Abstract
Diagnosis, detection and classification of tumors, in the brain MRI images, are important because misdiagnosis can lead to death. This paper proposes a method that can diagnose brain tumors in the MRI images and classify them into 5 categories using a Convolutional Neural Network (CNN). The proposed network uses a Convolutional Auto-Encoder Neural Network (CANN) to extract and learn deep features of input images. Extracted deep features from each level are combined to make desirable features and improve results. To classify brain tumor into three categories (Meningioma, Glioma, and Pituitary) the proposed method was applied on Cheng dataset and has reached a considerable performance accuracy of 99.3%. To diagnosis and grading Glioma tumors, the proposed method was applied on IXI and BraTS 2017 datasets, and to classify brain images into six classes including Meningioma, Pituitary, Astrocytoma, High-Grade Glioma, Low-Grade Glioma and Normal images (No tumor), the all datasets including IXI, BraTS2017, Cheng and Hazrat-e-Rassol, was used by the proposed network, and it has reached desirable performance accuracy of 99.1% and 98.5%, respectively.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Brain tumors are collection or mass of abnormal cells in brain which can be cancerous (malignant) or noncancerous (benign). Brain tumors are categorized as primary or secondary [23]. A primary brain tumor arises in the brain and most of them are benign, and a secondary brain tumor originates from cancer cells that spread to the brain from another organs, such as lung or breast [19, 39]. World Health Organization (WHO) classified brain tumors into over 120 type of tumors in 4 grades: grade I to VI (low dangerous to high dangerous), some types of brain tumors are more common, such as Glioma, Meningioma, Astrocytoma, and Pituitary [23]. Glioma is a type of brain tumor that starts in the glial cells of the brain or the spine and are the most common type of primary tumors with a count about 52 percent of all primary tumors. According to WHO, Glioma tumors can categorized to several level of malignancy. Grade I to grade IV such grade I tumors are approximately normal and grade II and grade III tumors are more malignant, respectively and also grade IV tumors are the most perilous named Glioblastoma Multiform (GBM) [6, 19]. Astrocytoma tumor begins in cells called astrocytes that support nerve cells. Some astrocytoma tumors grow very slowly and others can be aggressive cancers that grow quickly, so there are grading for this tumor type like Glioma [31]. Meningioma is one of brain tumor type that arise from meninges layer in brain tissue. Meninges layer covered the brain and spinal cord like a protector. This type mostly considered as benign tumors because these tumors spread slowly and have a distinct boundary [23, 31]. Pituitary tumors account for about 15 percent of all primary tumors and also they are benign but they can cause serious health problems due to their hurts on sensitive areas of the brain [31]. Medical image analysis especially tumor images analysis is an open field in image processing and machine vision. In recent years, with the advent of deep learning through multilayered neural networks, the accuracy of image processing has grown dramatically. Deep learning (DL) techniques, as a specialized form of machine learning approaches, motivate the benchmarks of classification and segmentation tasks in the field of computer vision [13, 34]. Deep learning employs deep architectures of learning or hierarchical features learning to performs end-to-end learning. Hierarchical architectures enable machines to process data with a non-linear approaches for feature extraction.
Among deep learning approaches, deep Convolutional Neural Networks (CNNs) play a pivotal role in the analysis of medical images in various research and clinical fields [27]. In recent years, the concept of deep CNNs have led to meaningful developments in medical image classification [17, 25, 26, 36, 44, 45]. In general, the machine learning algorithms have different performance in dealing with multiple representation of data, and because of this , Success of a machine learning algorithm depends on dataset representation [20, 43]. We used some feature learning techniques for construct a new representation to improve extracted features from proposed network and raise classification rate. The use of data labels makes a difference in the type of learning: supervised or unsupervised feature learning. [3, 20] In this research training an auto-encoder network with non-labeled data, leads to extracting global features from dataset, thus combining local and global extracted features represents new dataset for proposed network. Feature learning and high accuracy are the main advantages of deep CNNs which are accomplished with help of numerous layers and automated features extraction process and leads to more accurate and robust model [27]. Such architecture employs Convolutional filters as feature extractors to extract robust and high-level features, so that as one goes deeper within the network, one will generate deeper complex features.
Focusing on the brain, deep CNNs have demonstrated their potential for brain image analysis in several different domains, with classification efficiency at the detection of brain disease from Magnetic resonance imaging (MRI) images. Brain tumor classification is an intricate and a more challenging problem in the area of classification problem. The main challenges are referred to the following aspects:
-
1.
Brain tumors indicate high variations with respect to intensity, shape, and size [10];
-
2.
Tumors from various pathological types might demonstrate analogous appearances [11];
-
3.
Despite the need for large amount of labeled training data, medical image datasets are very limited. [41].
To tackle these challenges the researchers design systems, which assists doctors in diagnosing and detecting abnormalities and helps them to make accurate and rapid decisions [35, 37] Hence, the detection and classification of tumors by machine learning methods is also an open field for research and development because in previous classic methods there are still many challenges, including response time. Section 1.1 describes these problems by explaining the previous methods, but before that it should be mentioned that the proposed method can solve some of these challenges. We propose a deep architecture based on Convolutional Auto-encoder Neural Network (CANN) to diagnose brain tumors and to classify them into four types (Astrocytoma, Meningioma, Pituitary, and Glioma) and two grades (Low- and High-Grade Gliomas). Our proposed network based on feature learning techniques extracts some global and local features to detect tumors in the MRI brain image apart from physical features. In more details, the primitive contributions of this manuscript are listed as follows:
-
Feature extraction and feature selection, and the process of classification are executed in one step, such that tumor regions are not required to be selected and segmented.
-
Two different features or representation learning techniques are applied on the proposed network to determine the effect of feature learning and training procedure on the problem of interest.
-
In the proposed classification network, the brain tumors are diagnosed by extracting global features (high-level features) from the input MR brain images. Furthermore, they are classified and graded by extracting local features (low-level features).
-
The classification networks has been trained from Scratch without any pre-trained features. The Scratch framework makes model parameters fine-tuned which leads to simplicity of the proposed network structure in comparison to the pre-trained networks.
-
The proposed network does perform detection, classification, and gradation simultaneously, such that only one network is utilized to handle these three tasks.
The rest of this correspondence is structured as follows. The next sub-section focuses on other existing research and study performed in the field or area of brain tumor classification. Section 2 provides a detailed insight about the proposed framework. Section 3 is dedicated to learning mechanism. Experimental results and corresponding analysis generated from the proposed method with some discussions are described in Section 4. Finally, conclusions and directions for future work are drawn in Section 5.
1.1 Literature review
There have been several attempts to classify brain tumors using ML techniques [7, 10, 12, 15, 46], particularly those of the deep CNN approaches presented in [1, 2, 14, 16, 24, 32, 33, 40].
The proposed approach in [7] utilizes the concepts of wavelet and support-vector machine (SVM) to classify MR brain images as either normal or abnormal. This research has been able to represent a way for the efficiency of machine learning methods in classifying images of brain tumors, however, it has used very limited data to train the model. In [46], an approach is proposed for classification of different grades of Glioma together with a binary classification for high- and low-grade using SVM and k-Nearest Neighbors (k-NN). Despite the positive points, this method has been performed on a small amount of data in three time-consuming stage. Authors in [15] proposed a hybrid method to classify brain tumor as normal or abnormal MR brain images. This method uses Discrete Wavelet Transform (DWT) to obtain and Principle Component Analysis (PCA) to decrease features. Finally, it employs feed forward back-propagation Artificial Neural Network (fp-ANN) to classify images. In this research, training and predicting time have been increased relatively. In [10], authors introduced a method to improve the classification performance of brain tumors with three feature extraction methods, namely, intensity histogram, Gray Level Co-occurrence Matrix (GLCM), and Bag-of-Words (BoW) model. This enhancement is achieved by augmenting tumor region via image dilation as region-of-interest (RoI) and then by splitting augmented tumor region into ring-form sub-regions. Nevertheless, using region-of-interest in MRI images is not appropriate way to extract tumor, since all brain tissue has clues to detect a tumor, so extracting region-of-interest loses a lot of data. The proposed method in [12] uses multi-model radiomics imaging features to differentiate between HGG and LGG brain tumors. Extracting radiomics features from raw images using Gray-Level Co-occurrence Matrix(GLCM) then feature selection using correlation matrix had expected results.
In [16], authors proposed a deep learning-based CNN for automated grading of Gliomas (Grade II, Grade III, and Grade IV) in one task and classifying Low-Grade Glioma (LGG) and High-Grade Glioma (HGG) in another task using digital pathology images. Also split big dataset into some parts using tilling method was proposed as a pre-process, which is a time-consuming step. In [33], two types of neural networks (fully connected neural network and CNN) used to classify brain images with different types (Meningioma, Glioma, and Pituitary). As described in this article, the proposed CNN architecture was composed of two convolutional layers, two max-pooling layers followed by two fully connected layers. It has also been stated that employing Vanilla pre-processing has been impressive in classification accuracy. However, all of these steps slow down the process of tumor classification, and cascading network connections slows the process even further. In work [24], an approach that uses CNN to classify brain medical image into healthy and unhealthy, categorized as low- and high-grades, is proposed. An enhanced version of Alex Krizhevsky network (AlexNet) deep learning structure on MR brain images are used as their network architecture. This method can be named as one of the desirable methods. In [1] authors proposed a modified capsule networks (CapsNet) with access to the tumor surrounding tissues that merges both the tumor coarse boundaries as extra inputs and MR brain images for brain tumor classification. On the plus side, the researchers has not absorbed the tissue around the tumor in the brain images. In [2], a method based on CNN and Genetic algorithm is proposed to classify different types (Meningioma, Glioma and Pituitary) and grades (Grade II, Grade III, and Grade IV) of brain tumors. Unlike the existing methods that select a deep neural network architecture, the proposed structure of CNN has been evolved using GA. In this study, the number of mutations and cross-overs performed for the large problem space was not enough, so the model did not take into account all cases. Authors in [40] proposed a CNN architecture to classify brain tumors into different types (Meningioma, Glioma and Pituitary) and differentiate among the three Glioma grades (Grade II, Grade III, and Grade IV). To achieve the most suitable architecture, they evolved the network structure employing various configurations. The proposed CNN is satisfactory network for classification, but the extracted features could have been more acceptable. Recent work, [14], has been adopted a deep transfer learned CNN model and employed a pre-trained GoogLeNet to extract features from brain MR images. Doing so, the classification of three specific types of brain tumors (Glioma, Meningioma and Pituitary) has been addressed. In a recent work, [32], DWT and deep Neural Network (NN) are combined to classify normal and three types of brain tumors: Glioblastoma, Sarcoma and Metastatic Bronchogenic Carcinoma. In this study, like most mentioned studies, at first, the tumor is segmented in the image and then fed into the model. Segmentation is a time-consuming step to preparing images.
Considering the previous methods, several important points are understood. Most of these methods use limited datasets, and not all possibilities in the images may be considered. In some methods, primary tumor isolation, such as segmentation or selection of the tumor regions, is used for the feature extraction step, and some of the information in the brain tissue which is useful for tumor diagnosing, may be lost. Some researches have multi-step and time consuming methods, especially methods for manual feature extraction. However, recent neural network-based methods have performed well in feature extraction and classification. Deep neural network approaches extract better features from the input images, and resulting in more accurate output. Therefore, the proposed method is based on convolutional neural networks with an appropriate number of layers to learn the features to diagnose, classify and grade the tumor using a network in one step and achieve the desired result. Synchronization of tasks, reducing time, increasing classification accuracy and reducing missclassification rate are some of the achievements of the proposed method.
2 Methodologies
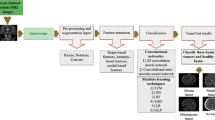
The block-diagram of the proposed framework is shown in Fig. 1. As it is obvious, the proposed framework can be divided into two main steps; a pre-processing followed by classification. In pre-processing, the skull stripping is performed to remove non-cerebral (non-brain) tissue from anatomical MR brain images. These de-skulled data are employed as initial-input to CNN. Then a CNN-based deep learning framework is applied to extract features from input data. The network itself composed of three subnetworks. Deep features at high- and low-level are respectively extracted by the first and second sub-networks. Both high- and low-level features are stacked together and fed into a proven classifier, third subnetwork. Finally, the classification task is accomplished and the outcomes are achieved. The hierarchy of the proposed method is summarized in the Fig. 2.
2.1 Pre-processing
Before feeding the images into the proposed classification network, a pre-processing is required to comply with the input formats. First, the original images are resized to 240 × 240 × 1 pixels to match the lowest image size exist in datasets. Secondly, to reduce over-fitting probability and enhance model performance, learn invariant features and improve the robustness of the model [42], the MR images are augmented. Although augmentation process bring some advantages to our model, feeding unrealistic images into the network may introduce undesirable knowledge to the learning process. Therefore, we limit the image augmentation to flipping, mirroring, shifting, scaling and rotation clockwise or counterclockwise, an example of these techniques are shown in Fig. 3. Employing augmentation process, we have increased the number of data, so that the final dataset consists of about 20,000 MR images. These images are used for training, validation and testing purposes.
In this paper, as the final stage of pre-processing, we use Brain extraction tool, namely BET, [38]. BET utilizes a deformable model that expands to fit the brain surfaces by the application of a set of locally adaptive model forces, an example is shown in Fig. 4. This technique is useful for the feature extraction stage because all the extracted features are related to the internal tissue of the brain.

Skull Stripping: a Image of brain with skull, b A de-skulled brain image [18]
2.2 CANN
Figure 5 illustrates the architecture of the proposed model for brain MR image classification. As it is clear, the model is based on deep convolutional auto-encoder-based neural network. In this sub-section, the architecture of CANN is briefly introduced. The sample image from brain MR images is fed into CANN for the aim of learning the feature description, that is utilized for classification task. The proposed CANN architecture consists of an encoder network and a corresponding decoder network. Each encoder includes convolution with a filter-bank, max-pooling and sub-sampling to generate a set of feature-map. The encoder network includes two convolutional layers followed by a middle layer and each encoder does execute convolution operation corresponding feature-map. These feature maps are then batch normalized [21]. Batch normalization is utilized to accelerate the convergence of the training procedure and decrease the probability of getting stuck in local minima. Since each encoder layer has its corresponding decoder layer, the decoder network further consists of two layers.
The convolutional auto-encoder [8] extract the output data, Y, to regenerate the input data, X and examine it in contrast with original input data. If time iteration goes to infinity, \(i \rightarrow \infty \), the cost function attains its optimal values. This translates to the fact that regenerated input data is able to estimate the original input data to a maximum range. The summary of proposed CANN can be seen in Algorithm 1 \(f\left (.\right )\) and \(\hat {f}\left (.\right )\) mathematically denote the convolutional encode and the convolutional decode functions, respectively. In Algorithm 1, the activation functions, η and ζ, are non-linear activation functions. The sigmoid, the hyperbolic tangent, and the rectified linear, namely Relu, function are three of which. In this paper, we implement Relu function that is defined as follows:
The minimization of error is accomplished by optimizing the following cost function
Employing stochastic gradient descent, the weight and error are minimized, and the convolutional auto-encoder layer is optimized. Finally, the trained parameters are utilized to output which are transmitted to the next layer.
The utilized CANN is analogous to the prevalent CNN, where the convolutional layer is followed by a pooling layer. Further, in CANN after each convolutional auto-encoder a max-pooling layer is used (see Fig. 5) and the resulting output is sub-sampled by a factor of 2.
where Xi,j denotes the i-th region of j-th input feature-map and Yi,j denotes i-th neuron of j-th output feature-map. The number of input and output feature-map are equal.
The proposed decoding technique is demonstrated in Fig. 5. In the decoder network, each decoder up-samples its feature-map employing the stored max-pooling indices from the related encoder feature-map. This phase provides sparse feature-map [4]. The corresponding feature-map is then convolved with a trainable decoder filter bank to regenerate the input image. Next, we briefly review the proposed CNN architecture.
2.3 CNN
As illustrated in Fig. 5, the deep CNN consists of 5 convolutional layers and 5 max-pooling layers, followed by two fully-connected layers. The fully-connected and soft-max layers are used to predict and classify output. The drop-out layer is used to avoid over-fitting.
Suppose that the proposed CNN composed of L layers, the output state of the ℓ-th layer is denoted Xℓ, \(\ell \in \left \{1,\ldots ,L\right \}\). Moreover, suppose that X0 refers to the input data. In each layer, there exist two trainable parameters, i.e. the weight matrix wℓ and the bias vector bℓ. As illustrated in Fig 5, the input data are fed into a convolutional layer. This layer performs a 2-D convolutional operation with a window of weights slides across an image, named convolutional kernel, wℓ. During the scanning process, these weights remain unchanged. Consequently, convolutional layers are able to learn robust features. The bias vector, bℓ, is then added to the outcome feature maps, where an element-wise non-linear activation \(\sigma \left (.\right )\) is normally performed afterwards. Eventually, to choose the superior features through non-overlapping square windows per feature map a max-pooling layer is typically followed. This process can be formulated as follows:
where ⊗ and p represent the convolution and max-pooling operation, respectively.
Multiple convolution and max-pooling layers are stacked together to build the hierarchical feature extraction architecture. Further, one fully-connected layer combines corresponding features into a 1-D feature vectors. Inputs to fully-connected layer is first processed with non-linear transformation via weight and bias vectors, \(\left \{w_{\ell }, b_{\ell }\right \}\), and the element-wise non-linear activation fuction in then followed:
Here, a piecewise rectified-linear non-linearity, i.e. ReLU, is applied (see 3)).
A soft-max layer, with the number of neurons equal with the number of classes to be classified, is the final classification layer. We generally need the network output to be a vector of label probabilities, that sum up to unity [29]. Thus, we use probability-based loss functions, e.g. cross entropy, for classification tasks. The soft-max function satisfies this constraint
Note that, applying soft-max, \(\text {ReLU}\left (x\right )\) is normalized to sum up unity.
The loss function is employed to measure the difference between the output of the CNN and the true image label, i.e. loss. Minimizing the value of loss function is the main aim of training the CNN. The cross-entropy loss function together with soft-max output activation is commonly utilized for classification tasks. The cross-entropy loss calculates the cross entropy between the predicted distribution of CNN and ground truth distribution as follows:
where s is the estimate for true distribution r, and n denotes the number of classes.
3 Training mechanism
In proposed framework, training mechanism composed of two training phases with an interruption among them to combine all features and extract more deeper ones. During training process two set of parameters, CANN and CNN parameters, should be initialized and updated. The former includes weight parameters of encoder and decoder, \(\left \{w, \hat {w}\right \}\), layers and the latter includes weights corresponding to CNN layers.
The first phase of training is related to training CANN network. This phase is performed by unlabeled training data to regenerate original input data and acquire best parameters corresponding to auto-encoder model. An auto-encoder model with optimized weights on convolutional layers is the production of this phase. The optimized model is now qualified to extract some more appropriate features and merge them with other features. Following this phase, the next phase of training is proceed with an interruption, which links middle layer of auto-encoder model and construct a new model. Current model is composed of an encoder part and a CNN architecture. Doing so, the optimized weights of the encoder part are loaded into current model and other weights are initialized in similar manner. The labeled data are then fed into the current model and next phase of training begins.
Supposed that \(\mathbf {X} = \left [{\mathbf {x}}_{i}\right ]\in {\mathbb {R}}^{n \times n}\), \(i=\left \{1,\ldots ,N\right \}\), are examples of unlabeled data which is used to train the CANN, where n denotes the dimension of input data and N denotes the number of training samples. Since the auto-encoder is an unsupervised learning and applies the back-propagation scheme to reconstruct the input data, every layer has an output. Supposed that \(\mathbf {Y} = \left [\mathbf {y}_{i}\right ]\in \mathbb {R}^{n \times n}\), \(i=\left \{1,\ldots ,N\right \}\), represents the output of last layer. At the end of training process the output feature maps would be a realization of the input data, i.e. Y≅X. Every training phase is carried out in two passes: Forward Pass and Backward Pass. Thus, we can describe the entire mechanism of training as follows:
-
(1)
First Phase of Training: Forward Pass
-
Step 1:
Initializing the parameter pair, \(\left \{w,\hat {w}\right \}\), randomly with uniform distribution.
-
Step 2:
Extracting features and lowering the dimension of each input feature maps using the following formula:
$$ {\mathbf{y}}_{i} = \sum\limits_{i=1}^{N}{\mathbf{x}}_{i} w $$(10) -
Step 3:
Assigning the max value to every stride on each input data layer to produce the output of max-pooling layer.
-
Step 4:
Expanding the input data of each layer by repeating rows and columns to generate the output of up-sampling layer.
-
Step 5:
Extracting features and expanding the feature maps in each layer convolutional layer of decoder unit using the following formula
$$ {\mathbf{y}}_{i} = \sum\limits_{i=1}^{N}{\mathbf{x}}_{i} \hat{w} $$(11)
-
(2)
First Phase of Training: Backward Pass
-
Step 1:
Calculation of the output error using corresponding loss function loss function;
-
Step 2:
Updating and optimizing the pair \(\left \{w,\hat {w}\right \}\) by the following cost function:
$$ \boldsymbol{\mathcal{J}}\left( \theta\right) = \underset{\mathbf{X}}{\sum}{\rho\left( \mathbf{X},\mathbf{Y}\right)},\quad {\pmb{\theta}}=\left\{w, \hat{w}\right\} $$(12)
-
(3)
Second Phase of Training: Forward Pass
-
Step 1:
Applying the following function on the input data of each layer to evolve them into feature maps.
$$ {\mathbf{y}}_{i} = \sum\limits_{i=1}^{N}{\mathbf{x}}_{i} {\pmb{\theta}} $$(13) -
Step 2:
Assigning the max value to every stride on each input data layer to produce the output of max-pooling layer.
-
Step 3:
Vectorizing the final feature map by flatten layers to pass to the former layers.
-
Step 4:
Employing drop-out layer. This layer help to remove some nodes together with their corresponding weights from final layer and decrease the probability of over-fitting in each iteration.
-
Step 5:
Applying (8) for normalization purpose.
-
(4)
Second Phase of Training: Backward Pass
-
Step 1:
Employing (9) to calculate the whole network errors and optimizing all weights.
-
Step 2:
Applying the SGD cost function on weights and optimizing parameters to improve the output of each layer during training process.
What achieved after executing this phase would be a model with the optimized parameters and minimum misclassified errors with ability to classify unlabeled test data.
It is worth mentioning that, the proposed network consists of 10 convolutional layers to extract features in all steps. Increasing or decreasing network convolutional layer deteriorates the classification accuracy, because a network with less layer does not train well and does not extract some more important features from input data images. Moreover, a network with more convolutional layer has more parameters, that increase the probability of over-fitting. As shown in Fig. 6, the proposed network with 10 convolutional layer has best performance in MR brain Images classification.
4 Experimental results and discussion
This research performs 3 brain tumor classification scenarios on 4 standard datasets including MRI images. All scenarios use deep feature learning extracted by proposed classifier network.
4.1 Datasets
In all body imaging techniques, Magnetic Resonance Imaging (MRI) is an efficient technique to illustrate (show) brain tissue. MRI system setting categorized into different class modality, such as T1 and T2 (also known as T1-Weighted and T2-Weighted). Also T1-CE and T2-CE are contract enhancement of T1 and T2 image modalities. Other changed modality known as Flair and Density Proton, which are less used. for current study, we used 4 different datasets, which have all images modalities, as follows:
-
IXI dataset [22], contains 582, 3D (256 × 256 × 140), MRI volumes from normal, healthy subjects;
-
BraTS 2017 datasets [5, 28, 30], contains 3D (240 × 240 × 155) MRI volumes of 285 high- and low-grade glioma cases;
-
Cheng datasets [9], contains 3064, 2D (512 × 512), MR images with three types of brain tumor, i.e. meningioma, glioma, and pituitary;
-
Dataset acquired from Hazrat-e-Rasool General Hospital [2], containing 230, 2D (256 × 256), MR images of three kinds of tumor, i.e. meningioma, glioma, and astrocytoma.
4.2 Evaluation
In this section, we describe the evaluation criteria and simulation results of the proposed architecture for three different scenarios, that had been carried out on the particular dataset, described on Section 4.1. The following subsections are detailed description and evaluation of the scenarios.
In this paper the performance of the proposed method is evaluated in terms of Accuracy, Precision, Recall and Fmeasure. The terms are defined as follows:
where TP (True Positive) denotes condition in which both prediction and actual value are positive or correct, TN (True Negative) represents the cases when both actual ad predicted value are negative or incorrect, FN (False Negative), denotes the cases in which the prediction values is incorrect and actual value is correct, and finally, FP (False Positive) represents cases, where the prediction is correct and the actual value is incorrect.
4.2.1 Scenario 1
The classification differences in the scenarios depend on the datasets used by the proposed network. The first scenario deals with classification of brain tumors into three types, Meningioma, Glioma, and Pituitary. The number of neurons in the last layer of the network varies according to the number of input datasets in the network. In the first scenario, only the Cheng dataset [9] was used, which consists of images of three tumors. Table 1 summarize system’s performance of the proposed classifiers as confusion matrix for this scenario. The predicted and actual values are respectively assigned to x-axis and y-axis. According to confusion matrix of the scenario, all of the performance measures are calculated in Table 2.
4.2.2 Scenario 2
The second scenario diagnoses between normal and abnormal brain images and differentiates between two Glioma grades, i.e. HGG and LGG. Data from [5, 22, 28, 30] are combined and are utilized in this scenario. Tables 3 and 4 demonstrates the confusion matrix and performance metrics for the second scenario, respectively.
4.2.3 Scenario 3
The last scenario gets involved with diagnosis (i.e. differentiates between normal and abnormal MR brain images), classification (i.e. classifies brain images as Meningioma, Astrocytoma, and Pituitary), and grading Gliomas (i.e. discriminates between High-Grade-Glioma (HGG) and Low-Grade-Glioma (LGG)). Data from [2, 5, 9, 22, 28, 30] are combined and are utilized in scenario 3. This scenario is the main purpose of this study, which is the basis for providing a method based on convolutional networks to be able to simultaneously diagnose a tumor in the brain and its type and grade. Finally, by combining the above datasets, the model is able to learn features from images of the six classes mentioned, so that it can correctly predict the label of brain images from the six available categories. The confusion matrix and performance metrics of the last scenario are shown in Tables 5 and 6, respectively.
The accuracy and loss variations during training and validation phases from the main scenario (scenario 3) are demonstrated in Fig. 7. As it is obvious in training process of the proposed network, the accuracy and loss function converge to specific values and the training process is accomplished properly. During the training phase the accuracy may decrease. This reduction might be due to the fact that cost function get trapped in local minima. In such cases, the network needs to be trained in the plenty of iterations, so that learning algorithm converges to global minima and parameters optimize. Consequently, the transient behavior of network should be ignored.
5 Conclusion
As mentioned in Section 2, basic machine learning methods, such as SVM, DWT, PCA or obsolete feature extraction methods, do not provide the expected results in brain images classification. However, neural network-based methods, especially convolutional networks, provide more accurate and desirable results. On the other hand, training neural networks from scratch requires large and numerous datasets, but in the field of medical images, data, especially images, are limited and scarce. Therefore, in this study, the number of images increased using the data augmentation method and the problem of data shortage for the network was solved (More data, convenient features, better learning). The proposed network, based on convolutional auto-encoder, uses modified brain images (de-skulled) for learning. Due to the number of layers and the network architecture, feature learning from MRI images is done correctly so that the classification accuracy is greatly increased. Deep feature learning utilized to use proposed network in multi task classification. The brain tumor diagnosis is a binary classification and needs some global features, while, the brain tumor classification or grading is a multi-class classification which needs local features and more details. Combining high and low level features leads to classifying all images in one step using one network. A small number of high and low level features extracted by sub-networks of proposed network are shown in Fig. 8:
As discussed above, the improvement of classification results is due to deep feature learning. According to the Section 4.2 (see Tables 2, 4 and 6), in all scenarios the proposed framework has performed the classification task with high precision. In Table 7, we draw a comparison between the proposed structure and some other previous literature with similar tasks. It is obvious that the proposed framework gives superior performance findings, compared to other existing method which uses different architecture and methods. This shows the reliability of the proposed system.
In summary in this paper, a new novel strategy is proposed to classify brain tumors into three types: Meningioma, Glioma, and Pituitary with accuracy of 99.3% in first study, diagnose between normal and abnormal brain images and differentiates between LGG and HGG with accuracy of 99.1% in second study, and further doing diagnosis task and classifying brain images into three types:Meningioma, Astrocytoma, and Pituitary, and two Glioma grades: LGG and HGG, with accuracy of 98.5% using a custom deep CNN structure. The performance criterion for the proposed approach has been compared with other state-of-the-art methods, and the results demonstrate the ability of the model for brain tumor classification task, in multi-task manner.
By examining Tables 2, 4 and 6, and the Precision and Recall criteria, the performance of the model can be examined. In the field of brain tumor analysis, a mistake may threaten a human life, the Recall criteria is particular importance. The value of this criterion is close to one, which means the model is able to identify tumor and classify it correctly and grade them in terms of being cancerous and dangerous. In all three scenarios, the Recall value is above 0.98, which means that the performance of the model is desirable and there is no risk to life. The accuracy of the classification models, which eventually turn into CAD systems, is very important because it is tried that despite these systems, doctors do not need to identify anomalies and illnesses. For all the convenience that these systems have for physicians, it is difficult to rely on these computer methods. Therefore, the method must be very accurate. For example, the proposed method has an Precision criterion of close to one in all three scenarios, which means that every time it predicts a label for a image, the label is really true and accurate (all images labeled as tumors are really tumors).
References
Afshar P, Plataniotis K, Mohammadi A (2019) Capsule networks for brain tumor classification based on mri images and coarse tumor boundaries. In: ICASSP 2019-2019 IEEE international conference on acoustics, speech and signal processing (ICASSP). IEEE, pp 1368–1372
Anaraki A, Ayati M, Kazemi F (2019) Magnetic resonance imaging-based brain tumor grades classification and grading via convolutional neural networks and genetic algorithms. Biocybern Biomed Eng 39(1):63–74
Argyriou A, Evgeniou T, Pontil M (2007) Multi-task feature learning. In: Advances in neural information processing systems. pp 41–48
Badrinarayanan V, Kendall A, Cipolla R (2017) Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell 39(12):2481–2495
Bakas S, Reyes M, Jakab A, Bauer S, Rempfler M, Crimi A, Shinohara RT, Berger C, Ha SM, Rozycki M et al (2018) Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats challenge. arXiv:1811.02629
Banerjee S, Mitra S, Shankar BU (2018) Automated 3d segmentation of brain tumor using visual saliency. Inform Sci 424:337–353
Chaplot S, Patnaik L, Jagannathan N (2006) Classification of magnetic resonance brain images using wavelets as input to support vector machine and neural network. Biomed Signal Process Control 1(1):86–92
Chen M, Shi X, Zhang Y, Wu D, Guizani M (2017) Deep features learning for medical image analysis with convolutional autoencoder neural network. IEEE Trans Big Data
Cheng J (2017) Brain tumor dataset. Web Site. http://figshare.com
Cheng J, Huang W, Cao S, Yang R, Yang W, Yun Z, Wang Z, Feng Q (2015) Enhanced performance of brain tumor classification via tumor region augmentation and partition. PloS One 10(10):e0140381
Cheng J, Yang W, Huang M, Huang W, Jiang J, Zhou Y, Yang R, Zhao J, Feng Y, Feng Q et al (2016) Retrieval of brain tumors by adaptive spatial pooling and fisher vector representation. PloS one 11(6):e0157112
Cho HH, Park H (2017) Classification of low-grade and high-grade glioma using multi-modal image radiomics features. In: 2017 39th Annual international conference of the IEEE engineering in medicine and biology society (EMBC). IEEE, pp 3081–3084
Criminisi A (2016) Machine learning for medical images analysis. Med Image Anal 33:91–93
Deepak S, Ameer P (2019) Brain tumor classification using deep cnn features via transfer learning. Comput Biol Med 103345:111
El-Dahshan E, Hosny T, Salem A (2010) Hybrid intelligent techniques for mri brain images classification. Digit Signal Process 20(2):433–441
Ertosun M, Rubin D (2015) Automated grading of gliomas using deep learning in digital pathology images: a modular approach with ensemble of convolutional neural networks. In: AMIA annual symposium proceedings, vol 2015. American Medical Informatics Association, p 1899
Esteva A, Kuprel B, Novoa R, Ko J, Swetter S, Blau H, Thrun S (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542(7639):115
Gonbadi FB, Khotanlou H (2019) Glioma brain tumors diagnosis and classification in mr images based on convolutional neural networks. In: 2019 9th International conference on computer and knowledge engineering (ICCKE). IEEE, pp 1–5
Grant R (2019) Medical management of adult glioma. In: Management of adult glioma in nursing practice. Springer, pp 61–80
Huang J, Rathod V, Sun C, Zhu M, Korattikara A, Fathi A, Fischer I, Wojna Z, Song Y, Guadarrama S et al (2017) Speed/accuracy trade-offs for modern convolutional object detectors. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp 7310–7311
Ioffe S, Szegedy C (2015) Batch normalization: accelerating deep network training by reducing internal covariate shift arxiv e-prints
IXI: brain-development website (2019) https://brain-development.org/ixi-dataset. Accessed: 2019-05-30
Johnson DR, Guerin JB, Giannini C, Morris JM, Eckel LJ, Kaufmann TJ (2017) 2016 updates to the who brain tumor classification system: what the radiologist needs to know. Radiographics 37(7):2164–2180
Khawaldeh S, Pervaiz U, Rafiq A, Alkhawaldeh R (2018) Noninvasive grading of glioma tumor using magnetic resonance imaging with convolutional neural networks. Appl Sci 8(1):27
Koitka S, Friedrich C (2016) Traditional feature engineering and deep learning approaches at medical classification task of imageclef 2016. In: CLEF (Working Notes). pp 304–317
Kumar A, Kim J, Lyndon D, Fulham M, Feng D (2016) An ensemble of fine-tuned convolutional neural networks for medical image classification. IEEE J Biomed Health Inform 21(1):31–40
Litjens G, Kooi T, Bejnordi B, Setio A, Ciompi F, Ghafoorian M, Van Der Laak J, Van Ginneken B, Sánchez C (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88
Liu L, Zheng G, Bastian JD, Keel MJB, Nolte LP, Siebenrock KA, Ecker TM (2016) Periacetabular osteotomy through the pararectus approach: technical feasibility and control of fragment mobility by a validated surgical navigation system in a cadaver experiment. Int Orthop 40(7):1389–1396
Liu M, Shi J, Li Z, Li C, Zhu J, Liu S (2016) Towards better analysis of deep convolutional neural networks. IEEE Trans Vis Comput Graph 23 (1):91–100
Lloyd CT, Sorichetta A, Tatem AJ (2017) High resolution global gridded data for use in population studies. Sci Data 4(1):1–17
Louis DN, Perry A, Reifenberger G, Von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW (2016) The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathologica 131(6):803–820
Mohsen H, El-Dahshan ESA, El-Horbaty ESM, Salem ABM (2018) Classification using deep learning neural networks for brain tumors. Appl Comput Inform J 3(1):68–71
Paul J, Plassard A, Landman B, Fabbri D (2017) Deep learning for brain tumor classification. In: SPIE, medical imaging, biomedical applications in molecular, structral, functional imaging, vol 10137, pp 10137–10142
Schmidhuber J (2015) Deep learning in neural networks: An overview. Neural Netw 61:85–117
Shen D, Wu G, Suk HI (2017) Deep learning in medical image analysis. Annu Rev Biomed Eng 19:221–248
Shen W, Zhou M, Yang F, Yu D, Dong D, Yang C, Zang Y, Tian J (2017) Multi-crop convolutional neural networks for lung nodule malignancy suspiciousness classification. Pattern Recogn 61:663–673
Shiraishi J, Li Q, Appelbaum D, Doi K (2011) Computer-aided diagnosis and artificial intelligence in clinical imaging. In: Seminars in nuclear medicine, vol 41. Elsevier, pp 449–462
Smith SM (2002) Fast robust automated brain extraction. Hum Brain Mapp 17(3):143–155
Sridhar K, Baskar S, Shakeel PM, Dhulipala VS (2019) Developing brain abnormality recognize system using multi-objective pattern producing neural network. J Ambient Intell Humaniz Comput 10(8):3287–3295
Sultan H, Salem N, Al-Atabany W (2019) Multi-classification of brain tumor images using deep neural network. IEEE Access
Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, Liang J (2016) Convolutional neural networks for medical image analysis: Full training or fine tuning? IEEE Trans Med Imaging 35(5):1299–1312
Wong KC, Syeda-Mahmood T, Moradi M (2018) Building medical image classifiers with very limited data using segmentation networks. Med Image Anal 49:105–116
Xie S, Sun C, Huang J, Tu Z, Murphy K (2018) Rethinking spatiotemporal feature learning: Speed-accuracy trade-offs in video classification. In: Proceedings of the European conference on computer vision (ECCV). pp. 305–321
Xu Y, Mo T, Feng Q, Zhong P, Lai M, Eric I, Chang C (2014) Deep learning of feature representation with multiple instance learning for medical image analysis. In: 2014 IEEE international conference on acoustics, speech and signal processing (ICASSP). IEEE, pp 1626–1630
Yu Y, Lin H, Meng J, Wei X, Guo H, Zhao Z (2017) Deep transfer learning for modality classification of medical images. Information 8(3):91
Zacharaki E, Wang S, Chawla S, Soo Yoo D, Wolf R, Melhem E, Davatzikos C (2009) Classification of brain tumor type and grade using mri texture and shape in a machine learning scheme. Magn Reson Med 62(6):1609–1618
Acknowledgements
We would like to thank Mr. Vahid Vahidpour for his valuable help and comments in this research project.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bashir-Gonbadi, F., Khotanlou, H. Brain tumor classification using deep convolutional autoencoder-based neural network: multi-task approach. Multimed Tools Appl 80, 19909–19929 (2021). https://doi.org/10.1007/s11042-021-10637-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-021-10637-1