model has better accuracy in identifying and sketching the contours of stomach (e.g., the first and fourth rows), which is consistent with the conclusions based on the above quantitative results. In the second row, only our model can delineate the outline of pancreas well, thus suggesting that our model has a better ability to capture long-range dependency given the fact that the shape of pancreas is long and narrow. In addition, as illustrated in the third row, our D-Former is able to identify the true region of liver, while the other three models incur some mistakes on the liver. This shows that our method is effective at exploiting the relations between the target organs’ patches and the other patches, owing to our model’s dynamic position encoding block. In a nutshell, the qualitative visualizations provide intuitive demonstrations of our model’s high segmentation accuracy, especially on some slices that are difficult to segment.
4.6 Ablation studies
We conduct ablation studies on the Synapse dataset to evaluate the effectiveness of our model design.
Effect of global scope module (GSM). To investigate the necessity of the Global Scope Module (GSM), we replace it by the Local Scope Module (LSM), with the other architectural components unchanged. As shown in Table 5, one can see that the GSM is beneficial to the segmentation accuracy, outperforming using only LSM modules by 0.0066 in the average DSC. This verifies the necessity to explore global interactions of patches across units.
Global scope multi-head self-attention (GS-MSA) vs. other self-attention. In order to confirm the effectiveness of our GS-MSA, we compare it with the shift window strategy proposed in Swin Transformer [29] which achieves state-of-the-art performance in multiple computer vision tasks. Similar to our GS-MSA design, the shift window strategy (SW-MSA) aims to introduce global attention. Table 6 shows that our global attention design surpasses that in Swin Transformer by 0.0133 in the average DSC.
Dynamic position encoding vs. other position encodings. We compare our dynamic position encoding (DPE) with other common position encoding methods, including the relative position encoding (RPE) [29, 48], absolute position encoding (APE) [22], and sinusoidal position encoding (SPE) [22]. The results are shown in Table 7. Compared to APE, SPE, and RPE, our DPE improves them by 0.0405, 0.0279, and 0.0248 in the average DSC, respectively.
Position of the dynamic position encoding block. We conduct experiments to examine the performances of different choices of positions to apply the dynamic position encoding block, including placing it (a) before the first LSM, (b) right after the first LSM, and (c) right after the first GSM, in every D-Former block, as illustrated in Fig. 5 taking D-Former block 3 as an example. Table 8 shows that introducing the position information before the first LSM provides the best segmentation outcomes.
The sizes of different architecture variants. To evaluate the performances of variants with different sizes, three variants of our D-Former are evaluated. Specifically, the architecture hyper-parameters of our model variants are:
-
D-Former-Small: C = 64, L = {2, 2, 2, 2},
-
D-Former-Base: C = 64, L = {2, 2, 6, 2},
-
D-Former-Large: C = 96, L = {2, 2, 6, 2},
where C is the channel number of the hidden layers and L is the total number of LSMs and GSMs in the encoder pathway. As shown in Table 9, D-Former-Large achieves the best performance in terms of the average DSC with 0.8883, improving by 0.0480 and 0.0142 comparing with D-Former-Small and D-Former-Base.
5 Conclusions
In this paper, we proposed a novel 3D medical image segmentation framework called D-Former, which utilizes the common U-shaped encoder–decoder design and is constructed based on our new Dilated Transformer. Our proposed D-Former model can achieve both good efficiency and accuracy, due to its reduced number of patches used in self-attention in local scope module (LSM) and its exploration of long-range dependency with a dilated scope of attention in global scope module (GSM). Moreover, we introduced the dynamic position encoding block, making it possible to flexibly learn vital position information within input sequences. In this way, our model not only reduces the model parameters and decreases the FLOPs, but also attains state-of-the-art semantic segmentation performance on the Synapse and ACDC datasets.
Data availability statements
The data that support the findings of this study are openly available at https://doi.org/10.7303/syn3193805 and https://acdc.creatis.insa-lyon.fr/#challenge/5846c3366a3c7735e84b67ec.
References
Christ PF, Ettlinger F et al. (2017) Automatic liver and tumor segmentation of CT and MRI volumes using cascaded fully convolutional neural networks. ArXiv:1702.05970
Pereira S, Pinto A (2016) Brain tumor segmentation using convolutional neural networks in MRI images. TMI 35(5):1240–1251
Brosch T, Tang LY, Yoo Y (2016) Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. TMI 35(5):1229–1239
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: CVPR. IEEE, pp 3431–3440
Korez R, Likar B, Pernuš F (2016) Model-based segmentation of vertebral bodies from MR images with 3D CNNs. In: MICCAI. Springer, pp 433–441
Zhou X, Ito T, Takayama R (2016) Three-dimensional CT image segmentation by combining 2D fully convolutional network with 3D majority voting. In: Deep learning and data labeling for medical applications. Springer, pp 111–120
Moeskops P, Wolterink JM (2016) Deep learning for multi-task medical image segmentation in multiple modalities. In: MICCAI. Springer, pp 478–486
Shakeri M, Tsogkas S, Ferrante E (2016) Sub-cortical brain structure segmentation using F-CNN’s. In: International symposium on biomedical imaging. IEEE, pp 269–272
Alansary A, Kamnitsas K, Davidson A (2016) Fast fully automatic segmentation of the human placenta from motion corrupted MRI. In: MICCAI. Springer, pp 589–597
Ronneberger O, Fischer P, Brox T (2015) U-Net: Convolutional networks for biomedical image segmentation. In: MICCAI, pp 234–241
Wang C, MacGillivray T, Macnaught G et al (2018) A two-stage 3D Unet framework for multi-class segmentation on full resolution image. ArXiv:1804.04341
Çiçek, Ö, Abdulkadir A, Lienkamp SS (2016) 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: MICCAI. Springer, pp 424–432
Kamnitsas K, Ledig C, Newcombe VF (2017) Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. MIA 36:61–78
Drozdzal M, Vorontsov E, Chartrand G (2016) The importance of skip connections in biomedical image segmentation. In: Deep learning and data labeling for medical applications. Springer, pp 179–187
Ghafoorian M, Karssemeijer N, Heskes T (2016) Non-uniform patch sampling with deep convolutional neural networks for white matter hyperintensity segmentation. In: International symposium on biomedical imaging. IEEE, pp 1414–1417
Brosch T, Tang LY, Yoo Y (2016) Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. TMI 35(5):1229–1239
Milletari F, Navab N, Ahmadi S-A (2016) V-Net: Fully convolutional neural networks for volumetric medical image segmentation. In: 3DV. IEEE, pp 565–571
Chen L-C, Papandreou G, Kokkinos I et al (2014) Semantic image segmentation with deep convolutional nets and fully connected CRFs. ArXiv:1412.7062
Chen L-C, Papandreou G, Kokkinos I (2017) DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. TPAMI 40(4):834–848
Chen L-C, Papandreou G, Schroff F, et al (2017) Rethinking atrous convolution for semantic image segmentation. ArXiv:1706.05587
Chen L-C, Zhu Y, Papandreou G (2018) Encoder-decoder with atrous separable convolution for semantic image segmentation. In: ECCV, pp 801–818
Vaswani A, Shazeer N, Parmar N (2017) Attention is all you need. In: NIPS, vol 30
Devlin J, Chang M-W, Lee K, et al (2018) Bert: pre-training of deep bidirectional Transformers for language understanding. ArXiv:1810.04805
Dosovitskiy A, Beyer L, Kolesnikov A, et al (2020) An image is worth 16x16 words: transformers for image recognition at scale. ArXiv:2010.11929
Touvron H, Cord M, Douze M (2021) Training data-efficient image transformers and distillation through attention. In: ICML. PMLR, pp 10347–10357
Carion N, Massa F, Synnaeve G (2020) End-to-end object detection with Transformers. In: ECCV. Springer, pp 213–229
Zhu X, Su W, Lu L, et al (2020) Deformable DETR: deformable transformers for end-to-end object detection. ArXiv:2010.04159
Wang X, Girshick R, Gupta A (2018) Non-local neural networks. In: CVPR. IEEE, pp 7794–7803
Liu Z, Lin Y, Cao Y, et al (2021) Swin transformers: hierarchical vision transformers using shifted windows. ArXiv:2103.14030
Wang W, Xie E, Li X, et al (2021) Pyramid vision transformers: a versatile backbone for dense prediction without convolutions. ArXiv:2102.12122
Zhang Z, Zhang H, Zhao L, et al (2021) Aggregating nested transformers. ArXiv:2105.12723
Zhou H-Y, Guo J, Zhang Y, et al (2021) nnFormer: interleaved transformers for volumetric segmentation. ArXiv:2109.03201
Sun Z, Cao S, Yang Y (2021) Rethinking transformer-based set prediction for object detection. In: ICCV, pp 3611–3620
Pan X, Xia Z, Song S (2021) 3D object detection with pointformer. In: CVPR. IEEE, pp 7463–7472
Yuan L, Chen Y, Wang T, et al (2021) Tokens-to-Token ViT: training vision Transformers from scratch on ImageNet. ArXiv:2101.11986
Yuan L, Hou Q, Jiang Z, et al (2021) VOLO: vision outlooker for visual recognition. ArXiv:2106.13112
Chen J, Lu Y, Yu Q, et al (2021) TransUNet: transformers make strong encoders for medical image segmentation. ArXiv:2102.04306
Hatamizadeh A, Tang Y, Nath V, et al (2021) UNETR: transformers for 3D medical image segmentation. ArXiv:2103.10504
Zhang Y, Liu H, Hu Q (2021) TransFuse: fusing transformers and CNNs for medical image segmentation. ArXiv:2102.08005
Xie Y, Zhang J, Shen C, et al (2021) CoTr: efficiently bridging CNN and transformer for 3D medical image segmentation. ArXiv:2103.03024
Cao H, Wang Y, Chen J, et al (2021) Swin-Unet: Unet-like pure Transformer for medical image segmentation. ArXiv:2105.05537
Lin A, Chen B, Xu J, et al (2021) DS-TransUNet: dual swin transformer U-Net for medical image segmentation. ArXiv:2106.06716
Huang X, Deng Z, Li D, et al (2021) MISSFormer: an effective medical image segmentation Transformer. ArXiv:2109.07162
El-Nouby A, Touvron H, Caron M, et al (2021) XCiT: cross-covariance image transformers. ArXiv:2106.09681
Wu Z, Liu Z, et al (2020) Lite Transformer with long-short range attention. ArXiv:2004.11886
Mehta S, Koncel-Kedziorski R, Rastegari M, Hajishirzi H (2020) DeFINE: DEep Factorized INput Token Embeddings for neural sequence modeling. ArXiv:1911.12385
Mehta S, Ghazvininejad M, Iyer S, et al (2020) DeLighT: very deep and light-weight transformer. CoRR
Shaw P, Uszkoreit J, Vaswani A (2018) Self-attention with relative position representations. ArXiv:1803.02155
Chu X, Tian Z, Zhang B, et al (2021) Conditional positional encodings for vision transformers. ArXiv:2102.10882
Chollet F (2017) Xception: deep learning with depthwise separable convolutions. In: CVPR. IEEE, pp 1251–1258
Diakogiannis FI, Waldner F, Caccetta P (2020) ResUNet-a: a deep learning framework for semantic segmentation of remotely sensed data. J Photogram Remote Sens 162:94–114
Ni Z-L, Bian G-B, Zhou X-H (2019) RAUNet: residual attention u-net for semantic segmentation of cataract surgical instruments. In: International conference on neural information processing. Springer, pp 139–149
Isensee F, Jaeger PF, Kohl SA (2021) nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 18(2):203–211
Cai S, Tian Y, Lui H (2020) Dense-UNet: a novel multiphoton in vivo cellular image segmentation model based on a convolutional neural network. Quant Imaging Med Surg 10(6):1275
Zhou Z, Siddiquee MMR, Tajbakhsh N (2018) UNet++: a nested U-Net architecture for medical image segmentation. In: Deep learning in medical image analysis and multimodal learning for clinical decision support. Springer, pp 3–11
Huang H, Lin L, Tong R (2020) UNet 3+: a full-scale connected UNet for medical image segmentation. In: IEEE international conference on acoustics, speech and signal processing, pp 1055–1059
Peng C, Zhang X, Yu G (2017) Large kernel matters—improve semantic segmentation by global convolutional network. In: CVPR. IEEE, pp 4353–4361
Chen L-C, Papandreou G, Kokkinos I (2017) DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. PAMI 40(4):834–848
Chen L-C, Zhu Y, Papandreou G (2018) Encoder-decoder with atrous separable convolution for semantic image segmentation. In: ECCV, pp 801–818
Roth HR, Shen C, Oda H (2018) A multi-scale pyramid of 3D fully convolutional networks for abdominal multi-organ segmentation. In: MICCAI, pp 417–425
Feng S, Zhao H, Shi F (2020) CPFNet: context pyramid fusion network for medical image segmentation. TMI 39(10):3008–3018
Heinrich MP, Oktay O, Bouteldja N (2019) OBELISK-Net: fewer layers to solve 3D multi-organ segmentation with sparse deformable convolutions. MIA 54:1–9
Li Z, Pan H, Zhu Y (2020) PGD-UNet: a position-guided deformable network for simultaneous segmentation of organs and tumors. In: International joint conference on neural networks. IEEE, pp 1–8
Han K, Xiao A, Wu E, et al (2021) Transformer in transformer. ArXiv:2103.00112
Zheng S, Lu J, Zhao H (2021) Rethinking semantic segmentation from a sequence-to-sequence perspective with transformers. In: CVPR. IEEE, pp 6881–6890
Valanarasu JMJ, Oza P, et al (2021) Medical transformer: gated axial-attention for medical image segmentation. ArXiv:2102.10662
Çiçek Ö, Abdulkadir A, Lienkamp SS (2016) 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: MICCAI. Springer, pp 424–432
Ba JL, Kiros JR, Hinton GE (2016) Layer normalization. ArXiv:1607.06450
Kauderer-Abrams E (2017) Quantifying translation-invariance in convolutional neural networks. ArXiv:1801.01450
Wang W, Chen C, Ding M (2021) TransBTS: multimodal brain tumor segmentation using Transformer. In: MICCAI. Springer, pp 109–119
Xu G, Wu X, Zhang X, et al (2021) LeViT-UNet: make faster encoders with transformer for medical image segmentation. ArXiv:2107.08623
Deng J, Dong W, Socher R (2009) ImageNet: a large-scale hierarchical image database. In: CVPR. IEEE, pp 248–255
Bottou L (2012) Stochastic gradient descent tricks. In: Neural networks: tricks of the trade. Springer, pp 421–436
Mishra P, Sarawadekar K (2019) Polynomial learning rate policy with warm restart for deep neural network. In: IEEE region 10 conference, pp 2087–2092
Jadon S (2020) A survey of loss functions for semantic segmentation. In: IEEE conference on computational intelligence in bioinformatics and computational biology, pp 1–7
Yi-de M, Qing L, Zhi-Bai Q (2004) Automated image segmentation using improved PCNN model based on cross-entropy. In: International symposium on intelligent multimedia, video and speech processing, pp 743–746
Fu S, Lu Y, Wang Y (2020) Domain adaptive relational reasoning for 3D multi-organ segmentation. In: MICCAI. Springer, pp 656–666
Schlemper J, Oktay O, Schaap M (2019) Attention gated networks: learning to leverage salient regions in medical images. MIA 53:197–207
Dixon WJ, Mood AM (1946) The statistical sign test. J Am Stat Assoc 41(236):557–566
Hsu H, Lachenbruch PA (2014) Paired t test. Statistics Reference Online, Wiley StatsRef
Acknowledgements
This research was partially supported by the National Key R &D Program of China under Grant No. 2019YFB1404802, the National Natural Science Foundation of China under Grants No. 62176231 and 62106218, the Zhejiang public welfare technology research project under Grant No. LGF20F020013, and the Wenzhou Bureau of Science and Technology of China under Grant No. Y2020082. D. Z. Chen’s research was supported in part by NSF Grant CCF-1617735.
Funding
This research was partially supported by the National Key R &D Program of China under Grant No. 2019YFB1404802, the National Natural Science Foundation of China under Grants Nos. 62176231 and 62106218, the Zhejiang public welfare technology research project under Grant No. LGF20F020013 and the Wenzhou Bureau of Science and Technology of China under Grant No. Y2020082. D. Z. Chen’s research was supported in part by NSF Grant CCF-1617735.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, Y., Liao, K., Chen, J. et al. D-former: a U-shaped Dilated Transformer for 3D medical image segmentation. Neural Comput & Applic 35, 1931–1944 (2023). https://doi.org/10.1007/s00521-022-07859-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-022-07859-1