Abstract
Early diagnosis of brain tumor using magnetic resonance imaging (MRI) is vital for timely medication and effective treatment. But, most people living in remote areas do not have access to medical experts and diagnosis facilities. Nevertheless, recent advancement in the Internet of Thing and artificial intelligence is transforming the healthcare system and has led to the development of the Internet of Medical Things (IoMT). An automated brain tumor classification system integrated with the IoMT framework can aid in remotely diagnosing brain tumors. However, the existing methods for brain tumor classification in MRI based on traditional machine learning and deep learning are compute-intensive. Deployment of these methods in the real-world clinical setup poses a serious challenge. Therefore, there is a requirement for robust and compute-efficient techniques for brain tumor classification. To this end, this paper presents a novel lightweight attention-guided convolutional neural network (AG-CNN) for brain tumor classification in magnetic resonance (MR) images. The designed architecture uses channel-attention blocks to focus on relevant regions of the image for tumor classification. Besides, AG-CNN uses skip connections via global-average pooling to fuse features from different stages. This approach helps the network extract enhanced features essential to differentiate tumor and normal brain MR images. To access the efficacy of the designed neural network, we evaluated it on four benchmark brain tumor MRI datasets. The comparison results with the existing state-of-the-art methods revealed the robustness and computational efficiency of the proposed AG-CNN model. The designed brain tumor classification pipeline can be easily deployed on a resource-constrained embedded platform and used in real-world clinical settings to quickly classify brain tumors in MR images.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Cancer results in one of the highest number of deaths across the globe. According to the world health organization (WHO) report, cancer was responsible for 10 million deaths in 2020 alone [1]. Of several tumors, the brain tumor is one of the most terrible diseases [2]. Therefore, early diagnosis and detection of brain tumors are extremely important. Manual detection and classification of brain tumors are challenging and prone to errors and thus require an expert radiologist to classify these tumors. Therefore, over the years, various machine learning and deep learning methods have been extensively studied and deployed to classify brain tumors from MRI automatically. Computer-aided diagnoses (CAD) of brain tumors using machine learning and deep learning methods have contributed immensely and may assist medical experts in detecting and classifying brain tumors. These techniques are highly reliable for their higher accuracy and consume less time.
One can define a brain tumor as an intracranial neoplasm within the brain or the central spinal canal caused due to the breakdown of the cell division control mechanism in a group of cells in the brain. Depending on their location, the brain tumors are classified broadly into meningioma, pituitary, and glioma. Meningioma occurs most often within the skull and outside the cell tissue and is mostly benign. Benign brain tumors can be fatal, and these tumors have a very low probability of turning into a malignant tumor-like meningioma [3]. The pituitary adenoma occurs at the pituitary’s location and is not malignant but can cause serious damage to the body’s control of hormones. It can lead to a lack of hormones in the body and vision loss. Glioma is a very common brain tumor in the supportive cells to the nerve cells, often known as the glial cells. It is one of the more common types of brain cancer and accounts for 30% of all primary tumors, and 80% of all malignant ones [4]. WHO divides gliomas into grades from 1 to 4, 1 being benign and 4 being malignant [5]. This classification system is one of the most accepted ones [6]. Grade 1 is the least dangerous tumor and can be cured, while grade 2 tumors tend to spread to areas other than their original location. Grade 3 and grade 4 tumors have varying appearances and need more sophisticated medical interventions to cure them.
Based on the learning scheme, the existing techniques for brain tumor classification can be classified broadly into handcrafted feature-based and deep learning-based methods. Handcrafted feature-based methods require extracting efficient handcrafted features by the experts [7]. These features, once extracted, are classified by traditional classifiers such as the support vector machine (SVM), the K-nearest neighbor (KNN), etc. Traditional machine learning-based brain tumor classification techniques have achieved satisfactory results, but they are time-consuming and require huge computational costs. On the other hand, the deep learning-based techniques using convolutional neural networks (CNNs) are end-to-end trainable and do not require handcrafted features. These methods are comparatively faster and more robust than the machine learning-based methods. Given the labeled images of normal and abnormal brain MR images, the CNNs can learn complex features and classify them directly without manual intervention. Such a scheme drastically reduces the time required to craft features and also helps to boost the accuracy of the model [8].
Training parameter-intensive deep CNN (DCNN) with high generalization requires large-scale brain tumor MRI datasets. However, the existing benchmark tumor MRI datasets are insufficient in size. Therefore, instead of training the CNNs from scratch, researchers have employed transfer learning techniques for brain tumor classification from small-scale MR images [9, 10]. Depending on their mode of operation, one can categorize the existing transfer learning-based scheme for brain tumor classification into feature extraction-based and fine-tuning-based methods. The former uses a pre-trained CNN such as DenseNet-121 initially trained on the large-scale ImageNet dataset to derive attributes from the brain MRI. The features thus obtained are classified using traditional machine learning classifiers such as the SVM, KNN, etc. [9]. The fine-tuning scheme, too, uses a seed CNN pre-trained on a large-scale source dataset. The scheme removes the softmax classifier layer from the off-the-self available seed model and appends a new softmax classifier to obtain the target CNN. In the next step, the framework transfers the learned parameters of the seed CNN to the target CNN. Finally, the framework fine-tunes the target CNN on the target brain MRI dataset [10]. Some existing works have also utilized the fine-tuned CNN as a feature extractor. These methods extract features from the CNN fine-tuned on the brain MRI dataset. Once extracted, these features are classified using the conventional machine-learning classifiers [2].
This work aims to classify MR images of brains into normal and tumor classes. To this end, we designed and implemented an attention-guided lightweight CNN named AG-CNN. We trained the designed AG-CNN from scratch on four benchmark MRI datasets. The designed CNN being lightweight is computationally efficient and requires less memory to load the model weights. We assessed the proposed CNN using five evaluation metrics: accuracy, precision, recall, specificity, and F1-score. The proposed model achieved competitive results on all the datasets with multi-fold improvement in the execution time due to fewer model parameters (floating-point operations) than the existing state-of-the-art models. In essence, the major contributions of this work are as follows:
-
Design and implement a novel lightweight attention-guided convolutional neural network (AG-CNN) for brain tumor classification in MR images. AG-CNN uses channel-attention blocks to focus on relevant regions of the MR images for tumor classification.
-
Extensive experimentation on four (BT-large-3c, BT-small-2c, BT-large-2c, and BT-large-4c) benchmark brain tumor MRI datasets. BT-small-2c and BT-large-2c consist of MR images of normal and tumor brain, BT-large-3c consists of MR images of meningioma, glioma, and pituitary brain tumor, and BT-large-4c consists of MR images of normal and three (meningioma, glioma, and pituitary) brain tumors.
-
Performance evaluation of the proposed AG-CNN model using precision, recall, specificity, and F1-score. Benchmarking the performance of the proposed brain tumor classification scheme against several state-of-the-art techniques.
-
The proposed scheme for brain tumor classification integrated with the IoMT framework may assist the medical practitioners and nursing staff in identifying the correct type of brain tumor in MR images and thus provide the patient with the best treatment.
In the subsequent sections of this paper, we briefly examine some related works on brain tumor classification, details of the existing publicly available brain tumor MRI datasets, proposed methodology, and performance evaluation outcomes. Section 2 reviews the existing state-of-the-art works on brain tumor classification in MR images. Section 3 describes the proposed methodology, including details of the image pre-processing step and neural network architecture. Details of the brain tumor MRI datasets, procedures used for training the proposed neural network, and the evaluation results of the model on four brain tumor MRI datasets form the content of Sect. 4. We provide results of comparing the designed CNN with the existing models and related discussions in Sect. 5. Finally, Sect. 6 concludes the paper with conclusive remarks.
2 Related work
There has been vast research done in machine learning and deep learning to develop methods for successfully classifying brain MR images into tumorous and normal. Convolutional neural networks (CNNs) have been used independently or in combination with other classification techniques to achieve high accuracy on various MRI datasets. The main hurdle is finding the right combination of CNN for feature extraction and classifiers to achieve maximum accuracy.
Researchers developed numerous techniques for brain tumor classification in the past several years using traditional machine learning. For instance, Jafari and Kasaei [11] presented a six-stage pipeline for automatic classification of brain MR images into normal, lesion benign, and malignant types. At first, the pipeline uses an image enhancement and restoration stage to enhance and restore the MR images. In the subsequent step, the pipeline partitions the image into meaningful regions using the seed region growing segmentation algorithm. In the third stage, the pipeline uses connected component labeling (CCL) to label each pixel according to its assigned component. The discrete wavelet transform (DWT) is employed in the fourth stage to derive useful attributes from the meaningful regions of the brain MRI. The principal component analysis (PCA) is used in the fifth stage to reduce the dimension of DWT features and obtain relevant features. The final stage of the pipeline employs the neural network classifier to classify the subjects as normal or abnormal (benign or malignant). In another related work, El-Dahshan et al. [12] introduced a three-stage hybrid technique to classify brain tumors in MR images. In the first stage, the pipeline employs DWT to derive features from the brain MR images. In the subsequent stage, the pipeline reduces the dimensions of the DWT features obtained in the previous step through PCA. The pipeline classifies the PCA-reduced features using the artificial neural network (ANN) and the K-nearest neighbor (K-NN) into normal or abnormal MR images in the third stage. Zacharaki et al. [13] also proposed a machine learning technique to classify and grade brain tumors in MR images. Their proposed pipeline has used shape, statistical characteristics of the tumor, the mean and variance of image intensities, and texture feature calculated using a bank of 40 Gabor filters with eight orientation and five frequency scales. Relevant features are selected using the forward selection method based on a ranking criterion from the high-dimensional feature vector. Subsequently, the important subset of features is selected using the SVM recursive feature elimination (SVM-RFE) feature selection algorithm from the reduced feature vectors. Finally, the framework classifies the selected features using the SVM classifier with the Gaussian kernel. Besides, the technique introduced by Saritha et al. [14] has employed wavelet entropy-based spider web plots for feature extraction and a probabilistic neural network to classify tumors in brain MR images. Ismael and Abdel-Qader [15] introduced a framework for brain tumor classification in MR images that uses a hybrid method comprising statistical methods for feature extraction and neural network algorithms. Initially, the framework uses DWT and Gabor filters to extract features from the brain tumor segments. The framework extracts and fuses statistical features derived from the DWT and Gabor convolved images in the subsequent step. Finally, the fused features are classified using the neural network classifier. The brain tumor classification scheme proposed by Mohsen et al. [16] has also used the DWT to extract features from the brain MR images and the PCA to lower the dimensions of the features. Once reduced, the PCA-reduced DWT features were classified using the deep neural network (DNN) into normal, glioblastoma, sarcoma, and metastatic bronchogenic carcinoma tumors. Ayadi et al. [17] introduced a hybrid feature extraction scheme using DWT and bag-of-words (BoW) to distinguish between normal and unhealthy brain MR images. Once extracted, the features were classified using different classifiers: SVM, KNN, random forest (RF), and AdaBoost. In their other work [18], the authors employed a hybrid of dense speeded-up robust features (DSURF) and histogram of oriented gradients (HoG) for feature extraction from the brain MR images. The features extracted from the brain MR images are classified into meningioma, pituitary, and glioma tumors, using the SVM classifier. The scheme proposed by Anjum et al. [19] has introduced a novel feature extractor named the reconstruction independent component analysis (RICA) for multi-class (pituitary, meningioma, and glioma) brain tumor detection in MR images. Once extracted from the brain MR images, the features were classified using the linear and nonlinear SVM and linear discriminant analysis (LDA) classifier. The computer-aided diagnosis (CAD) system for the classification of brain tumors in MRI proposed by Ghahfarrokhi and Khodadadi [20] has utilized chaos theory to estimate the complexity measures such as Lyapunov exponent (LE), approximate entropy (ApEn), and fractal dimension (FD). The framework has employed the gray-level co-occurrence matrix (GLCM) and DWT for feature extraction and the SVM, K-NN, and pattern net for feature classification. In their recent work, Amin et al. [21] analyzed the performance of the local binary pattern (LBP), and Gabor wavelet transform (GWT) feature extractor coupled with the SVM, K-NN, decision tree (DT), random forest (RF), and naïve Bayes (NB) classifier for the classification of brain tumors in MR images.
Due to their widespread popularity in numerous computer vision tasks, researchers developed several deep learning algorithms for brain tumor classification in MRI. In one of such works, Kokkalla et al. [22] introduced dense inception residual network for multi-class brain tumor classification. The network replaces the final classifier layer of Inception ResNet v2 CNN with a dense network for enhanced feature extraction and a softmax layer for the multi-class brain tumor classification. In another work, Ma and Zhang [23] introduced a novel lightweight neural network for brain tumor detection. The designed network achieved a better trade-off between accuracy and efficiency than the traditional parameter-intensive neural network. In one of the works [24], the authors introduced a convolutional auto-encoder neural network (CANN) to obtain and learn deep features to classify three brain tumor classes. Masood et al. [25] proposed a custom Mask Region-based convolution neural network (Mask RCNN) with DenseNet-41 backbone for automatic segmentation and classification of brain tumors in MR images, while Isunuri and Kakarla [26] proposed a seven-layer CNN to classify three-class brain MR images. Their designed CNN employed separable convolution to optimize computation time. Abiwinanda et al. [27] also introduced a lightweight CNN to detect three types of brain tumors, namely glioma, meningioma, and pituitary, in MRI. Besides, Kakarla et al. [28] proposed a lightweight eight-layer average-pooling CNN for three-class brain tumor classification. The designed architecture trained using the N-Adam optimizer with a sparse-categorical cross-entropy loss function takes lesser computation time and attains competitive performance. The brain tumor classification scheme introduced by Alhassan and Zainon [29] has used an image pre-processing step and a CNN with a hard swish-based rectified linear unit (ReLU) activation function to improve the robustness and the learning speed of the pipeline. Essentially, the pipeline first uses the normalization technique and histogram of oriented gradients (HoG) to enhance the raw MR images. Once available, the pipeline passes the HoG processed MR images as input to the designed CNN that classifies them into three tumor classes: meningiomas, gliomas, and pituitary. Sultan et al. [30] proposed a 16-layer CNN to classify different brain tumor types (meningioma, glioma, and pituitary) and grades (Grade II, Grade III, and Grade IV). A cross-channel normalization layer is used in the designed CNN to normalize the input layer by scaling and adjusting the related activations. In their recent work, Wozniak et al. [31] proposed a novel correlation learning mechanism (CLM) for deep neural network architectures that combines CNN with classic architecture. Their proposed CLM scheme helps the CNN obtain the right set of filters for pooling and convolution layers and helps the main neural network classifier converge faster with better accuracy. In [32], the authors proposed a highly generalizable two-channel deep neural network architecture for tumor classification. Besides, the neural network uses a novel pooling method to extract local features from convolution blocks of InceptionResNetV2 and Xception networks. Also, the attention mechanism in their proposed network helps it focus on tumor regions and thus better classify the type of tumor present in the images. The deep learning algorithm introduced by Kumar et al. [33] has used the ResNet-50 CNN and global average pooling to resolve the vanishing gradient and overfitting problems when training parameter-intensive CNN on a small-scale brain tumor MRI dataset. The techniques introduced by Irmak [34] have used a CNN for multi-classification of brain tumors for their early diagnosis. The authors proposed three different CNNs for brain tumor detection, brain tumor type (normal, glioma, meningioma, pituitary, and metastatic) classification, and tumor grade (Grade II, Grade III, and Grade IV) classification. Diaz-Pernas et al. [35] presented a multi-scale DCNN for automatic segmentation and classification of brain tumors in MRI. Inspired by the human visual system, the designed model processes the input image in three spatial scales along different processing pathways and classifies the MRIs into meningioma, glioma, and pituitary tumor. Liu et al. [36] introduced a novel neural network named global average pooling residual network (G-ResNet) to classify brain tumor images. The G-ResNet uses the ResNet34 CNN as the backbone classifier. However, instead of the flatten layer, the G-ResNet uses the global average pooling layer that reduces the model parameters and thus helps it mitigate over-fitting. Also, the G-ResNet concatenates features from the different layers of the backbone ResNet34 CNN to obtain enhanced features for better classification. Besides, to increase the penalty for misclassification, the framework used the sum of interval and cross-entropy loss to optimize the loss during G-ResNet training. The deep learning framework for automated classification of brain tumors from MRI proposed by Deepak and Ameer [37] has employed a lightweight CNN for feature extraction and the conventional support vector machine (SVM) for classification. Gu et al. [38] proposed a method for brain tumor classification in MR images using convolutional dictionary learning with local constraint (CDLLC). CDLLC integrates multilayer dictionary learning into a CNN structure to explore the discriminative information for enhanced tumor classification.
Training parameter-intensive CNN on small-scale brain tumor MRI datasets leads to over-fitting; therefore, researchers have extensively used deep learning transfer-learning for brain tumor classification in MRI. For instance, Panwar et al. [39] utilized the deep learning transfer learning scheme using ImageNet pre-trained AlexNet CNN to classify three types of brain tumors in MR images. Though the framework achieved competitive accuracy, the AlexNet CNN with 120 million (M) parameters and 120 megabytes (MB) of memory footprint is compute-intensive and thus restricts the use of the framework in real-world brain tumor diagnosis. The scheme introduced by Anjum et al. [40] has suggested using deep features extracted from the ImageNet pre-trained ResNet-101 CNN and the traditional machine learning classifiers such as KNN and SVM for the classification of the brain tumor in MR images. Tandel et al. [41] proposed an MRI-based noninvasive brain tumor grading method using deep learning and machine learning techniques. Their proposed framework used five CNNs, namely AlexNet, VGG16, ResNet18, GoogleNet, and ResNet50, for feature extraction from the brain MR images. The features once extracted were classified using five traditional machine learning classifiers, namely the support vector machine (SVM), K-nearest neighbors (K-NN), naïve Bayes (NB), decision tree (DT), and linear discriminant analysis (LDA) using fivefold cross-validation. In one of the studies, Polat and Gungen [42] also proposed a solution to classify brain tumors in MR images using transfer learning. The brain tumor classification scheme introduced by Lu et al. [43] has employed transfer-learning-based feature extraction using ImageNet pre-trained MobileNetV2 and three feed-forward network classifiers, namely the extreme learning machine (ELM), Schmidt neural network, and random vector functional-link net for classification. The framework utilized the chaotic bat algorithm to optimize the weights and biases of the three feed-forward network classifiers and boost their recognition accuracy. Noreen et al. [44] employed the deep learning fine-tuning scheme using the ImageNet pre-trained Inception-v3 and DenseNet201 CNN for three-class brain tumor classification. The brain tumor classification method introduced by Kaur and Gandhi [45] has utilized the transfer-learning scheme using ImageNet pre-trained DCNNs, namely AlexNet, Resnet50, GoogLeNet, VGG-16, Resnet101, VGG-19, Inceptionv3, and InceptionResNetV2. Khan et al. [46] introduced an automated multi-modal scheme using deep learning for brain tumor type classification. The framework initially uses edge-based histogram equalization and discrete cosine transform (DCT) techniques to enhance MR images. In the subsequent step, the framework utilized the transfer-learning-based feature extraction scheme using the ImageNet pre-trained VGG16 and VGG19 CNN. The framework selects relevant features from the high-dimensional deep features using the correntropy-based joint learning approach with the extreme learning machine (ELM). Finally, the selected features are classified using the ELM classifier. In another work, Kang et al. [9] also adopted the transfer-learning-based feature extraction scheme to extract features from the brain MR images using several ImageNet pre-trained deep CNNs. Various machine learning classifiers then evaluate the extracted deep features. Once evaluated, the framework concatenates the deep features from the top three CNNs and feeds the features to several machine learning classifiers. Mehrotra et al. [47] investigated the transfer-learning-based fine-tuning scheme using five ImageNet pre-trained CNN, namely AlexNet, GoogLeNet, ResNet50, ResNet101, and SqueezeNet, to classify brain tumors in MR images into malignant and benign. The brain tumor classification pipeline introduced by Pashaei et al. [48] has utilized deep features extracted from the MR images using CNN and a kernel ELM (KELM) to classify the deep features into meningioma, glioma, and pituitary tumor in T1-weighted contrast-enhanced MRI (CE-MRI) images. Deepak and Ameer [10] employed the deep learning transfer learning-based scheme using ImageNet pre-trained GoogLeNet for brain tumor classification in MR images. The proposed scheme prunes the softmax classifier layer and appends a new softmax classifier layer. It subsequently transfers the learned parameters of pre-trained GoogLeNet to the target CNN and fine-tunes the target CNN on the target brain MRI dataset. Sekhar et al. [2] introduced a brain tumor classification scheme that employed fine-tuned GoogLeNet features classified using the SVM and the K-NN classifier.
Numerous works also exist in the literature that has utilized advanced neural networks and optimization techniques for brain tumor classification in MRI. For instance, Anaraki et al. [49] combined CNNs and the genetic algorithm (GA) to classify different grades of Glioma using MRI. Unlike existing trial and error-based methods of selecting a deep neural network architecture, the GA in their proposed framework automatically identifies the optimal CNN architecture. Rammurthy and Mahesh [50] proposed the Whale Harris Hawks optimization (WHHO) for brain tumor detection using MRI. Huang et al. [51] introduced an innovative differential feature map (DFM) block that magnifies tumor regions. Besides, DFM blocks combined with squeeze-and-excitation (SE) blocks form a differential feature neural network (DFNN). In the initial step, the proposed framework applies an automatic image rectification method to align the symmetric axes of the brain MR images parallel to the perpendicular axis. In the subsequent step, the framework classifies the aligned brain MR images using DFNN into abnormal and normal classes. Sert et al. [52] employed an image super-resolution algorithm first to increase the resolution of the MR images. The maximum fuzzy entropy segmentation (MFES) is subsequently employed to segment the brain tumor from the MR image. Finally, the framework utilized the ImageNet pre-trained ResNet CNN for feature extraction and the SVM classifier to classify the MR images into benign and malignant brain tumors. Abd-Ellah et al. [53] proposed a novel two-phase multi-model automatic diagnosis system for brain tumor detection and localization. In the first phase, the system classifies the brain MRIs into normal and abnormal classes using deep features extracted using a CNN and the error-correcting output codes support vector machine (ECOC-SVM) classifier. While in the second phase, the system localizes the tumor within the abnormal MRIs using a fully designed five-layer region-based CNN (R-CNN). Finally, compared to the conventional CNN, Afshar et al. [54] adopted the capsule network (CapsNets) for brain tumor classification.
3 Proposed methodology
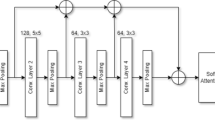
We aim to design a CNN that can automatically classify a brain MR image either into two classes (normal and tumor), three classes (meningioma, glioma, and pituitary tumor), or four classes (normal, meningioma, glioma, and pituitary tumor). The classification of MR images into normal or tumor class comes under binary classification tasks. In contrast, classifying brain MR images into three tumor classes or normal & three tumor classes are a multi-class classification task. The training pipeline of the proposed framework for brain MR image classification starts with pre-processing. From the original brain MR images, as shown in Fig. 1, the pre-processing operation crops the relevant image pixels removing the redundant black pixels. In the subsequent step, the cropped image is resized to 128 \(\times\) 128 \(\times\) 1-pixels dimension and passed to the proposed AG-CNN model. The proposed AG-CNN consists of multiple convolutional and max-pooling layers. These layers help the model extract discriminative features from the input MR images. Besides, the channel-attention blocks in the proposed AG-CNN help the network focus only on the relevant area of the brain for better classification. Also, in addition to the traditional CNN, the proposed AG-CNN uses high-level feature concatenation via skip-connections using the global average pooling (GAP) layer. High-level feature concatenation helps the network learn enhanced features leading to better classification of different brain tumors and normal brain MR images. The fully connected layers in the design CNN transform the concatenated features into high-level features using the nonlinear rectified linear unit (ReLU) activation function. Finally, the designed CNN passes the transformed features through a 3-way softmax classifier, classifying the input MR image into meningioma, glioma, and pituitary tumor classes. It is worth mentioning that in the case of the binary classification task (tumor or normal), the network uses a 2-way softmax classifier, and in the case of the four-class multi-classification task (meningioma tumor, glioma tumor, pituitary tumor, and normal), the network uses 4-way softmax classifier after the fully connected layer. Once trained, the designed AG-CNN can make predictions on the unseen brain MR images. The subsequent sections provide details of various steps employed in the proposed scheme for brain tumor classification.
3.1 Image pre-processing
As shown in Fig. 1, the raw brain MR image contains undesired spaces and areas that lead to poor classification performance of the CNN trained on these images. Thus, removing the unwanted regions and cropping the images becomes essential for retaining only useful information. In this work, we used the extreme point calculation scheme initially introduced by Kang et al. [9] to segment the useful areas from the original brain MR images. Figure 2 shows the sequence of steps to crop the MR images using the extreme point calculation method. Initially, on the original brain MR images, we apply Gaussian smoothing using filters of kernel size 7 \(\times\) 7 to eliminate the noisy pixels from the input image. Next, the smoothed MR image is passed through the Otsu threshold operation to convert them into binary images. The subsequent step of pre-processing detects the outer edge contours. Using the largest contour, we calculate the four extreme points (extreme top, extreme bottom, extreme right, and extreme left) of the binary image. Finally, we crop the images based on the four points. The cropped MR image, as shown in Fig. 2 contains only useful regions, removing the redundant black regions in the original image. It is important to note that before passing the cropped images to the AG-CNN model, they are resized to 128 \(\times\) 128 \(\times\) 1-pixels.
Once cropped and resized, the brain MR images are standard normalized before being utilized for training the proposed AG-CNN model. The data normalization process follows a three-step procedure, wherein the first step divides the images by 255. This step brings the pixel intensities in the range of 0 to 1. Subsequently, the pre-processing step normalizes the scaled images by subtracting the pixel intensities from the mean pixel intensities computed on the complete dataset. The step eliminates individual image differences and facilitates better network convergence during training. Once mean-normalized, we divide the pixel intensities of the mean-normalized images by the standard deviation computed on the complete dataset. Furthermore, we used different on-the-fly data augmentation strategies (horizontal flip, random rotation, zooming, shearing, and horizontal and vertical shift) to overcome overfitting limitations when training the CNN on small-scale brain MRI datasets.
3.2 Neural network architecture
Figure 3 shows the architectural details of the proposed lightweight AG-CNN model. The proposed model consists of seven convolutional layers (Conv_1, Conv_2, Conv_3, Conv_4, Conv_5, Conv_6, and Conv_7), four max-pooling layers (MP_1, MP_2, MP_3, and MP_4), two channel-attention blocks (CAB_1 and CAB_2), two global-average pooling layers (GAP_1 and GAP_2), followed by an additional layer, fully connected layer, and a softmax classifier layer at the end. From the brain tumor MR images, the convolutional layer in the initial and the later stages of the designed CNN hierarchically extracts low and high-level details, respectively. On the other hand, the max-pooling layers in the designed model reduce the spatial dimensions of the feature maps obtained from the convolutional layers. The channel-attention blocks perform the selection of relevant feature maps. Finally, the feature vector addition obtained by skip connections via GAP enhances the feature extraction capability of the network. The softmax classifier subsequently classifies the aggregated one-dimensional enhanced feature vector into three tumorous classes. Table 1 depicts the configuration details of the designed AG-CNN model. The designed lightweight model has 1.31 million parameters and thus can be deployed on low-cost portable embedded platforms.
The first convolutional layer (Conv_1) has utilized 32 filters of kernel size 7 \(\times\) 7. We initialized the filters with weights computed using 32 real Gabor filters of kernel size 7 \(\times\) 7. Intuitively, a 2D Gabor filter is a Gaussian wave modulated sinusoidal signal of a particular frequency and orientation [55], and defined as:
where
and
Equation (1) can further decomposed into real and imaginary components, expressed by Eqs. (4) and (5), respectively.
We used filter coefficients of 32 real Gabor filters of kernel size 7 \(\times\) 7 generated using Eq. (4) by varying the hyperparameters. The convolutional layer (Conv_2), on the other hand, has utilized He_uniform initialized 96 filters of kernel size 5 \(\times\) 5. The remaining five convolutional layers viz. Conv_3, Conv_4, Conv_5, Conv_6, and Conv_7 have utilized He_uniform initialized filters of kernel size 3 \(\times\) 3. After each convolutional layer, the combination of convolutional and max-pooling layer, and the fully connected layer, the proposed AG-CNN has utilized batch-normalization to accelerate training and faster convergence.
The convolutional and fully connected layers in the designed AG-CNN model utilized the rectified linear unit (ReLU) activation function for the nonlinear transformation of the features. In contrast to other activation functions, ReLU is compute-efficient and does not require any sophisticated mathematical operations. Besides, the designed model has used a dropout regularization layer with a probability of 0.2 after each batch-normalization layer. The dropout regularization helps the model learn more robust features by dropping out arbitrary neurons.
The feature maps in the traditional CNNs have three dimensions: channel, height, and width. Intuitively, many channels in the feature map lead to information redundancy. Redundant channels in the feature maps may impact the performance of CNNs. Therefore, the designed AG-CNN has used the channel attention block (CAB). The CAB generates a channel attention matrix that highlights the inter-channel relationships among the channels of the feature maps and assigns higher weights to the important channels and vice-versa. Assigning higher weights to significant channels helps the AG-CNN model focus more on the selected channels of the feature maps.
In the literature, numerous techniques exist to compute the channel attention efficiently. For channel computation, most existing works use pooling strategies that squeeze the spatial dimension of the input feature. For instance, the CAB proposed by Hu et al. [56] has used global average pooling, while the technique introduced by Zhou et al. [57] has suggested using the average pooling. Also, the CAB proposed by Ling et al. [58] has used both global average pooling and global max-pooling layers to squeeze the spatial dimensions of the input feature maps parallelly. Besides, to directly learn the attention masks, the techniques suggested by Hu et al. [56] and Woo et al. [59] have used the fully connected layer with the sigmoid activation function after pooling layers. Such operations do not consider the inter-channel relationship and thus may not be useful for brain tumor classification. Therefore, following the strategy suggested by Ling et al. [58], this work has adopted a cross-matrix multiplication to obtain the channel relationship matrix for the attention mask, as shown in Fig. 4.
Given an input feature map \(F_{I}\) \(\in\) \(R^{C \times H \times W}\), the CAB, as shown in Fig. 4, first compresses the feature map along the spatial axis simultaneously using both global-average pooling (GAP) and global-max pooling (GMP) to get C \(\times\) 1 \(\times\) 1-dimensional feature vector \(F^{C}_{\mathrm{avg}}\) and \(F^{C}_{\mathrm{max}}\), respectively. The feature vectors \(F^{C}_{\mathrm{avg}}\) and \(F^{C}_{\mathrm{max}}\) are subsequently passed through a convolutional layer with kernel size 1 \(\times\) 1 and ReLU activation, followed by batch-normalization (BN), and another convolutional layer with kernel size 1 \(\times\) 1 and ReLU activation to obtain the intermediate feature maps \(F^{C}_{\mathrm{conv}\_\mathrm{avg}}\) and \(F^{C}_{\mathrm{conv}\_\mathrm{max}}\), respectively. Finally, we summed up the feature maps \(F^{C}_{\mathrm{conv}\_\mathrm{avg}}\) and \(F^{C}_{\mathrm{conv}\_\mathrm{max}}\) element-wise to obtain the aggregated feature \(F^{C}_{\mathrm{sum}}\), mathematically expressed as:
where \(F^{C}_{\mathrm{sum}}\) \(\in\) \(R^{C \times 1 \times 1}\) and \(\oplus\) means element-wise summation. After that, we first reshape and subsequently transpose the aggregated feature vector \(F^{C}_{\mathrm{sum}}\) to get another feature vector with dimension 1 \(\times\) 1 \(\times\) C. The aggregated feature vector and its transposed is then multiplied and passed through the softmax activation function to get the channel attention matrix \(A_{C}\) as mathematically expressed in Eq. (7).
In Eq. (7), \(A_{C}\) \(\in\) \(R^{C \times C}\) is a two-dimensional matrix representing the inter-channel relationship among the channels of the input feature maps and \(\otimes\) means matrix multiplication. Once the channel attention matrix is available, we multiplied it first with the input feature map \(F_{I}\). In the subsequent step, the multiplied feature map is reshaped and added to the input feature map to get the channel-refined feature map \(F_{C}\) \(\in\) \(R^{C \times H \times W}\), as mathematically expressed below:
where \(\oplus\) means element-wise summation and \(\otimes\) means matrix multiplication.
4 Experiments and results
In this section, we report the experimental analysis results of the proposed framework on four brain tumor MRI datasets. At first, we provide details of the four benchmark brain tumor datasets, namely BT-large-3c, BT-small-2c, BT-large-2c, and BT-large-4c. The section also provides strategies for training the neural network and details of different performance evaluation metrics.
4.1 Dataset details
Brain tumor datasets consisting of MR images are not as common. Besides, the number of images is less for successfully training and testing deep learning models for multi-class brain tumor classification. The experiments in this work consider four different types of brain tumor MRI datasets. These datasets are open source, and several researchers have utilized them to validate the efficiency of the brain tumor classification pipeline. For ease in referencing, we have named the datasets BT-large-3c, BT-small-2c, BT-large-2c, and BT-large-4c.
In the BT-large-3c dataset, there are 3064 T1-weighted contrast-enhanced (CE) MRI slices of 233 different patients [60]. The imbalanced dataset contains 708, 930, and 1426 tumor images of meningioma, pituitary, and glioma, respectively, in .mat file format. Also, the dimension of the images in the dataset is 512 \(\times\) 512, and the pixel size is 49mm \(\times\) 49mm. Table 2 provides details of the dataset, and Fig. 5 shows sample MR images of different tumor classes from the dataset. We followed the standard fivefold cross-validation testing protocol to validate the proposed model on the BT-large-3c dataset for a fair comparison with the existing works in the literature.
The BT-small-2c dataset contains 253 MR images and comes pre-divided into two categories: tumor and normal MR images [61]. There are 155 tumor MR images in the dataset, while the remaining 98 are normal MR images of the brain. Following the procedure adopted by Kang et al. [9], we randomly divided the dataset into an 80:20 train-test ratio. We trained the model with 202 MR images and tested it on 51 MR images. Like BT-small-2c, the BT-large-2c dataset contains MR images of normal and tumorous brain [62]. There are 3000 images in the dataset containing 1500 images with tumors and the remaining 1500 images without tumors. To evaluate the proposed model’s performance on the BT-large-2c dataset, we divided it into 80:20 ratio, where 80% of the total dataset (2400 MR images) were used for training and the remaining 20% of the total dataset (600 MR images) to test the model. In contrast to the BT-small-2c and BT-large-2c, the BT-large-4c dataset contains MR images of normal and three different types of brain tumors (meningioma, pituitary, and glioma) [63]. The dataset contains 3264 T1-weighted MR images. Like BT-large-2c, to evaluate the proposed CNN, we divided the BT-large-4c dataset into a training set containing 80% of the total images and a test set containing 20% of the total images in the dataset. Table 3 presents the details of the BT-small-2c, BT-large-2c, and BT-large-4c datasets. Figures 6, 7, and 8 show samples images from the BT-large-4c, BT-small-2c, and BT-large-2c dataset, respectively.
4.2 Neural network training
This work aims to classify brain MR images into normal and tumor classes. To this end, on the MR images, we first apply an input-pre-processing step to crop the region of interest (ROI). The input to the designed AG-CNN model is cropped MR image resized to 128 \(\times\) 128 \(\times\) 1-pixels. Given the input images, the model automatically extracts essential attributes that can discriminate between normal and tumorous brain MR images during training. Besides, the CAB in the designed network helps the AG-CNN model select the most suitable filters to focus on the important spatial regions in the brain MR images.
We used the Keras functional API with the TensorFlow backend to design and compile the proposed neural network. The model is trained using the Adam optimizer with the categorical cross-entropy loss function for network parameter optimization. We started network training with an initial learning rate of 0.001 and subsequently reduced the learning rate by a factor of 0.5 using the step-decay learning rate scheduler after every 25 epochs. On each of the four brain tumor MRI datasets, the model is trained for 200 epochs using training MR images in batches of 16. The model takes around 38 seconds to complete one epoch and thus confirms the structural simplicity and compute-efficiency of the designed AG-CNN model. Model training and testing have been carried out on the Ubuntu desktop machine with Nvidia GeForce GTX 1080Ti GPU with 3584 CUDA cores and 11GB GDDR5 VRAM.
4.3 Performance evaluation
We evaluated the proposed AG-CNN model using five performance metrics: classification accuracy, precision, recall, specificity, and F1-score. The expressions for calculating the metrics are as follows:
In Eqs. (9)–(13), TP stands for the number of true positive samples while FP is the number of false-positive samples in the confusion matrix. TN represents the number of true negatives, and FN stands for false-negative samples in the confusion matrix.
4.3.1 Performance evaluation on the BT-large-3c dataset
On the BT-large-3c dataset, following the fivefold cross-validation scheme, the AG-CNN model achieved a mean recognition accuracy of 97.23%. Besides, the classification report of the AG-CNN model as summarized in Table 4 shows that the model attained an average precision, recall, specificity, and F1-score of 97.07%, 96.95%, 98.57%, and 97.00%, respectively. Figure 9 shows the normalized confusion matrix of the proposed AG-CNN model on the BT-large-3c dataset. The trained AG-CNN model correctly classified most of the glioma (98% accuracy) and pituitary (98% accuracy) brain tumor MR images. However, the model correctly classified only 95% of the samples from the meningioma tumor. Out of the 5% misclassified brain MR image samples from the meningioma tumor, the model classified 2% samples into the glioma tumor and the remaining 3% into the pituitary tumor. Low classification accuracy for the meningioma class may be due to class imbalance in the dataset as it contains 1426, 930, and 708 brain MR image samples from the glioma, pituitary, and meningioma tumor. The AG-CNN model trained on the BT-large-3c dataset tends to be more biased towards the majority glioma class, causing bad classification of the minority meningioma class. Finally, the receiver operating characteristic (ROC) curve shown in Fig. 10 shows that on the BT-large-3c dataset, the trained AG-CNN model attained an AUC (Area under the ROC curve) of 0.996, 0.996, and 0.995 corresponding to meningioma, glioma, and pituitary brain tumor class, respectively.
4.3.2 Performance evaluation on the BT-large-4c dataset
On the BT-large-4c dataset, following the 80:20 train-test validation scheme, the AG-CNN model achieved a test recognition accuracy of 95.71%. Table 5 summarizes the classification report of the proposed AG-CNN model on the BT-large-4c brain tumor MRI dataset. On the 20% of the randomly selected test MR images from the dataset, the trained AG-CNN model achieved an average precision, recall, specificity, and F1-score of 95.77%, 96.33%, 98.52%, and 95.98%, respectively. Upon closely examining the confusion matrix results of the AG-CNN model on the BT-large-4c dataset, as shown in Fig. 11, one can find that the proposed model correctly classified all the sample brain tumor MR images from the pituitary class. Also, the model correctly classified 99%, 95%, and 91% of the brain tumor images from the normal, meningioma, and glioma classes, respectively. Out of 5% misclassified brain tumor MR images from the meningioma class, the model classified 4% samples into the pituitary class and the remaining 1% samples into the glioma class. Besides, out of the 9% misclassified samples from the glioma class, the trained AG-CNN model classified 5% images into meningioma, 2% images into pituitary, and the remaining 1% into the normal class. Finally, the receiver operating characteristic (ROC) curve shown in Fig. 12 shows that on the BT-large-4c dataset, the trained AG-CNN model attained a very good AUC (Area under the ROC curve) of 0.990, 0.959, 0.991, and 0.993 corresponding to meningioma, glioma, pituitary, and normal brain MR image class, respectively.
4.3.3 Performance evaluation on the BT-small-2c dataset
We performed the next set of experiments on the BT-small-2c dataset. For a fair comparison, we followed the evaluation procedure adopted by Kang et al. [9] and trained and tested the AG-CNN model on an 80:20 train-test split. On the randomly selected 20% test brain MR images from the BT-small-2c dataset, the proposed AG-CNN model achieved a recognition accuracy of 96.08%. Besides, the classification report of the AG-CNN model as summarized in Table 6 shows that the trained CNN model attained an average precision, recall, specificity, and F1-score of 95.89%, 95.89%, 95.89%, and 95.89%, respectively. Figure 13 shows the normalized confusion matrix of the proposed AG-CNN model on the BT-small-2c dataset. A closer look at the results of Fig. 13 shows that the trained model correctly classified 97% of the brain tumor MR images while misclassifying the remaining 3% samples into the normal class. Besides, the model classified 95% of the normal brain MR images into the normal class and the remaining 5% into the tumor class. Also, on the BT-small-2c dataset, the receiver operating characteristic (ROC) curve shown in Fig. 14 shows that the trained AG-CNN model attained an AUC (Area under the ROC curve) of 0.969.
4.3.4 Performance evaluation on the BT-large-2c dataset
We performed the final set of experiments on the BT-large-2c dataset. For training and testing the proposed model on this dataset, we followed the evaluation procedure adopted by Kang et al. [9] and trained and tested the AG-CNN model on an 80:20 train-test split. On the randomly selected 20% test brain MR images from the BT-large-2c dataset, the proposed AG-CNN model achieved a recognition accuracy of 99.83%. Besides, the classification report of the AG-CNN model as summarized in Table 7 shows that the trained CNN attained an average precision, recall, specificity, and F1-score of 99.84%. Figure 15 shows the normalized confusion matrix of the proposed AG-CNN model on the BT-large-2c dataset. The trained model correctly classified all the brain MR images from the tumor class and misclassified one MR image from the normal class into the tumor class. The classification results reflect the high generalization ability of the proposed AG-CNN model. The good performance of the trained AG-CNN model on the BT-large-2c dataset is also confirmed by the shape of the ROC curve (in Fig. 16), with an area under the curve (AUC) equal to 1.00.
5 Comparison with state-of-the-art methods and discussions
We compared the performance of the proposed method with several existing state-of-the-art methods for brain tumor classification on all four MRI datasets, namely BT-large-3c, BT-large-4c, BT-small-2c, and BT-large-2c. Table 8 reports the comparison results of the proposed AG-CNN model with several state-of-the-art deep learning models on the BT-large-3c dataset. Among the state-of-the-art models listed in Table 8, the deep learning technique introduced by Sekhar et al. [2] using the fine-tuned GoogLeNet CNN from the MR images and classified using the K-NN classifier attained the highest mean fivefold cross-validation accuracy of 98.30%. Also, the fine-tuned GoogLeNet attained a fivefold cross-validation accuracy of 94.90%, while the deep features classified using the SVM classifier achieved a mean classification score of 97.60%. Meanwhile, the lightweight CNN introduced by Deepak and Ameer [37] has attained a mean fivefold cross-validation accuracy of 94.26% when trained from scratch. In contrast, the deep features extracted from the pre-trained CNN and classified using the SVM classifier achieved a mean fivefold cross-validation accuracy of 95.82%. The brain tumor classification scheme introduced by Afshar et al. [54] has achieved mean recognition accuracy of 90.89% using the CapsNet neural network. Also, the lightweight eight-layer average-pooling CNN introduced by Kakarla et al. [28] has attained a competitive recognition accuracy of 97.42%. Besides, the deep learning scheme using ResNet-50 CNN and global average pooling introduced by Kumar et al. [33] has attained a mean fivefold cross-validation accuracy of 97.48% on the small-scale BT-large-3c brain tumor MRI dataset. Nevertheless, the proposed AG-CNN model with a mean fivefold cross-validation accuracy of 97.23% also achieved competitive performance. Also, compared to the existing models, the proposed AG-CNN is lightweight and can be deployed for the real-time classification of brain tumors in MR images on low-cost embedded devices.
Table 9 details the results of comparing the accuracy of the AG-CNN model with the related works in the literature on the BT-large-4c dataset. On this dataset, the ensemble of DenseNet-169 + MnasNet CNN features classified using the SVM classifier with radial basis function (RBF) kernel has reported a test recognition accuracy of 93.72% [9]. However, on the BT-large-4c dataset, the proposed AG-CNN model achieved a state-of-the-art test recognition accuracy of 95.71%. The results demonstrate the usefulness of the proposed AG-CNN model trained from scratch compared to the existing transfer learning scheme. Besides the technique using deep features extracted from the ensemble of several parameter-intensive CNN classified using the softmax and the traditional machine learning classifiers such as Gaussian naïve Bayes (NB), AdaBoost, K-NN, RF, ELM, and SVM with linear, sigmoid, and RBF kernel requires high classification time than the proposed lightweight AG-CNN model. The overall classification time of the proposed brain tumor classification pipeline is comparatively lesser than the existing method.
We also compared the performance of the proposed AG-CNN model with several brain tumor classification schemes on the BT-small-2c dataset. On the BT-small-2c dataset, as illustrated in Table 10, the deep features extracted from the brain MRI images using the ImageNet pre-trained DenseNet-169 CNN and classified using the SVM with RBF kernel achieved the highest test recognition accuracy of 98.04% [9]. Besides, the fine-tuned DenseNet-169 achieved a test recognition accuracy of 96.08%. On this dataset, the proposed AG-CNN model, too, achieved a test recognition accuracy of 96.08%. A reduction in the recognition accuracy of the proposed model on the BT-small-2c dataset may be due to its small size. The dataset has just 253 brain MR images, and thus, the model might have over-fitted the dataset when trained from scratch.
Analyzing the comparison results of Table 11, one can find that on the BT-large-2c brain MRI dataset, the designed AG-CNN model with a test recognition accuracy of 99.83% sets the new state-of-the-art. The previous best technique using fine-tuned ensembled of DenseNet-121 + ResNeXt-101 + MnasNet introduced by Kang et al. [9] has attained a test recognition accuracy of 98.83%. Besides, the deep features extracted using the ensemble of DenseNet-121 and MnasNet and classified using the K-NN and SVM classifier with the linear, sigmoid, and RBF kernel achieved a test recognition accuracy of 98.50%. Also, deep features extracted from the brain tumor MR images using the ImageNet pre-trained DenseNet-121, ResNeXt-101, and MnasNet CNN and classified using the extreme learning machine (ELM) classifier achieved a test recognition accuracy of 98.67%. The results demonstrate the advantage of the AG-CNN model over the deep-learning-based feature extraction scheme.
The designed lightweight attention-guided CNN (AG-CNN) presented in this work is more efficient in recognition accuracy and computational efficiency than the other related methods available in the literature. The channel-attention block and high-level feature concatenation via global average pooling (GAP) helped the designed AG-CNN model achieve competitive performance on the BT-large-3c and BT-small-2c brain tumor MRI dataset. On the BT-large-4c and BT-large-2c brain MRI datasets, the proposed AG-CNN model sets the new state-of-the-art accuracy. Also, the designed model, compared to the popular CNN such as GoogLeNet, DenseNet-169, AlexNet, VGG-16, etc., is compute-efficient and requires a low memory footprint. One can easily deploy the designed model on a low-cost portable embedded platform for the real-time classification of brain tumors in MR images.
In summary, the CNNs have demonstrated excellent performance in the brain tumor classification in MR images. There are still several problems encountered by CNNs, such as the loss of spatial information due to max-pooling operations in the CNNs and poor generalization when used with input data from different machines (different vendors, models, etc.). The use of max-pooling layers in CNN following the convolution layer progressively reduces the spatial dimensions of the feature maps as only the most active neurons are chosen to move to the next layer. Reduction in the spatial dimensions of the feature maps reduces the number of parameters in the network and consequently reduces the network’s computational complexity and memory requirements. Additionally, the max-pooling operation may also help to reduce over-fitting and can help CNN to learn invariant features. However, for medical images encoding spatial information is necessary. Therefore, there is a requirement for an efficient pooling scheme that can capture spatial information in medical images. Over the years, researchers have developed several pooling methods for different approaches [64]. Among them, the spatial pyramid pooling and its variants have proved to capture spatial or structural information in images. Thus, one can use these pooling methods with CNN in the medical image classification task. Alternatively, one can use advanced neural networks such as CapsNet, which uses routing-by-agreement to retain valuable spatial information [65]. The CapsNet have shown promising results in several medical image classification task [54, 66].
6 Conclusions
This work presented the design and implementation of a robust and compute-efficient pipeline for classifying brain tumors in magnetic resonance (MR) images. The proposed pipeline has utilized an input pre-processing to crop relevant regions from the brain MR images and a novel lightweight channel-attention guided convolutional neural network named AG-CNN to classify the cropped brain MR images into normal and tumor classes. The designed AG-CNN has employed a channel-attention block and feature fusion using skip-connection via global average-pooling (GAP) to select relevant feature maps and enhanced features, respectively, for better classification of brain tumors in MR images. The proposed AG-CNN model evaluated on four brain MRI datasets using different performance metrics has attained competitive accuracy. Besides, the designed lightweight model is compute-efficient and has a low memory footprint. Overall, the designed classification pipeline is robust and efficient. It can be deployed on a low-cost embedded device to quickly classify brain MR images in real-world clinical settings. Future research in this domain shall deal with designing a deep-learning-based integrated framework for brain tumor segmentation and classification in MR images.
Data availability
The codes and data generated and analyzed during the current study are available from the corresponding author on reasonable request.
References
Ferlay J, Ervik M, Lam F, Colombet M, Mery L, Piñeros M, Znaor A, Soerjomataram I, Bray F (2020) Global cancer observatory: cancer today. Lyon, France: international agency for research on cancer pp 1–6
Sekhar A, Biswas S, Hazra R, Sunaniya AK, Mukherjee A, Yang L (2021) Brain tumor classification using fine-tuned googlenet features and machine learning algorithms: Iomt enabled cad system. IEEE J Biomed Health Inform
Marosi C, Hassler M, Roessler K, Reni M, Sant M, Mazza E, Vecht C (2008) Meningioma. Crit Rev Oncol/Hematol 67(2):153–171
Weller M, Wick W, Aldape K, Brada M, Berger M, Pfister SM, Nishikawa R, Rosenthal M, Wen PY, Stupp R et al (2015) Glioma. Nat Rev Dis Primers 1(1):1–18
Kleihues P, Burger PC, Scheithauer BW (1993) The new who classification of brain tumours. Brain Pathol 3(3):255–268
Banan R, Hartmann C (2017) The new who 2016 classification of brain tumors-what neurosurgeons need to know. Acta Neurochir 159(3):403–418
Zacharaki EI, Wang S, Chawla S, Soo Yoo D, Wolf R, Melhem ER, Davatzikos C (2009) Classification of brain tumor type and grade using mri texture and shape in a machine learning scheme. Magn Reson Med Off J Int Soc Magn Reson Med 62(6):1609–1618
Işın A, Direkoğlu C, Şah M (2016) Review of mri-based brain tumor image segmentation using deep learning methods. Procedia Comput Sci 102:317–324
Kang J, Ullah Z, Gwak J (2021) Mri-based brain tumor classification using ensemble of deep features and machine learning classifiers. Sensors 21(6):2222
Deepak S, Ameer P (2019) Brain tumor classification using deep cnn features via transfer learning. Comput Biol Med 111:103345
Jafari M, Kasaei S (2011) Automatic brain tissue detection in mri images using seeded region growing segmentation and neural network classification. Australian J Basic Appl Sci 5(8):1066–1079
El-Dahshan ESA, Hosny T, Salem ABM (2010) Hybrid intelligent techniques for mri brain images classification. Digit Signal Process 20(2):433–441
Zacharaki EI, Wang S, Chawla S, Yoo DS, Wolf R, Melhem ER, Davatzikos C (2009) Mri-based classification of brain tumor type and grade using svm-rfe. In: 2009 IEEE international symposium on biomedical imaging: from nano to macro, IEEE, pp 1035–1038
Saritha M, Joseph KP, Mathew AT (2013) Classification of mri brain images using combined wavelet entropy based spider web plots and probabilistic neural network. Pattern Recognit Lett 34(16):2151–2156
Ismael MR, Abdel-Qader I (2018) Brain tumor classification via statistical features and back-propagation neural network. In: 2018 IEEE international conference on electro/information technology (EIT), IEEE, pp 0252–0257
Mohsen H, El-Dahshan ESA, El-Horbaty ESM, Salem ABM (2018) Classification using deep learning neural networks for brain tumors. Fut Comput Inform J 3(1):68–71
Ayadi W, Elhamzi W, Charfi I, Atri M (2019) A hybrid feature extraction approach for brain mri classification based on bag-of-words. Biomed Signal Process Control 48:144–152
Ayadi W, Charfi I, Elhamzi W, Atri M (2020) Brain tumor classification based on hybrid approach. The Visual Computer pp 1–11
Anjum S, Hussain L, Ali M, Abbasi AA (2020) Automated multi-class brain tumor types detection by extracting rica based features and employing machine learning techniques. In: Machine Learning in Clinical Neuroimaging and Radiogenomics in Neuro-oncology, Springer, pp 249–258
Ghahfarrokhi SS, Khodadadi H (2020) Human brain tumor diagnosis using the combination of the complexity measures and texture features through magnetic resonance image. Biomed Signal Process Control 61:102025
Amin J, Sharif M, Raza M, Saba T, Anjum MA (2019) Brain tumor detection using statistical and machine learning method. Comput Methods Progr Biomed 177:69–79
Kokkalla S, Kakarla J, Venkateswarlu IB, Singh M (2021) Three-class brain tumor classification using deep dense inception residual network. Soft Comput 25(13):8721–8729
Ma L, Zhang F (2021) End-to-end predictive intelligence diagnosis in brain tumor using lightweight neural network. Appl Soft Comput 111:107666
Bashir-Gonbadi F, Khotanlou H (2021) Brain tumor classification using deep convolutional autoencoder-based neural network: multi-task approach. Multimed Tools Appl 80(13):19909–19929
Masood M, Nazir T, Nawaz M, Mehmood A, Rashid J, Kwon HY, Mahmood T, Hussain A (2021) A novel deep learning method for recognition and classification of brain tumors from mri images. Diagnostics 11(5):744
Isunuri BV, Kakarla J (2021) Three-class brain tumor classification from magnetic resonance images using separable convolution based neural network. Concurrency and Computation: Practice and Experience p e6541
Abiwinanda N, Hanif M, Hesaputra ST, Handayani A, Mengko TR (2019) Brain tumor classification using convolutional neural network. In: World congress on medical physics and biomedical engineering 2018, Springer, pp 183–189
Kakarla J, Isunuri BV, Doppalapudi KS, Bylapudi KSR (2021) Three-class classification of brain magnetic resonance images using average-pooling convolutional neural network. Int J Imaging Syst Technol
Alhassan AM, Zainon WMNW (2021) Brain tumor classification in magnetic resonance image using hard swish-based relu activation function-convolutional neural network. Neural Computing and Applications pp 1–13
Sultan HH, Salem NM, Al-Atabany W (2019) Multi-classification of brain tumor images using deep neural network. IEEE Access 7:69215–69225
Woźniak M, Siłka J, Wieczorek M (2021) Deep neural network correlation learning mechanism for ct brain tumor detection. Neural Computing and Applications pp 1–16
Bodapati JD, Shaik NS, Naralasetti V, Mundukur NB (2021) Joint training of two-channel deep neural network for brain tumor classification. Signal Image Video Process 15(4):753–760
Kumar RL, Kakarla J, Isunuri BV, Singh M (2021) Multi-class brain tumor classification using residual network and global average pooling. Multimed Tools Appl 80(9):13429–13438
Irmak E (2021) Multi-classification of brain tumor mri images using deep convolutional neural network with fully optimized framework. Iranian Journal of Science and Technology, Transactions of Electrical Engineering pp 1–22
Díaz-Pernas FJ, Martínez-Zarzuela M, Antón-Rodríguez M, González-Ortega D (2021) A deep learning approach for brain tumor classification and segmentation using a multiscale convolutional neural network. In: Healthcare, Multidisciplinary Digital Publishing Institute, vol 9, p 153
Liu D, Liu Y, Dong L (2019) G-resnet: Improved resnet for brain tumor classification. In: international conference on neural information processing, Springer, pp 535–545
Deepak S, Ameer P (2020) Automated categorization of brain tumor from mri using cnn features and svm. J Ambient Intell Human Comput pp 1–13
Gu X, Shen Z, Xue J, Fan Y, Ni T (2021) Brain tumor mr image classification using convolutional dictionary learning with local constraint. Frontiers in Neuroscience 15
Panwar SA, Munot MV, Gawande S, Deshpande PS (2021) A reliable and an efficient approach for diagnosis of brain tumor using transfer learning. Biomed Pharmacol J 14(1):283–294
Anjum S, Hussain L, Ali M, Alkinani MH, Aziz W, Gheller S, Abbasi AA, Marchal AR, Suresh H, Duong TQ (2021) Detecting brain tumors using deep learning convolutional neural network with transfer learning approach. International Journal of Imaging Systems and Technology
Tandel GS, Tiwari A, Kakde O (2021) Performance optimisation of deep learning models using majority voting algorithm for brain tumour classification. Computers in Biology and Medicine p 104564
Polat Ö, Güngen C (2021) Classification of brain tumors from mr images using deep transfer learning. The Journal of Supercomputing pp 1–17
Lu SY, Wang SH, Zhang YD (2020) A classification method for brain mri via mobilenet and feedforward network with random weights. Pattern Recognit Lett 140:252–260
Noreen N, Palaniappan S, Qayyum A, Ahmad I, Imran M, Shoaib M (2020) A deep learning model based on concatenation approach for the diagnosis of brain tumor. IEEE Access 8:55135–55144
Kaur T, Gandhi TK (2020) Deep convolutional neural networks with transfer learning for automated brain image classification. Mach Vis Appl 31(3):1–16
Khan MA, Ashraf I, Alhaisoni M, Damaševičius R, Scherer R, Rehman A, Bukhari SAC (2020) Multimodal brain tumor classification using deep learning and robust feature selection: a machine learning application for radiologists. Diagnostics 10(8):565
Mehrotra R, Ansari M, Agrawal R, Anand R (2020) A transfer learning approach for ai-based classification of brain tumors. Mach Learn Appl 2:100003
Pashaei A, Sajedi H, Jazayeri N (2018) Brain tumor classification via convolutional neural network and extreme learning machines. In: 2018 8th international conference on computer and knowledge engineering (ICCKE), IEEE, pp 314–319
Anaraki AK, Ayati M, Kazemi F (2019) Magnetic resonance imaging-based brain tumor grades classification and grading via convolutional neural networks and genetic algorithms. Biocybernet Biomed Eng 39(1):63–74
Rammurthy D, Mahesh P (2020) Whale harris hawks optimization based deep learning classifier for brain tumor detection using mri images. Journal of King Saud University-Computer and Information Sciences
Huang Z, Xu H, Su S, Wang T, Luo Y, Zhao X, Liu Y, Song G, Zhao Y (2020) A computer-aided diagnosis system for brain magnetic resonance imaging images using a novel differential feature neural network. Comput Biol Med 121:103818
Sert E, Özyurt F, Doğantekin A (2019) A new approach for brain tumor diagnosis system: single image super resolution based maximum fuzzy entropy segmentation and convolutional neural network. Med Hypotheses 133:109413
Abd-Ellah MK, Awad AI, Khalaf AA, Hamed HF (2018) Two-phase multi-model automatic brain tumour diagnosis system from magnetic resonance images using convolutional neural networks. EURASIP J Image Video Process 1:1–10
Afshar P, Mohammadi A, Plataniotis KN (2018) Brain tumor type classification via capsule networks. In: 2018 25th IEEE international conference on image processing (ICIP), IEEE, pp 3129–3133
Verma K, Khunteta A (2017) Facial expression recognition using gabor filter and multi-layer artificial neural network. 2017 international conference on information. Communication, Instrumentation and Control (ICICIC), IEEE, pp 1–5
Hu J, Shen L, Sun G (2018) Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7132–7141
Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A (2016) Learning deep features for discriminative localization. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2921–2929
Ling H, Wu J, Huang J, Chen J, Li P (2020) Attention-based convolutional neural network for deep face recognition. Multimed Tools Appl 79(9):5595–5616
Woo S, Park J, Lee JY, Kweon IS (2018) Cbam: Convolutional block attention module. In: Proceedings of the European conference on computer vision (ECCV), pp 3–19
Cheng J (2017) brain tumor dataset. https://doi.org/10.6084/m9.figshare.1512427.v5, https://figshare.com/articles/dataset/brain_tumor_dataset/1512427
Chakrabarty N (2019) Brain MRI Images for Brain Tumor Detection Dataset. https://www.kaggle.com/navoneel/brain-mri-images-for-brain-tumor-detection, Accessed date: September 2021
Hamada A (2020) Br35H Brain Tumor Detection 2020 Dataset. https://www.kaggle.com/ahmedhamada0/brain-tumor-detection, Accessed date: September 2021
Sartaj B, Ankita K, Prajakta B, Sameer D (2020) Brain Tumor Classification (MRI) Dataset. https://www.kaggle.com/sartajbhuvaji/brain-tumor-classification-mri, Accessed date: September 2021
Nirthika R, Manivannan S, Ramanan A, Wang R (2022) Pooling in convolutional neural networks for medical image analysis: a survey and an empirical study. Neural Computing and Applications pp 1–27
Sabour S, Frosst N, Hinton GE (2017) Dynamic routing between capsules. Advances in neural information processing systems 30
Deepika J, Rajan C, Senthil T (2022) Improved capsnet model with modified loss function for medical image classification. Signal, Image and Video Processing pp 1–9
Acknowledgements
The authors would like to thank the Director, CSIR-CEERI, Pilani, for providing the necessary infrastructure and technical support. We would also like to acknowledge the consistent encouragement and motivation by the Head of the Intelligent Systems Group (ISG) at CSIR-CEERI, Pilani.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Saurav, S., Sharma, A., Saini, R. et al. An attention-guided convolutional neural network for automated classification of brain tumor from MRI. Neural Comput & Applic 35, 2541–2560 (2023). https://doi.org/10.1007/s00521-022-07742-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-022-07742-z