Abstract
Medical image segmentation (MIS) is a key technique in computer-aided diagnosis. With the development of deep learning, especially convolutional neural networks, the performance of MIS has been significantly improved, however, some mainstream convolution-based methods still suffer from inaccurate target boundaries and imprecise segmentation results. At the same time, transformer-based methods have gradually achieved better segmentation results. To overcome the challenges of traditional methods, an accurate MIS model (CascadeMedSeg) is proposed in this paper, which combines a pyramid vision transformer (PVT) and multi-scale fusion. This network model follows a standard encoder-decoder segmentation architecture, where PVT is used as an encoder. PVT, designed as a pure Transformer backbone for pixel-level dense prediction tasks, can consistently generate a global receptive field and, as an encoder, flexibly learn multi-scale features of medical images. Two additional modules, namely Enhanced Attention Fusion (EAF) and Edge-Enhanced Segmentation (EES) are introduced. The EAF module fuses up-sampled and skip-connected features using an attention mechanism that enhances the perception of channel and positional information. The EES module enhances the boundary features of the network through the aggregation of multi-level features of the encoder and a dynamic boundary detection operator used to obtain a boundary mask and embed it into the decoder. Extensive experiments on five datasets show that CascadeMedSeg exhibits improved performance over several state-of-the-art methods. The MIoU values for the Kvasir-SEG, CVC-ClinicDB, ISIC 2018, and BUSI datasets are 88.16, 89.79, 86.32, and 66.69%, respectively.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
As an indispensable part of modern medicine, medical images allow physicians to visualize physiological structures and pathological changes, and play a crucial role in the early detection, accurate diagnosis and treatment of diseases [1, 2]. Segmentation is one of the key technologies in the field of medical image processing [3,4,5], and its purpose is to separate specific areas in the image from the background. This process enables more detailed patient disease analysis and provides a reliable basis for clinical diagnosis and pathology research [6,7,8].
Methods based on Convolutional Neural Networks (CNNs) are widely used for medical image segmentation (MIS) tasks [9,10,11,12]. Due to the outstanding performance of UNet and its variations [13] in MIS, CNN networks with a U-shaped encoder-decoder structure have become prevalent [15, 15, 17]. Many researchers have also incorporated attention mechanisms into CNNs [17, 18] to enhance segmentation by emphasizing relevant channels and suppressing of irrelevant ones. While CNN-based models have demonstrated satisfactory performance in MIS, and because of convolutional layers possess translation equivariance but can not scale with a large receptive field [19, 20]. Consequently, they have limitations in capturing distant dependencies between pixels [21].
Recently, many researchers have adopted Transformer structures for visual information processing tasks in order to capture remote dependencies and have achieved performance comparable to that of CNNs [22,23,24]. The Vision Tranformer (ViT) [25] was a groundbreaking approach that divided the image into small patches and utilized transformers to form a classification network. However, the ViT requires more complex processing and greater computational resources than CNNs. The Swin Transformer (SwinT) [22] and the Pyramid Vision Transformer (PVT) [23, 26] required reduced spatial attention, which resulted in a substantial decrease in the computational load incurred while still achieving excellent capture of remote dependencies. However, the self-attention mechanism used in transformers limits their ability to learn local contextual relationships between pixels [27]. During the decoding stage of MIS, the ability of the network to recover detailed features is particularly important [28, 29]. Transformer excels at capturing remote dependencies, whereas, CNN excels at maintaining local details [30]. Researchers have investigated combining both networks to maximize their strengths in MIS tasks.
Based on the above analysis, this paper proposes a highly accurate MIS model called CascadeMedSeg, which combines PVT and multi-scale fusion techniques to realize effective MIS. First, a PVT is used as an encoder to extract multi-level features hierarchically. Then, an Enhanced Attention Fusion (EAF) module is designed to fuse skip-connected encoder features with decoder up-sampled features and enhance the network’s perception of channel and location information. Finally, an Edge-Enhanced Segmentation (EES) module targets enhanced edge modeling, aggregates multi-level features, and extracts boundary information using a learnable boundary detection operator embedded into the decoder to enhance boundary features.
The main contributions of this paper are summarized as follows.
-
(1)
In the proposed MIS model, CascadeMedSeg, a PVT and multi-scale fusion are employed to enhance the network’s performance compared to traditional approaches based on UNet. The network utilizes a PVT during the encoding phase to extract robust and rich multi-scale features.
-
(2)
An EAF module is designed to improve the perception of location information when cross-channel interactions are applied during feature fusion.
-
(3)
We design an EES module, that realizes boundary detection and enhancement by combining the multi-level features produced by the encoder and using a trainable boundary operator to calculate the gradient information in the image.
-
(4)
Extensive experiments demonstrate that the proposed CascadeMedSeg outperforms other state-of-the-art models.
2 Related work
2.1 Medical image segmentation
Deep learning models have been widely used in MIS, which is an important task in computer-aided diagnostic methods [31]. Among these methods, CNN-based deep learning techniques have demonstrated outstanding performance in MIS. Specifically, UNet++ [15] uses a series of dense skip connections to extract features at different scales, thus reducing the semantic gap between the encoder and decoder. Although this method improves feature extraction, its complex structure leads to high computational costs and lengthy training times.
UNet3+ [15] directly combines high- and low-level semantics, enhancing segmentation performance; however, it also has a high computational complexity and hardware requirements. ResUNet [15] constituted a novel approach to U-Net by integrating residual concatenation from ResNet [32] to address the issue of gradient vanishing. The Attention U-Net [13] incorporated attention gating units into the original UNet to enhance the latter’s sensitivity to pixels in the foreground target region during segmentation; however, this increases computational complexity and may slow down the training. The DoubleU-Net [9] featured two encoders and decoders and employed atrous spatial pyramid pooling to capture contextual information. DCSAU-Net [33] utilized primary feature conservation to capture essential features from the input image, and a compact split-attention block to output feature maps with different combinations of receptive field sizes; however, its segmentation accuracy does not perform well on some datasets. However, since convolution is essentially a local operation, CNN-based approaches for MIS methods may result in incomplete segmentation masks.
Transformer-based methods have recently demonstrated significant success in MIS. For instance, Chen et al. [34] introduced TransUNet, which integrated the intricate spatial information from CNN features with the comprehensive context captured by a transformer. Another approach, Swin-UNet [35] developed by Cao et al., utilized the Swin transformer as an encoder to extract contextual features, which were subsequently up-sampled using a symmetric Swin transformer decoder; however, it uses transformers in both the encoder and decoder, which does not lead to performance improvement [10]. UCTransNet [36] utilizes multi-scale channel cross-fusion with a transformer and channel-wise cross-attention modules(CTrans module), replacing the original skip connections. In SSFormer [27], a PVT was used as an encoder, while focus on local features was achieved through a progressive locality decoder, which improved the neural network’s capacity to process detailed information. In CTO [37], the encoder network employs a well-known CNN backbone structure to capture local semantic information and a lightweight ViT auxiliary network to integrate long-range dependencies, however, their number of parameters is enormous.
2.2 Vision transformer
The Transformer is a deep neural network that uses a self-attention mechanism and was originally designed for Natural Language Processing (NLP). Inspired by the Transformer’s powerful representation function, researchers proposed extending the Transformer to computer vision tasks to capture remote dependencies, resulting in higher accuracy. The ViT [25] can classify images directly using a purely self-concerned Transformer. Compared to other network types, the Transformer-based model performs equally well, if not better, on various vision benchmarks. Subsequently, various Transformer-based models have been proposed to improve Transformer performance in computer vision tasks. The Swin Transformer (SwinT) [22] with sliding window reduces calculation effort while capturing remote dependencies accurately. The Detection Transformer (Detr) [38] is an ensemble-based target detector that employs transformers on top of a convolutional backbone. The Data-efficient Image Transformer (DeiT) [39] is a visual transformer for image classification tasks that was trained using a Transformer-specific teacher-student approach. The PVT [23, 26] reduces spatial attention and makes it an effective pillar of intensive forecasting tasks by employing a pyramid structure. It can handle various downstream tasks, such as classification and detection. In this paper, we attempt to use PVT v2 [26] as the fundamental unit of an U-structure encoder to balance accuracy and efficiency in MIS. Furthermore, experimental results from multiple datasets show that our method is effective. Finally, our method applies to a broader range of scenarios, including but not limited to endoscopy, ultrasound, dermoscopy, and Magnetic Resonance Imaging (MRI).
3 Methods
3.1 Network architecture
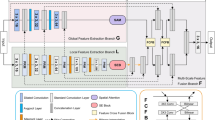
The proposed basic framework of CascadeMedSeg utilizes an encoder-decoder architecture, as presented in Fig. 1. CascadeMedSeg mainly comprises a PVT encoder, an EAF module, and an EES module. Specifically, using a PVT v2-b2 to extract features, we can model global relationships to extract more contextual information.
The EAF module enhances the network’s perception of channel and location information by combining up-sampled and jump-connected features. The EES module aggregates multi-scale features from the encoder, extracts contour and boundary masks from the image, and embeds them into the decoder to enhance the boundary features.
3.2 Transformer encoder
Transformers have great potential to solve the problem of complex scale variations in medical image segmentation and perform multi-scale feature processing [40]. By capturing remote dependencies and global contextual information, the robustness and generalization ability of the model are enhanced. Our approach uses a PVT v2 as an encoder to obtain hierarchical features through four stages. It is worth noting that PVT v2 is an improved version of the original PVT, which requires less calculation and provides more powerful feature extraction. The encoder comprises four stages. Each stage includes overlapping patch embedding and a transformer encoder. The number of output channels for each stage is \({C_i} \in \{ 64,128,320,512\}\), and the number of layers in the transformer encoder for each stage is \({L_i} \in \{ 3,3,6,3\}\). Given an input image of size \({I \in {\mathbb {R}}^{3 \times H \times W}}\). In the stage1, it is first divided into \({\frac{{HW}}{{4 ^2}}}\) patches with the size of \({3 \times 3 \times 4}\) through overlapping patch embedding. Then, it passes through a transformer encoder with \({L_1}\) layers and the output is reshaped into a feature map \({F_1}\) with the size of \({\frac{H}{{4}} \times \frac{W}{{4}} \times {C_1}}\). Similarly, using the feature map from the previous stage as input, the following feature maps are generated: \({F_2}, {F_3}\), and \({F_4}\), with sizes of \({\frac{H}{{8}} \times \frac{W}{{8}} \times {C_2}}\), \({\frac{H}{{16}} \times \frac{W}{{16}} \times {C_3}}\), and \({\frac{H}{{32}} \times \frac{W}{{32}} \times {C_4}}\), respectively.
3.3 Enhanced attention fusion module
The low-level features extracted by the encoder encompass detailed information, but lack semantic content. High-level features have more semantic information but do not reflect details clearly. In MIS tasks, reducing the semantic gap between encoder and decoder features and realizing effective fusion are important means to improve segmentation performance.
Common techniques used to reduce the semantic gaps between encoder and decoder features, such as feature fusion through element addition or channel concatenation, tend to impair predictions around target boundaries [41]. In this paper, we propose the EAF module for dynamic learning and enhancing the multi-scale feature representation of medical images. As shown in Fig. 2, two branching paths exist after the initial fusion of encoder and decoder features. The channel attention branch learns the correlations between different channels and dynamically enhances the feature dimensions that are essential to the current task. Meanwhile, the multi-head hybrid convolutional branch captures a range of spatial features at multiple scales, thus enhancing the ability of the network to extract local information.
Specifically, features from the encoder and decoder are added pixel by pixel. Adaptive average pooling for the channel attention branch is utilized to gather global information about the features. Then, the attention weights are applied to the input feature mapping to emphasize or suppress the feature responses of different channels [42]. The process can be expressed mathematically as:
Where GAP means the Global Average Pooling layer; \({Conv1{D_{k \times k}}}\) denotes a one-dimensional convolution with convolution kernel size k; \({\sigma }\) means the Sigmoid activation function, \({\otimes }\) denotes element-by-element multiplication (Hadamard product); \({k = \left\lfloor {\frac{{\left| {{{\log }_2}(C) + \beta } \right| }}{\gamma }} \right\rfloor }\), \({\beta }\)=1, \({\gamma }\)=2, \({\left\lfloor {...} \right\rfloor }\)denotes downward rounding; \({\mathrm{ }{F_{t\_i}}\mathrm{ }}\) denotes the result of pixel-by-pixel summation of encoder and decoder features.
To capture spatial features at different scales, multiple convolutions with different kernel sizes are applied to different parts of the input feature map. This enhances the spatial information of the input features while effectively filtering out the background information. The fusion of features obtained at different granularity levels and interacting with the information improves the information representation [43]. The process can be expressed as:
Where \({{x_1}}\), \({{x_2}}\),..., \({{x_n}}\) designates the splitting of input feature \({{F_{t\_i}}}\) into multiple heads in the channel dimension; n denotes the number of channels of \({{F_{t\_i}}}\); \({D{W_{(2n + 1) \times (2n + 1)}}}\) denotes a depth-separable convolution with a kernel size \({2n + 1}\); cat denotes a splice along the channel dimension; and \({Con{v_{1 \times 1}}}\)denotes a \({1 \times 1}\) convolutional layer.
Finally, the outputs of the two branches are fused, which combines the dynamics of the adjusted features with the residual branches of the original features, thus realizing smoother feature fusion.
3.4 Edge-enhanced segmentation module
The boundaries of a lesion region provide important clues to the location of objects, so they can be utilized to significantly improve the performance of semantic segmentation algorithms. However, the boundaries of lesion regions are usually complex and diverse [44]. Inspired by [37], we employ the EES module to forecast boundary masks for medical images and improve network features related to boundaries.
Due to the complexity of boundary information in medical images, single-scale features cannot be used, as they do not contain sufficient information for this representation. Using features from any scale directly to generate a boundary mask cannot meet the requirements for precision and accuracy.
As shown in Fig. 3, the EES module aims to improve edge modeling by initially combining the features generated by the encoders at the four scales. This enhances the model’s ability to represent multi-scale information, thus generating an informative and multi-granular representation of features. Then, these representations are used for subsequent boundary detection, so predictions are based on a comprehensive understanding of both overall semantics and local details, enhancing the effectiveness of boundary detection. The boundary detection module uses a trainable boundary operator to calculate the gradient features of the image and obtain precise boundary information.
Specifically, the feature maps at different resolutions \({F_i}\) are uniformly up-sampled to \(\frac{H}{4} \times \frac{W}{4} \times {C_1}\) to ensure consistent feature map size. Splicing in the channel dimension and using convolution for feature fusion allows for a better combination of the fine boundary information contained in the low-level features and the semantic information present in the high-level features. The detailed boundary information is added to the deeper features to provide well-informed and rich feature representations for subsequent boundary prediction.
After obtaining the hybrid feature MF, two dynamic boundary operators are utilized to capture the corresponding edge information in the horizontal and vertical directions, respectively, and the response to the boundary in the feature map is enhanced by calculating the gradient amplitude.
where \({{W}_x}\) and \({{W}_y}\) are depth-separable convolutional kernels of \({3\times 3}\) size, and “\({*}\)” denotes the convolution operation.
The final extracted boundary information is pooled to match different layer feature sizes and then multiplied element-by-element with the features fused by the EAF module to directly enhance the boundary features of the network.
where \({MaxPool_m}\) denotes the max pooling layer; \(m \in \{2,4,8\} \); \(i \in \{2,3,4\} \).
3.5 Loss function
To supervise the CascadeMedSeg network’s training, binary cross-entropy (Bce) [34] and Dice loss [34] are adopted.
Bce loss is commonly used in binary classification problems and is defined as:
Where \({{y_i}}\) and \({{p_i}}\) denote the fundamental truth value and predicted probability label of the i-th pixel, respectively; N denotes the product of the height and width of the image.
The Dice loss is defined as follows:
Where \({{y_i}}\) and \({{p_i}}\), and N are as defined in Eq. (7), and the \({\varepsilon }\) represents a smoothing function, adding a small constan \({\varepsilon }\) (set to \({10^{ - 4}}\) in this paper) to both the numerator and denominator prevents the denominator from being zero, even in extreme cases where the prediction and ground truth do not overlap at all or are both zero. Therefore, this prevents division by zero errors and ensures numerical stability.
Finally, the complete loss function [34] can be expressed as:
Where \({L_{Total}}\) is the total loss, \({{L_{Bce}}}\) and \({{L_{Dice}}}\) denote Bce loss and Dice loss between the segmentation result and segmentation label.
4 Experiments
In this section, the proposed CascadeMedSeg network is evaluated using five datasets, namely Kvasir-SEG [45], CVC-ClinicDB [46], ISIC 2018 [47, 48], BUSI [49], and ACDC [50, 51]. We also perform a series of experiments using these datasets. Finally, the efficiency of the proposed method is compared with that of other state-of-the-art (SOTA) methods.
4.1 Experimental data
The proposed CascadeMedSeg was implemented using the PyTorch DL framework. All experiments were conducted using PyTorch 1.90 and CUDA 11.1.1, and the hardware environment included an i7-10700F CPU and a NVIDIA 3090 GPU. In this paper, the pre-trained PVT v2-b2 was used as the backbone network.
Kvasir-SEG dataset: This is an endoscopic dataset for pixel-level segmentation of colon polyps, consisting of 1000 images of gastrointestinal polyps and their corresponding segmentation masks. The number of training, validation, and test sets in the experiment is 800, 100, and 100, respectively.
CVC-ClinicDB dataset: This data consists of 612 still images extracted from colonoscopy videos from 29 different sequences. Each frame image is accompanied by ground truth that identifies the region covered by polyps in the image. The dataset is split into 490 training samples, 61 validation samples, and 61 testing samples.
ISIC 2018 dataset: This is a publicly available dataset of skin lesion images containing 2594 dermoscopic images of different types, sizes, and colors of skin lesions from 2056 unique patients with segmentation masks. The number of training, validation and test sets are 2074, 260, and 260, respectively.
BUSI dataset: The BUSI is a classification and segmentation dataset containing breast ultrasound images. The dataset consists of 780 images which are classified into three categories: normal, benign, and malignant. The benign and malignant breast ultrasound images also contain detailed segmentation annotations corresponding to chest tumors. The number of training, validation, and test sets are 624, 78, and 78, respectively.
ACDC dataset: The ACDC consists of 100 cardiac MRI images collected from different patients. Each scan contains three organs: namely the right ventricle (RV), the left ventricle (LV), and the myocardium (Myo). The dataset is split into 70 training samples, 10 validation samples, and 20 testing samples.
All experiments were performed on the same training, validation and testing datasets. For the Kvasir-SEG, CVC-ClinicDB, ISIC 2018, and BUSI datasets, the image resolution was set to 352\(\times \)352, while for the ACDC dataset, the resolution was set to 224\(\times \)224. We use the Adam optimizer and the ReduceLROnPlateau scheduler with a learning rate of \({1 \times 10^{ - 4}}\). The model is trained for 150 epochs. The batch size for the ACDC dataset is set to 24, while the batch size for other datasets is set to 16. The other SOTA models were trained on the same datasets using their default parameters.
4.2 Quantitative evaluation
A total of four metrics were used to evaluate the proposed model, where mDice, mIoU, Recall, and Precision were used as evaluation metrics on the Kvasir-SEG, CVC-ClinicDB, ISIC 2018, and BUSI datasets, and only mDice scores were considered for the ACDC dataset [34, 35].
The equations for calculating the specific metrics are as follows.
where n denotes the total number of samples; True Positive (TP), False Negative (FN), False Positive (FP), and True Negative (TN) follow their usual definitions.
4.3 Results
4.3.1 Experimental results on Kvasir-SEG and CVC-ClinicDB datasets
Through training, validation and testing, we verified the performance of the proposed method on the Kvasir-SEG and CVC-ClinicDB datasets. As shown in Tables 1 and 2, CascadeMedSeg yielded the best values for the chosen metrics. The methods selected for comparison were U-Net [13], UNet++ [17], Attention U-Net [17], DoubleU-Net [9], TransUNet [34], DCSAU-Net [33], and CTO [37]. Tables 1 and 2 show the quantitative comparison results for the Kvasir-SEG and CVC-ClinicDB datasets.
For Kvasir-SEG, CascadeMedSeg achieved the best results and significantly improved polyp segmentation performance compared to the other algorithms. Compared with TransUNet, the CascadeMedSeg method improved the mDice and mIoU scores on the Kvasir-SEG dataset by 0.95% and 1.46%, respectively.
Some indicative visual results are presented in Fig. 4. The proposed CascadeMedSeg method generated prediction masks that were closer to the ground truth labels. With better segmentation results in some cases, CascadeMedSeg can segment target boundaries more accurately, regardless of whether a small, large, or multiple targets are being segmented.
Apart from the normal experiments conducted using the Kvasir-SEG and CVC-ClinicDB datasets, we also performed cross-validation tests to verify the generalization ability of the models, i.e. training on the Kvasir-SEG dataset and testing on the CVC-ClinicDB dataset and vice versa. The results of the generalization test are given in Tables 3 and 4. Table 3 shows the results of training on the Kvasir-SEG dataset and testing on the CVC-ClinicDB dataset, where CascadeMedSeg outperformed the other models, achieving an mDice value of 93.19%, mIoU of 87.57%, and accuracy 93.42%. Table 4 shows that trained on the CVC-ClinicDB dataset and tested on the Kvasir-SEG dataset, CascadeMedSeg achieved mDice of 88.67%, mIoU 82.56%, and a precision of 96.01%, again outperforming the other models. These results demonstrate the stronger generalization ability of the CascadeMedSeg. Due to its larger size, training on the Kvasir-SEG dataset resulted in a more robust model than training on the CVC-ClinicDB dataset.
4.3.2 Experimental results on the ISIC 2018 dataset
Table 5 shows the quantitative results of the ISIC-2018 dataset for the lesion boundary segmentation task. According to Table 5, CascadeMedSeg has an increase of 1.81% over DCSAU-Net in mIoU, and 3.03% over UNet++ in mDice. Among other metrics, our model achieved a highly competitive recall rate of 93.36% and an Precision of 92.20%.
To provide a more intuitive demonstration of the superiority of CascadeMedSeg, we also visualized the segmentation results of the proposed and other SOTA methods, as depicted in Fig. 5. It is evident that the proposed method can identify the location of the lesion more accurately, and its output is closer to the Ground Truth.
To more intuitively show the characteristics of different levels of features, we perform thermal visualization mapping on these feature maps in the ISIC 2018 dataset. To visualize the intermediate features in Fig. 6, the average of all channels in the feature map is calculated, and a heatmap is generated.
4.3.3 Experimental results on the BUSI dataset
We used the Breast Ultrasound Image dataset to compare the performance of CascadeMedSeg with that of the other SOTA networks. The results of the comparison between the models are shown in Table 6, and show that the mDice score of CascadeMedSeg was 75.19%, which was 4.06% higher than that of TransUNet; its mIoU score was 66.69%, i.e. 0.72% higher than that of DoubleU-Net. Overall, the proposed model exhibited the highest scores in most evaluation metrics, including Recall.
To provide a more intuitive demonstration of the superiority of the proposed method, the segmentation results of CascadeMedSeg and other SOTA methods are presented in Fig. 7. The results of CascadeMedSeg are shown in column 10, demonstrating more accurate tumor predictions with fewer incorrectly identified regions.
4.3.4 Experimental results on the ACDC dataset
The experimental results of CascadeMedSeg and other modeling methods on the ACDC dataset are shown in Table 7. It can be seen that CascadeMedSeg achieved an mDice of 91.49%, with the mDice organ subindices of RV, Myo and LVs of 89.96%, 88.82% and 95.70%, respectively. All values are obviously improved compared to the classical network models and the latest methods currently proposed for ACDC.
4.4 Ablation study
To evaluate the influence of each CascadeMedSeg module on its overall performance, we performed ablation experiments on the ACDC dataset. Specifically, we measured the impact of individually designed modules on the model performance by adding them to the baseline network in sequence. In this case, the pre-trained PVT v2-b2 model was used as the baseline model (PVT Encoder), on which the following three models were based: (1) CascadeMedSeg w/o EAF; (2) CascadeMedSeg w/o EES; and (3) CascadeMedSeg(Ours). The results of the ablation experiments are shown in Table 8.
As shown in Table 8, our implementation is compared with various structures. We attempted to verify their effectiveness by removing EAF or EES. Both EAF and EES are required for the model, and removing either one leads to a decrease in performance. The mDice of CascadeMedSeg w/o EAF improved by 0.86% compared to PVT Encoder, while the mDice of CascadeMedSeg w/o EES improved by 0.84%. The results demonstrate that the designed EAF and EES modules significantly enhance the network’s performance. Furthermore, compared to PVT Encoder, the mDice of CascadeMedSeg (Ours) increased by 1.63%. This demonstrates that integrating the proposed EAF and EES modules into the network improves its performance.
5 Conclusions
In this paper, a MIS method called CascadeMedSeg is proposed. This framework utilizes the PVT v2 backbone as an encoder to extract more powerful and robust features. In addition, it incorporates the novel EAF and EES modules, with the former efficiently integrating the encoder and decoder features and reducing the issue of information inconsistency. At the same time, the latter uses a dynamic boundary detection operator to extract the boundary mask and embed it in the decoder to enhance the image boundary features. The model is evaluated on five different MIS datasets and generalization tests are conducted. The results indicate that the CascadeMedSeg architecture outperforms other SOTA models and demonstrates high generalization performance. In the future, we will focus on optimizing the architecture of CascadeMedSeg to improve its performance and make it suitable for a wider range of MIS tasks.
Data availibility
All data, models, or code generated or used during the current study are available from the corresponding author on reasonable request.
References
Gu, R., Wang, G., Song, T., Huang, R., Aertsen, M., Deprest, J., Ourselin, S., Vercauteren, T., Zhang, S.: Ca-net: Comprehensive attention convolutional neural networks for explainable medical image segmentation. IEEE Transactions on Medical Imaging 40(2), 699–711 (2021). https://doi.org/10.1109/TMI.2020.3035253
Bhandary, S., Kuhn, D., Babaiee, Z., Fechter, T., Benndorf, M., Zamboglou, C., Grosu, A.-L., Grosu, R.: Investigation and benchmarking of u-nets on prostate segmentation tasks. Computerized Medical Imaging and Graphics 107, 102241 (2023). https://doi.org/10.1016/j.compmedimag.2023.102241
Lin, G., Chen, M., Tan, M., Chen, L., Chen, J.: A dual-stage transformer and mlp-based network for breast ultrasound image segmentation. Biocybernetics and Biomedical Engineering 43(4), 656–671 (2023). https://doi.org/10.1016/j.bbe.2023.09.001
Yu, Z., Lee, F., Chen, Q.: Hct-net: hybrid cnn-transformer model based on a neural architecture search network for medical image segmentation. Applied Intelligence 53(17), 19990–20006 (2023). https://doi.org/10.1007/s10489-023-04570-z
Wu, H., Zhang, Z., Zhang, Y., Sun, B., Zhang, X.: Acx-unet: a multi-scale lung parenchyma segmentation study with improved fusion of skip connection and circular cross-features extraction. Signal, Image and Video Processing 18(1), 525–533 (2024). https://doi.org/10.1007/s11760-023-02770-1
Alam, M.S., Wang, D., Liao, Q., Sowmya, A.: A multi-scale context aware attention model for medical image segmentation. IEEE Journal of Biomedical and Health Informatics 27(8), 3731–3739 (2022). https://doi.org/10.1109/JBHI.2022.3227540
Sinha, A., Dolz, J.: Multi-scale self-guided attention for medical image segmentation. IEEE Journal of Biomedical and Health Informatics 25(1), 121–130 (2020). https://doi.org/10.1109/JBHI.2020.2986926
Huang, R., Lin, M., Dou, H., Lin, Z., Ying, Q., Jia, X., Xu, W., Mei, Z., Yang, X., Dong, Y., et al.: Boundary-rendering network for breast lesion segmentation in ultrasound images. Medical Image Analysis 80, 102478 (2022). https://doi.org/10.1016/j.media.2022.102478
Jha, D., Riegler, M.A., Johansen, D., Halvorsen, P., Johansen, H.D.: Doubleu-net: A deep convolutional neural network for medical image segmentation. In: International Symposium on Computer-Based Medical Systems (CBMS), pp. 558–564 (2020). IEEE. https://doi.org/10.1109/CBMS49503.2020.00111
Rahman, M.M., Marculescu, R.: Medical image segmentation via cascaded attention decoding. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), pp. 6222–6231 (2023). https://doi.org/10.1109/WACV56688.2023.00616
Hemelings, R., Elen, B., Stalmans, I., Van Keer, K., De Boever, P., Blaschko, M.B.: Artery-vein segmentation in fundus images using a fully convolutional network. Computerized Medical Imaging and Graphics 76, 101636 (2019). https://doi.org/10.1016/j.compmedimag.2019.05.004
Ning, Z., Zhong, S., Feng, Q., Chen, W., Zhang, Y.: Smu-net: Saliency-guided morphology-aware u-net for breast lesion segmentation in ultrasound image. IEEE Transactions on Medical Imaging 41(2), 476–490 (2021). https://doi.org/10.1109/TMI.2021.3116087
Ronneberger, O., Fischer, P., Brox, T.: U-net: Convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention, pp. 234–241 (2015). Springer. https://doi.org/10.1007/978-3-319-24574-4_28
Zhou, Z., Rahman Siddiquee, M.M., Tajbakhsh, N., Liang, J.: Unet++: A nested u-net architecture for medical image segmentation. In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, pp. 3–11 (2018). Springer. https://doi.org/10.1007/978-3-030-00889-5_1
Xiao, X., Lian, S., Luo, Z., Li, S.: Weighted res-unet for high-quality retina vessel segmentation. In: International Conference on Information Technology in Medicine and Education (ITME), pp. 327–331 (2018). IEEE. https://doi.org/10.1109/ITME.2018.00080
Huang, H., Lin, L., Tong, R., Hu, H., Zhang, Q., Iwamoto, Y., Han, X., Chen, Y.-W., Wu, J.: Unet 3+: A full-scale connected unet for medical image segmentation. In: IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pp. 1055–1059 (2020). IEEE. https://doi.org/10.1109/icassp40776.2020.9053405
Oktay, O., Schlemper, J., Folgoc, L.L., Lee, M., Heinrich, M., Misawa, K., Mori, K., McDonagh, S., Hammerla, N.Y., Kainz, B., et al.: Attention u-net: Learning where to look for the pancreas. arXiv abs/1804. 03999 (2018)
Hu, J., Shen, L., Sun, G.: Squeeze-and-excitation networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 7132–7141 (2018)
Khan, S., Naseer, M., Hayat, M., Zamir, S.W., Khan, F.S., Shah, M.: Transformers in vision: A survey. ACM Computing Surveys (CSUR) 54(10s), 1–41 (2022). https://doi.org/10.1145/3505244
Wu, B., Xu, C., Dai, X., Wan, A., Zhang, P., Yan, Z., Tomizuka, M., Gonzalez, J., Keutzer, K., Vajda, P.: Visual transformers: Token-based image representation and processing for computer vision. arXiv abs:2006/03677 (2020)
Xie, J., Zhu, R., Wu, Z., Ouyang, J.: Ffunet: A novel feature fusion makes strong decoder for medical image segmentation. IET Signal Processing 16(5), 501–514 (2022). https://doi.org/10.1049/sil2.12114
Liu, Z., Lin, Y., Cao, Y., Hu, H., Wei, Y., Zhang, Z., Lin, S., Guo, B.: Swin transformer: Hierarchical vision transformer using shifted windows. In: Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), pp. 10012–10022 (2021). https://doi.org/10.1109/ICCV48922.2021.00986
Wang, W., Xie, E., Li, X., Fan, D.-P., Song, K., Liang, D., Lu, T., Luo, P., Shao, L.: Pyramid vision transformer: A versatile backbone for dense prediction without convolutions. In: Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), pp. 568–578 (2021). https://doi.org/10.1109/ICCV48922.2021.00061
Cheng, B., Misra, I., Schwing, A.G., Kirillov, A., Girdhar, R.: Masked-attention mask transformer for universal image segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 1290–1299 (2022). https://doi.org/10.1109/CVPR52688.2022.00135
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer, M., Heigold, G., Gelly, S., et al.: An image is worth 16x16 words: Transformers for image recognition at scale. arXiv abs/2010.11929. (2020)
Wang, W., Xie, E., Li, X., Fan, D.-P., Song, K., Liang, D., Lu, T., Luo, P., Shao, L.: Pvt v2: Improved baselines with pyramid vision transformer. Computational Visual Media 8(3), 415–424 (2022). https://doi.org/10.1007/s41095-022-0274-8
Wang, J., Huang, Q., Tang, F., Meng, J., Su, J., Song, S.: Stepwise feature fusion: Local guides global. In: Medical Image Computing and Computer Assisted Intervention, pp. 110–120 (2022). Springer. https://doi.org/10.1007/978-3-031-16437-8_11
Badrinarayanan, V., Kendall, A., Cipolla, R.: Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Transactions on Pattern Snalysis and Machine Intelligence 39(12), 2481–2495 (2017). https://doi.org/10.1109/TPAMI.2016.2644615
Howard, A.G., Zhu, M., Chen, B., Kalenichenko, D., Wang, W., Weyand, T., Andreetto, M., Adam, H.: Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv abs1704/04861 (2017)
Zhang, W., Fu, C., Zheng, Y., Zhang, F., Zhao, Y., Sham, C.-W.: Hsnet: A hybrid semantic network for polyp segmentation. Computers in Biology and Medicine 150, 106173 (2022). https://doi.org/10.1016/j.compbiomed.2022.106173
Khalifa, A.F., Badr, E.: Deep learning for image segmentation: A focus on medical imaging. Comput. Mater. Contin 75(1), 1995–2024 (2023). https://doi.org/10.32604/cmc.2023.035888
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 770–778 (2016)
Xu, Q., Ma, Z., Na, H., Duan, W.: Dcsau-net: A deeper and more compact split-attention u-net for medical image segmentation. Computers in Biology and Medicine 154, 106626 (2023). https://doi.org/10.1016/j.compbiomed.2023.106626
Chen, J., Lu, Y., Yu, Q., Luo, X., Adeli, E., Wang, Y., Lu, L., Yuille, A.L., Zhou, Y.: Transunet: Transformers make strong encoders for medical image segmentation. arXiv abs 2102/04306 (2021)
Cao, H., Wang, Y., Chen, J., Jiang, D., Zhang, X., Tian, Q., Wang, M.: Swin-unet: Unet-like pure transformer for medical image segmentation. In: European Conference on Computer Vision, pp. 205–218 (2022). Springer. https://doi.org/10.1007/978-3-031-25066-8_9
Wang, H., Cao, P., Wang, J., Zaiane, O.R.: Uctransnet: rethinking the skip connections in u-net from a channel-wise perspective with transformer. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 36, pp. 2441–2449 (2022). https://doi.org/10.1609/aaai.v36i3.20144
Yi, L., Dong, Z., Xiao, F., Yufan, C., Kwang-Ting, C., Hao, C.: Rethinking boundary detection in deep learning models for medical image segmentation. In: Information Processing in Medical Imaging., pp. 730–742 (2023). Springer. https://doi.org/10.1007/978-3-031-34048-2_56
Carion, N., Massa, F., Synnaeve, G., Usunier, N., Kirillov, A., Zagoruyko, S.: End-to-end object detection with transformers. In: European Conference on Computer Vision, pp. 213–229 (2020). Springer. https://doi.org/10.1007/978-3-030-58452-8_13
Touvron, H., Cord, M., Douze, M., Massa, F., Sablayrolles, A., Jégou, H.: Training data-efficient image transformers & distillation through attention. In: International Conference on Machine Learning, pp. 7358–7367 (2021)
Zhou, S., Nie, D., Adeli, E., Yin, J., Lian, J., Shen, D.: High-resolution encoder-decoder networks for low-contrast medical image segmentation. IEEE Transactions on Image Processing 29, 461–475 (2019). https://doi.org/10.1109/TIP.2019.2919937
Lin, T.-Y., Dollár, P., Girshick, R., He, K., Hariharan, B., Belongie, S.: Feature pyramid networks for object detection. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 2117–2125 (2017). https://doi.org/10.1109/CVPR.2017.106
Wang, Q., Wu, B., Zhu, P., Li, P., Zuo, W., Hu, Q.: Eca-net: Efficient channel attention for deep convolutional neural networks. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 11534–11542 (2020). https://doi.org/10.48550/arXiv.1910.03151
Lin, W., Wu, Z., Chen, J., Huang, J., Jin, L.: Scale-aware modulation meet transformer. In: Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), pp. 5992–6003 (2023). https://doi.org/10.1109/ICCV51070.2023.00553
Tang, F., Xu, Z., Huang, Q., Wang, J., Hou, X., Su, J., Liu, J.: Duat: Dual-aggregation transformer network for medical image segmentation. In: Chinese Conference on Pattern Recognition and Computer Vision (PRCV), pp. 343–356 (2023). Springer. https://doi.org/10.1007/978-981-99-8469-5_27
Jha, D., Smedsrud, P.H., Riegler, M.A., Halvorsen, P., De Lange, T., Johansen, D., Johansen, H.D.: Kvasir-seg: A segmented polyp dataset. In: MultiMedia Modeling: International Conference, pp. 451–462 (2020). Springer. https://doi.org/10.1007/978-3-030-37734-2_37
Bernal, J., Sánchez, F.J., Fernández-Esparrach, G., Gil, D., Rodríguez, C., Vilariño, F.: Wm-dova maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Computerized Medical Imaging and Graphics 43, 99–111 (2015). https://doi.org/10.1016/j.compmedimag.2015.02.007
Codella, N.C., Gutman, D., Celebi, M.E., Helba, B., Marchetti, M.A., Dusza, S.W., Kalloo, A., Liopyris, K., Mishra, N., Kittler, H., et al.: Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In: International Symposium on Biomedical Imaging, pp. 168–172 (2018). IEEE. https://doi.org/10.1109/ISBI.2018.8363547
Tschandl, P., Rosendahl, C., Kittler, H.: The ham10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Scientific Data 5(1), 1–9 (2018). https://doi.org/10.1038/sdata.2018.161
Al-Dhabyani, W., Gomaa, M., Khaled, H., Fahmy, A.: Dataset of breast ultrasound images. Data in Brief 28, 104863 (2020). https://doi.org/10.1016/j.dib.2019.104863
Bernard, O., Lalande, A., Zotti, C., Cervenansky, F., Yang, X., Heng, P.-A., Cetin, I., Lekadir, K., Camara, O., Ballester, M.A.G., et al.: Deep learning techniques for automatic mri cardiac multi-structures segmentation and diagnosis: is the problem solved? IEEE Transactions on Medical Imaging 37(11), 2514–2525 (2018). https://doi.org/10.1109/10.1109/TMI.2018.2837502
Wang, H., Xie, S., Lin, L., Iwamoto, Y., Han, X.-H., Chen, Y.-W., Tong, R.: Mixed transformer u-net for medical image segmentation. In: IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pp. 2390–2394 (2022). IEEE. https://doi.org/10.1109/ICASSP43922.2022.9746172
Funding
This work is supported by the in part by the National Natural Science Foundation of China (Grant No. 82100052), the Key Scientific and Technological Projects in Henan Province, China (Grant No. 232102311012) and the Cross Project of Medical and Engineering in Henan University(Grant No. YGJC2022002)
Author information
Authors and Affiliations
Contributions
Junwei Li contributed to the conception of the study and reviewed draft; Shengfeng Sun performed the experiment and written draft preparation; Shijie Li contributed significantly to methodology analysis; Ruixue Xia helped perform the analysis with constructive discussions.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no Conflict of interest to declare that are relevant to the content of this paper.
Ethical approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, J., Sun, S., Li, S. et al. CascadeMedSeg: integrating pyramid vision transformer with multi-scale fusion for precise medical image segmentation. SIViP (2024). https://doi.org/10.1007/s11760-024-03530-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11760-024-03530-5