Abstract
Water distribution network (WDN) is one of the most essential infrastructures all over the world and ensuring water quality is always a top priority. To this end, water quality sensors are often deployed at multiple points of WDNs for real-time contamination detection and fast contamination source identification (CSI). Specifically, CSI aims to identify the location of the contamination source, together with some other variables such as the starting time and the duration. Such information is important in making an efficient plan to mitigate the contamination event. In the literature, simulation-optimisation methods, which combine simulation tools with evolutionary algorithms (EAs), show great potential in solving CSI problems. However, the application of EAs for CSI is still facing big challenges due to their high computational cost. In this paper, we propose DLGEA, a deep learning guided evolutionary algorithm to improve the efficiency by optimising the search space of EAs. Firstly, based on a large number of simulated contamination events, DLGEA trains a deep neural network (DNN) model to capture the relationship between the time series of sensor data and the contamination source nodes. Secondly, given a contamination event, DLGEA guides the initialisation and optimise the search space of EAs based on the top K contamination nodes predicated by the DNN model. Empirically, based on two benchmark WDNs, we show that DLGEA outperforms the CSI method purely based on EAs in terms of both the average topological distance and the accumulated errors between the predicted and the real contamination events.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Water distribution networks (WDNs) are essential infrastructures for modern life and economy. To ensure the water quality, sensors are deployed at multiple points of WDNs for collecting real-time data regarding water quality. These data are used for contamination detection and fast contamination source identification (CSI). Specifically, CSI [21] aims to identify the location of the contamination source, together with some other variables of the contamination event, e.g. starting time and duration. Such information is important in making efficient plans to mitigate the contamination event. CSI problems are difficult due to a number of reasons. First, it is infeasible to do experimental analysis in real-world WDNs. Second, the water quality data are often sparse as the deployment of water quality sensors is limited in WDNs. Third, the dynamics of water demand patterns add uncertainty to the analysis. Fourth, it is important that the contamination source can be identified within a short period of time.
As an alternative to the experimental analysis in real-world WDNs, simulation tools are often adopted for solving CSI problems, e.g. the hydraulic modelling and simulation tool EPANET [26] developed by the US environmental protection agency. A WDN is modelled by setting up the topology, hydraulic and water quality parameters, which can then be used to run simulations to observe the hydraulic and water quality changes. It is possible to explore the contamination configurations by simulating a large number of contamination events and searching for the one that generates the most similar water quality data as to the real contamination event. In other words, we need to identify the event which minimises the difference between the simulated water quality data and the observed water quality data. As such, CSI is transformed into an optimisation problem.
In the literature, simulation-optimisation methods, which combine simulation tools with evolutionary algorithms (EAs), show great potential in solving CSI problems [9, 16, 22, 30, 33, 38]. EAs are population-based algorithms widely adopted for solving optimisation problems [1]. However, the application of EAs for CSI is still facing big challenges due to their high computational cost. To this end, we propose DLGEA, a deep learning guided evolutionary algorithm to improve the searching efficiency of EAs. In particular, DLGEA trains a deep neural network (DNN) model based on a large number of samples generated by EPANET to infer the ranking of all the nodes in a WDN by their probability of being the contamination source node. By prioritising the search space of the set of top K nodes with the highest probabilities, the computation of EAs can be optimised. By carrying out a set of experiments with dynamic water demand patterns, we show that DLGEA outperforms the CSI method purely based on EAs [38] in terms of the average topological distance as well as the accumulated errors between the predicted and the real contamination events.
In summary, the contributions of this paper are as follows:
-
We propose a deep learning guided evolutionary algorithm DLGEA for CSI which takes advantage of the learning capabilities of deep neural networks to optimise the search space of evolutionary algorithms, and we show that DLGEA outperforms the state-of-the-art CSI method purely based on EAs through a set of experiments with two benchmark WDNs.
-
We investigate the application of two types of DNN models, i.e. convolutional neural networks and recurrent neural networks, for contamination source node localisation, and show that the DNN models are able to localise the real source node with an accuracy above 85% for a medium-size WDN and above 69% for a large-scale WDN when the top 5 nodes with the highest probability are considered.
-
The evaluations are performed using two benchmark WDNs, respectively, with 97 nodes and 1786 nodes. We release the datasets used in this paper as well as the source codeFootnote 1 of DLGEA to facilitate the reproducibility of our work.
The rest of the paper is organised as follows: Sect. 2 discusses the related work. Section 3 gives a formal description of the CSI problem. Section 4 presents the proposed deep learning guided evolutionary algorithm DLGEA for CSI. Section 5 describes the dataset used in this paper. Section 6 illustrates the experiment preparation and the evaluation metrics, and analyses the experiment results. Finally, in Sect. 7, we conclude the paper with possibilities of future work.
2 Related work
In the early stage, researchers regarded the CSI problem as an inverse problem and tried to solve it by analysing the inner hydraulic model and water quality model of the WDN directly. For example, Shang et al. proposed a particle backtracking algorithm which aims to describe the relationship between water quality at input and output locations and by the relationship, the inverse problem can be solved [28]. Laird at al. presented an origin tracking algorithm for solving the inverse problem of contamination source identification based on a nonlinear programming framework [14] and later extended their work by incorporating a mixed-integer quadratic program to address non-unique solution problems [13]. However, these methods require complicated analysis and are difficult to be applied to large-scale WDNs.
Machine learning methods have also been investigated for CSI problems. For example, Huang and McBean proposed to combine a screening approach and a maximum likelihood method to identify the location and time of a contamination event based on limited sensor data [10]. Perelman and Ostfeld proposed a solution based on topological clustering and Bayesian Networks (BNs) [20]. It first applies the clustering method proposed in [19] to formulate a simplified representation of WDNs based on nodal connectivity properties. With evidence from the clusters, information is then combined through probabilistic inference using BNs to find the most likely source of contamination and its propagation in the network. Wang and Harrison proposed to use Markov Chain Monte Carlo (MCMC) to enable probabilistic inference of contamination events [31], and combine support vector regression (SVR) to speed up the likelihood evolution during the MCMC chain evolution [32].
More recently, simulation-optimisation methods are gaining more popularity. The idea is that the accumulated errors between the observed concentration data and the simulated concentration data should be minimum if the simulated contamination event is the same as the real contamination event. Although some other optimisation techniques were considered such as the reducing gradient method [5], a commonly adopted approach for CSI is combining the simulation tool EPANET with variants of EAs. For example, Preis and Ostfeld integrated a genetic algorithm (GA) with EPANET [22], in which GA provides heuristic information for searching while EPANET is used to evaluate the fitness of a candidate solution as to the real event. Praveen et al. conducted experiments to show the effectiveness of simulation-optimisation methods by taking dynamic water demand into account [30]. Liu et al. developed an EA based adaptive dynamic optimisation procedure that uses multiple population-based search to provide a real-time response to contamination events [16]. Hu et al. developed a MapReduce based Parallel Niche Genetic Algorithm (MR-PNGA) which explores the cloud resources for performance improvement [9]. Yan et al. investigated different optimization algorithms for CSI such as cultural algorithms [34] and variants of genetic algorithms [33, 35,36,37,38,39].
All these works show great potential of simulation-optimisation methods in solving CSI problems. However, the simulation part of these EA-based approaches is usually expensive and the search space of contamination events is also huge. Therefore the total computational cost is high and it is hard to find the optimal solution within a short period of time. To the best of our knowledge, there is no previous work that has investigated the application of deep neural networks for CSI problems by using the predictions from neural networks as a prior to optimise the search space and speed up the search process.
3 Problem statement
Given a WDN, a contamination event can be characterised by a set of variables including the location of the contamination source, the starting time, the duration of the contamination and the concentration of the contaminants at the source node. Formally, we define a contamination event s as:
where n indicates the contamination source node in the WDN, \(t_{0}\) indicates the starting time of the contamination, t indicates the duration of the contamination and \(\rho \) indicates the concentration of the contaminants at the source node. For example, a contamination event \(s = [10, 3, 2, 100]\) indicates that some contaminants are injected into the WDN at node 10, starting from 3:00, lasting for 2 h and the contaminant concentration at the source node is 100mg/L.
When a contamination event encoded by s occurs in a WDN, the water quality at different parts of the WDN will change according to the topology, water hydraulic and quality model of the WDN. Suppose we have the following mapping:
where \({\mathcal {F}}\) is a function of the WDN that maps contamination events \({\mathcal {S}}=\{s\}\) to concentration matrices \({\mathcal {C}}=\{C\}\) in which \({\mathcal {C}} \subseteq {\mathbb {R}}^{T \times M}\) and a concentration matrix C captures the spatio-temporal contaminant concentration data at all T time steps from all M sensor nodes. Note that the set of sensor nodes is a subset of all the nodes in the WDN. Suppose the WDN in total has N nodes, we have \(M \le N\). In this paper, we rely on the water distribution network modelling and simulation tool EPANETFootnote 2 to realise such a mapping. That is, we can simulate a contamination event s in a WDN using EPANET, and the tool will return the contaminant concentration data for all M sensor nodes at all T time steps.
Given a contamination event s occurring in a WDN, the aim of CSI is to find a solution \(s^*\) that minimises the errors between the simulated concentration data based on \(s^*\) and the real concentration data obtained from the sensors when s occurred. Accordingly, the objective of CSI can be formulated as follows ( [9, 22, 38]):
where \(C^{s}(t,m)\) and \(C^{s^*}(t,m)\) are the observed and simulated contaminant concentration data at time step t from node m, respectively. As such, the CSI problem is transformed into an optimisation problem, which can be handled by EAs.
4 Deep learning guided evolutionary algorithm
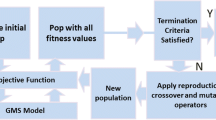
As illustrated in Sect. 2, the CSI methods purely based on EAs suffer from high computational cost in finding the optimal solution. To alleviate this problem, in this paper, we propose DLGEA, a deep learning guided evolutionary algorithm for CSI to improve the searching efficiency of EAs by leveraging the knowledge from the historical contamination data. The framework of DLGEA is shown in Fig. 1, which consists of two stages. Given a contamination event occurred in a WDN, the first stage, called DNN Based Search Space Division, uses a pre-trained DNN model to estimate the likelihood (probability) of each node in the WDN being the contamination source. Based on the probability ranking of all the nodes returned by the DNN model, the set of possible solutions (i.e. all the nodes in a WDS) is divided to construct two populations, with the nodes of higher probabilities in one population and the rest of the nodes in the other population. The second stage, called Dual-Population Evolution, applies a simulation-optimisation approach to identify all the components of the contamination event as defined in (1) based on the two populations constructed from the first stage. The details of these two stages are illustrated in the rest of this section.
4.1 DNN based search space division
A WDN often consists of a large number of nodes such as pipe junctions, water sources and water tanks. When a contamination event occurs, we assume in this paper that the contamination is injected from one of the nodes in the WDN as in ([9, 16, 22, 30, 38]). The goal of the first stage of DLGEA is to estimate the probability of each node in the WDN being the contamination source node. For this purpose, a deep neural network (DNN) model is trained using the historical contamination data.
Specifically, given a set of contamination events, the inputs for the DNN model are sequences of readings from all the sensor nodes in the WDN, and the outputs of the DNN model are the probability of each node being the source of contamination. For real-world WDNs, contamination events rarely occur and thus we cannot get sufficient data for training DNN models. To solve this problem, we turn to EPANET to generate training data, which will be illustrated in Sect. 5.
Given a new contamination event, the trained DNN model can then be used to infer the probability of each node in the WDN being the contamination source node based on the water quality data collected from the sensor nodes. We pool the top K candidate source nodes to construct one population and the rest of the nodes to construct another population for the second stage which aims at identifying the source node and recover the other variables of the contamination events such as the starting time, duration, etc.
4.2 Dual-population evolution
EA based CSI methods often follow a simulation-optimisation approach and encode contamination events as individuals with components such as the source node, the starting time, etc. [16, 22, 30, 33]. Similarly, we model a contamination event encoded by (1) as an individual in a population and its fitness value f is defined according to the objective function (3), which is formulated as follows:
As mentioned in Sect. 3, matrix \(C^{s^*}\) is the time-series concentration data of candidate contamination events. In specific, for each given individual, the original network model will be configured to initiate the corresponding candidate event and then simulated with EPANET. Accordingly, a water quality report will be generated, recording the concentration data at all time steps from all the sensor nodes, which is then transformed into matrix \(C^{s^*}\). Notice that in our setting, an individual with a smaller fitness value is considered to be a better solution.
Most of the EA based CSI methods in the literature (e.g. [9, 16, 33, 38]) treat all the nodes in a WDN equally by putting them in the same population for optimisation. This means that the EAs have to search a large solution space and in turn require intensive computation. To speed up the searching process, we propose a dual-population evolution method in which the search of the solution space is performed independently within the two populations constructed by the first stage. The intuition is that there is a large probability that the real source node is within the top K nodes localised by the DNN model and by prioritising towards these top K candidate source nodes the search space of the EA based CSI methods can be optimised. The detailed process is shown in Algorithm 1.

We run two EAs separately with two populations of equal size. One population is constructed from only the top K candidate nodes while the other population is constructed from the rest \(N-K\) nodes. The intuition is that with fewer candidate nodes (\(K \ll N\)), the search space of \(EA_1\) can be largely reduced, which is likely to find the optimal solution much faster if the real contamination source node is in the set of K candidate nodes. On the other hand, in case the real contamination source node is not in the K candidate nodes, \(EA_2\) may still have certain probability of finding the optimal solution. For both \(EA_1\) and \(EA_2\), after a fixed number of iterations, the individuals are pooled together and the one with the best fitness value will be selected as the final solution. Note that the computational resources are evenly split between the two EAs as they are running in parallel with the same size of population and the same number of iterations (generations).
In addition, as CSI problems are usually multimodal problems, niching methods can be combined with EAs to maintain the diversity of populations and achieve better solutions [38]. In this paper, two niching EA based CSI methods are used as baselines. The first one realises the process of niching based on the concept of fitness sharing [18]. The second one we implemented follows the work presented by Yan et al. [39].
The main idea of fitness sharing is to transform the raw fitness value of an individual into a shared one based on the similarities between the individual and its neighbours. Suppose we have two individuals i: \([n_{i}, t_{0i}, t_{i}, \rho _{i}]\ \) and j: \( [n_{j}, t_{0j}, t_{j}, \rho _{j}] \). The shared fitness value of individual i is:
where \(\mu \) is the population size and f(i) is the raw fitness value of individual i. The sharing function based on individual distance is defined as:
If \(n_{i}=n_{j}\), the distance between them can be calculated as follows:
If the source nodes are not the same, i and j are considered not in the same niche and their distance is set to a pre-defined value \(d_{share}\).
By applying the steps above, the raw fitness values of the individuals are transformed into shared ones, which helps to maintain the diversity of the population.
The second niching strategy used in this paper is from Yan et al. [39]. The main idea is that an individual is retained only if it has the lowest fitness value among all the individuals with the same source node, otherwise it will be replaced by a new individual which is randomly initialised and evaluated. By doing so, the diversity of the source nodes will be maintained in the evolving process.
5 DataSet
Following the CSI literature (e.g. [9, 16, 22, 30, 33, 38]), the data used in this paper are from the simulations with EPANET. Specifically, we evaluate DLGEA using two benchmark water distribution networks as shown in Figs. 2 and 3. The first network, denoted as \(\mathbf{WDN }_1\), is an example network provided by EPANET and has been widely used in the literature for studying CSI problems (e.g. [16, 22, 27, 30, 38]). It comprises of 97 nodes (\(N=97\)) and 117 pipes, together with a water hydraulic model and a water quality model. The second network, denoted as \(\mathbf{WDN }_2\), is the Wolf-Cordera model [15] which comprises of 1786 nodes and 1985 pipes, together with a water hydraulic model and a water quality model. Based on these two networks, EPANET is able to simulate the water hydraulic and quality changes with different settings of simulation duration (e.g. minutes, hours, days) and data recording rate (e.g. minutes, hours).
In this paper, given a contamination event, we run a simulation of 24 h with EPANET using the default water demand pattern and record the contaminant concentration data at all nodes every 5 min (\(T=1\)). In this way, we generate 10,000 contamination events for \(\mathbf{WDN }_1\) and 100,000 for \(\mathbf{WDN }_2\). However, in real-world systems, sensor nodes are relatively sparse and thus we select 4 and 6 nodes (\(<7\%\) nodes) as the sensor nodes for \(\mathbf{WDN }_1\) (\(M=4, 6\)) following the literature [16, 17, 22, 27]. Similarly, we select 10 and 30 nodes (\(<2\%\) nodes) as the sensor nodes for \(\mathbf{WDN }_2\) (\(M=10, 30\)). After looking into the sensor data of the 10,000 contamination events for \(\mathbf{WDN }_1\) and the 100,000 contamination events for \(\mathbf{WDN }_2\), we notice that some of the contamination events do not have any influence on any of the sensor nodes within certain time limits (e.g. 30 and 60 min), i.e. the concentration data recorded by the sensor nodes are all zeros. These contamination events are excluded from our experiment as they are not detectable.
As a result, we obtain four datasets under different sensor nodes selections and time limits for both \(\mathbf{WDN }_1\) and \(\mathbf{WDN }_1\), as shown in Table 1. For \(\mathbf{WDN }_1\), the first dataset consists of 3140 contamination events each of which is represented by a time series of contamination concentration collected from the four sensor nodes (\(M=4\)) within a time limit of 30 min (\(T=6\)) after the first time the contamination is detected by any of the four sensor nodes. The second dataset, consisting of 3092 contamination events, is collected from the same set of four sensor nodes within a time limit of 60 min (\(T=12\)). The third and fourth datasets for \(\mathbf{WDN }_1\) are collected from six sensor nodes (\(M=6\)) with a time limit of 30 min (\(T=6\)) and 60 min (\(T=12\)), respectively. Similarly, the four datasets for \(\mathbf{WDN }_2\) are also collected within a time limit of 30 min and 60 min but from a larger number of sensor nodes, i.e. \(M=10\) and \(M=30\).
From Table 1 we can see that the dataset size under the setting of \(T=12\) is smaller than that of \(T=6\). A larger T means that we require sensor readings collected over a longer time period since the first time the contamination is detected by any of the sensor nodes. The reason why increasing T results in less events is due to the fact that the simulation is of 24 h. This means when a contamination event is detected at a time point which is too close to the 24 h limit it will not have sufficient number of records to cover T. Such events will be removed. For the same contamination event, a larger T means more information is provided.
In order to reflect the differences between the simulation model and the real-world WDN, we consider two types of uncertainties. The first type of uncertainty comes from the fact that the demand pattern of each node in the WDN may change over time. To capture such uncertainties, we introduce a Gaussian noise into the nodal demands as follows:
where \(\alpha (t,n)\) indicates the water demand at time step t at node n in the default demand pattern and \({\mathcal {N}}\) is a Gaussian distribution used to simulate the demand changes. The demand changes are re-sampled for each simulation. Following [30], \(\theta \) is set to 0.25.
The second type of uncertainty is caused by the fact that the sensor measurements may not be accurate. Accordingly, we introduce a random noise into the nodal concentration data as follows:
where c(t, m) indicates the concentration value at time step t at sensor node m obtained from EPANET, \({\mathcal {U}}\) is a uniform distribution used to simulate the possible noise in the sensor measurements. Following [24], \(\delta \) is set to 0.05.
6 Experiments and result analysis
6.1 Experiment setup
To evaluate DLGEA, we carry out two sets of experiments. The first set of experiments is based on the dataset obtained from \(\mathbf{WDN }_1\) consisting of 97 nodes and the second is based on the dataset obtained from \(\mathbf{WDN }_2\) consisting of 1786 nodes, as described in Sect. 5.
6.1.1 DNN based search space division
Convolutional neural networks (CNNs) and Recurrent neural networks (RNNs) are two popular DNN models that have achieved the state-of-the-art performance in many applications such as computer vision [12], speech and audio processing [4, 25] and natural language processing [2, 3]. In this paper, we investigate these two types of DNN models for search space division as described in Sect. 4.1. Specifically, we built a four-layer CNN model and a four-layer RNN model with LSTM (Long Short-Term Memory) [7] units. Given a set of contamination events, the inputs for the CNN and LSTM models are sequences of readings from all the sensor nodes in the WDN, and the outputs are the probability of each node in the WDN being the source of contamination. We use 60% of the contamination events from each of the datasets shown in Table 1 for model training and the rest 40% are evenly split into a validation set and a test set.
For the design of the CNN model, considering that the size of the data samples is relatively small (\(4 \times 12\) or \(30 \times 12\)), we use 4 convolutional layers. The number of filters chosen for each of the four convolutional layers increases linearly with a factor of 2, which follows the common practice in the domains such as image classification [12] and audio processing [11]. The first dimension of the filter corresponds to the sensor nodes. Since the sensor nodes are not directly dependent, the convolution operation is not applied along this dimension. The second dimension of the filter corresponds to the readings of each sensor node. To capture their time dependencies, we experimented with convolution sizes of 2 which are commonly used in the literature. In specific, the architecture of the four-layer CNN model used in this paper is as follows: the number of filters for the four CNN layers are, respectively, 16, 32, 64 and 128, with a kernel size of \(1 \times 2\) and the Relu activation function; each of the four CNN layers is followed by a dropout layer with a dropout rate of 0.25; the last dropout layer is followed by a Dense layer with 256 hidden units with a Relu activation function, and finally another Dense layer with 97 hidden units with a Softmax activation function for \(\mathbf{WDN }_1\) and 1786 hidden units for \(\mathbf{WDN }_2\) to represent the probability of each node in the WDN being the source of contamination.
For the design of the LSTM model, we follow the previous work on anomaly detection for water distribution systems [23]. The architecture of the four-layer LSTM model used in this paper is as follows: each of the four LSTM layers has 128 hidden units and is followed by a dropout layer with a dropout rate of 0.25; the last dropout layer is followed by a Dense layer with 256 hidden units with a Relu activation function, and finally another Dense layer with 97 hidden units with a Softmax activation function for \(\mathbf{WDN }_1\) and 1786 hidden units for \(\mathbf{WDN }_2\) to represent the probability of each node in the WDN being the source of contamination.
The categorical crossentropy is used as the loss function for both the CNN and LSTM models. A batch size of 128 and the Adam optimizer are used with a learning rate of 0.001 to minimise the loss function. These hyper-parameters are chosen experimentally.
Note that the focus of this paper is not on comparing the performance of different DNN models, but to show the effectiveness of applying DNN models to the problem of contamination source identification.
6.1.2 Dual-population evolution
As illustrated earlier in Sect. 4.2, an individual in EAs is represented as a vector containing four variables, i.e. the source node, starting time, duration and concentration. The first three variables are discretised following [16] and concentration is also discretised to simplify the experiment settings. Details of the individual representation are shown in Table 2.
As one of the most popular EAs, genetic algorithms (GAs) [8] have been widely investigated for solving CSI problems (e.g. [33, 36, 38, 39]). In this paper, two niching genetic algorithms (NGAs) [39] are used as the EAs for dual-population evolution as described in Sect. 4.2. Specifically, the population size is set to 40, and the evolving process is stopped once 100 generations of population are evaluated. Tournament selection, uniform mutation and single point crossover are used as selection, mutation and crossover operators, respectively. For tournament selection, the best one out of three random individuals is selected for crossover and mutation operations. For single point crossover, the crossover rate is 0.8 and the value of the crossover point is chosen from 1 to 3 since there are four variables (genes) in each solution (chromosome). Suppose we have two individuals \(i_1=[n_1,{t_0}_1,t_1,\rho _1]\) and \(i_2=[n_2,{t_0}_2,t_2,\rho _2]\) for crossover and the crossover point is 1, then the new generated individuals will be: \(i_1'=[n_1, {t_0}_2, t_2, \rho _2]\), \(i_2'=[n_2, {t_0}_1, t_1, \rho _1]\) . The uniform mutation is performed on one of the four variables, which means for the individual to be mutated, the position is randomly selected from 1 to 4 first and the corresponding variable at that position will be re-initialised following the bounds with a mutation rate set to 0.8. The sharing radius \(d_{share}\) used for fitness sharing is set to 100. In addition, the elitist strategy is adopted and two elites is kept during the evolving process. All the parameters are set experimentally.
In this paper, we use two NGAs as baseline CSI methods. To demonstrate the efficiency of the proposed DLGEA presented in Sect. 4, we evaluate the performance of DLGEA with 300 randomly selected contamination events, respectively, from the test set of \(\mathbf{WDN }_1\) under the setting of (\(M=4, T=12\)) and \(\mathbf{WDN }_2\) under the setting of (\(M=30, T=12\)). Instead of running experiments for a single contamination event multiple times, we carry out experiments on a larger number of events to show the effectiveness of the proposed CSI method.
6.2 Evaluation metrics
As illustrated in Sect. 4, DLGEA has outputs in both stages, i.e. DNN based search space division and dual-population evolution. Accordingly, two sets of metrics are used for evaluation. The first set of metrics is based on accuracy to evaluate the percentage of instances in which the real source node is found. The second set of metrics is based on topological distance and nodal concentration similarity.
In the stage of search space division, the DNN models estimate the probability of each node in a WDN being the source node. Depending on the topology of the WDN, some closely located nodes may have similar reactions in response to certain contamination events. In such cases, it is likely that these nodes would have comparable probabilities of being the source node. Therefore, in stead of evaluating the accuracy of the top 1 node predicted by the DNN models, we look at the the top K accuracy. Given W contamination events, the top K accuracy \({\mathrm {Acc}}_K\) achieved by a DNN model is defined as follows:
where \(s_w\) indicates a contamination event; \(\varGamma ^{s_w} \in \{0,1\}^{N\times N\times K}\) is a three dimensional matrix representing a prediction of the event in which its first dimension represents the index of the real source node with respect to the contamination event \(s_w\), the second dimension represents the index of the predicted source node and the third dimension indicates the ranking of the predictions in terms of their probability being the source node. For example, \(\varGamma _{5,16,2}^{s_3}=1\) means that the index of the real source node is 5 and Node 16 is ranked at the second highest position. However, this prediction will not be counted as a correct prediction due to the restriction bounded by \(\mathbb {1}(i==j)\).
For dual-population evolution, we use two metrics for evaluation. The first metric is called average topological distance (ATD) [29] that quantifies the predictions of the source node by considering the topological properties of the WDN. Given W contamination events, ATD can be defined as follows:
where \(A_{i,j}\) contains the minimum topological distance between the nodes indexed by i and j.
The second metric is the average accumulated error (AAE) between the changes of the contamination concentration returned by the predicted contamination event and that returned by the real contamination event, which is defined as follow:
6.3 Result analysis
6.3.1 DNN based search space division
For the stage of DNN based search space division, \({\mathrm {Acc}}_1\) and \(Acc_5\) are used for evaluating the performance of the DNN models, i.e. the percentage of instances in which the real contamination source node is found in the top 1 and top 5 nodes returned by the DNN model. Tables 3 and 4, respectively, shows the test accuracy of the top 1 and top 5 nodes returned by the CNN and LSTM models in the case of \(\mathbf{WDN }_1\) when there are 4 sensor nodes and 6 sensor nodes deployed and in the case of \(\mathbf{WDN }_2\) when there are 10 sensor nodes and 30 sensor nodes deployed.
It can be seen that for both \(\mathbf{WDN }_1\) and \(\mathbf{WDN }_2\) in general the test accuracy increases when T is larger. This may be due to two reasons. One reason is that larger T means more information is provided. The other reason is that larger T results in less events as explained in Sect. 5. On the other hand, when more sensors are deployed more contamination events could be detected. For example, in the case of \(\mathbf{WDN }_1\), out of 10000 contamination events, 3140 events can be detected with 4 sensor nodes within 6 time steps, whereas 4468 events can be detected with 6 sensor nodes within 6 time steps. Since source identification is often initiated after contamination is detected, the test accuracy shown in Tables 3 and 4 only considers the cases when a contamination event can be detected by the sensors within the time limit. Therefore, the test accuracy between different numbers of sensors are not directly comparable.
For \(\mathbf{WDN }_1\), the CNN model in general achieves better performance than the LSTM model. As for \(\mathbf{WDN }_2\), the CNN model achieves higher \(Acc_1\) whereas the LSTM model achieves higher \(Acc_5\). In both cases, among the top 5 nodes, a test accuracy of more than 85.0% is achieved. This shows that using the DNN models there is a big probability that the real contamination source node could be localised within a small set of candidate nodes compared to the total number of nodes in the WDNs. It has to be emphasised that this paper is not intended to provide a comprehensive comparison of different DNN models for the task of source node localisation but rather to demonstrate the effectiveness of using DNN models to localise the contamination source nodes.
6.3.2 Dual-population evolution
Tables 5 and 6 show the performance of two groups of CSI methods. The first group is based on NGAs (GA with the niching method from [39] is denoted as NGA1 and GA with fitness sharing is denoted as NGA2) and the second group is based on DLGEA which combines the pre-trained CNN model with NGAs (denoted as CNN+NGA1 and CNN+ NGA2). It can be seen that by using the pre-trained CNN model to instruct the composition of two populations, for NGA1, the performance has been improved by more than 33.2% (89.0%) in terms of ATD and by more than 34.7% (81.3%) in terms of AAE in the cases of \(\mathbf{WDN }_1\) (\(\mathbf{WDN }_2\)) while for NGA2, the performance has been improved by more than 23.9% (80.0%) in terms of ATD and by more than 16.6% (77.4%) in terms of AAE in the cases of \(\mathbf{WDN }_1\) (\(\mathbf{WDN }_2\)).
Moreover, we further look into the metric of ATD by examining the distribution of the source nodes in the predicted contamination events in terms of their topological distance from the real source nodes. \(\mathbf{TD }=\mathbf{0} \) indicates that the predicted source node is the real source node, and \(\mathbf{TD }=\mathbf{1} \) indicates that the predicated source node is the direct neighbour of the real source node. It is shown in Tables 5 and 6 that CNN+NGA1 and CNN+NGA2 achieve higher accuracy for both \(\mathbf{TD }=\mathbf{0} \) and \(\mathbf{TD }=\mathbf{1} \) than NGA1 and NGA2. In particular, for more than half of the events, CNN+NGA1 and CNN+NGA2 are able to find the real source nodes.
In addition, to evaluate the computational time of DLGEA, we adopt the metric of the first hitting time (FHT) [6, 40]. FHT of EAs is the time used to find an optimal solution for the first time in each single run. In our case, FHT is quantified by the number of individuals that have been evaluated as the time used for individual evaluations is the same for all the contamination events in the same WDN. Moreover, considering that the optimal solutions obtained by different CSI methods may be different, we also report the first hitting time of the comparable optimal solution, denoted as FHT*. The comparable optimal solution here refers to the best solution that can be achieved by every CSI method. Table 7 shows the average FHT and FHT* of all the contamination events for both \(\mathbf{WDN }_1\) and \(\mathbf{WDN }_2\) with respect to each CSI method. It can be seen that for both \(\mathbf{WDN }_1\) and \(\mathbf{WDN }_2\), the average FHT of NGAs gets shorter when the CNN model is applied. In terms of FHT*, the improvement is more significant.
To provide an intuitive understanding, Fig. 4 shows the evolution of the best solutions returned by NGA1, NGA2, CNN+NGA1 and CNN+NGA2 for \(\mathbf{WDN }_1\) and \(\mathbf{WDN }_2\) in terms of the average of the best fitness values of (a) all the 300 randomly selected contamination events, (b) the contamination events in which the real source node is in the top 5 candidate nodes returned by the CNN model, and (c) the contamination events in which the real source node is not in the top 5 candidate nodes returned by the CNN model.
For \(\mathbf{WDN }_1\), it can be seen that CNN+NGA1 and CNN+NGA2 converge faster than NGA1 and NGA2 in all the three cases. In the cases of Fig. 4a1 and b1, the optimal solutions obtained by NGA2 and CNN+NGA2 have similar fitness values while the optimal solution obtained by CNN+NGA1 outperforms the one obtained by NGA1. In the case of Fig. 4c1 where the CNN model fails to identify the real source node, NGA2 in general found better solutions than CNN+NGA2 while CNN+NGA1 found better solutions than NGA1. An explanation is that even when the top 5 nodes returned by the CNN model does not include the real source node, it is still likely that the real source node is close to the 5 nodes returned by the CNN model and NGA1 could benefit more from such information. Another observation is that the fitness value of the individuals in Fig. 4c1 is much smaller than that in Fig. 4a1 and b1. This indicates that in the cases where the real source node was not in the top 5 nodes returned by the CNN model the differences of the fitness values of different solutions are smaller. Similar trends can be observed in the results of \(\mathbf{WDN }_2\) as shown in Fig. 4a2, b2 and c2 except that the fitness value of the optimal solution obtained by CNN+NGA1 and CNN+NGA2 in (a2) and (b2) is much smaller than that obtained by NGA1 and NGA2. As shown in all the six sub-figures in Fig. 4, CNN+NGA1 and CNN+NGA2 converge constantly faster than NGA1 and NGA2. Moreover, CNN+NGA1 and CNN+NGA2 are able to find relatively better sub-optimal solutions in about 20 generations whereas NGA1 and NGA2 need more than 40 generations to converge and may not even get a solution close to the one found by CNN+NGA1 and CNN+NGA2.
From both the evaluation of ATD and AEE as well as the evolution of the best fitness values, we can see that the improvement of CNN+NGA1 and CNN+NGA2 is more significant in \(\mathbf{WDN }_2\) with 1786 nodes compared to \(\mathbf{WDN }_1\) with 97 nodes, which indicates that DLGEA has potential in large-scale WDN applications.
6.4 Sensitivity analysis
To investigate the effectiveness of DLGEA under different layouts of sensor nodes and different assignments of K (the top K source nodes predicted by the DNN models), we carry out another two sets of experiments.
For the layout of sensor nodes, we randomly generate 100 layouts of 4 sensor nodes for \(\mathbf{WDN }_1\). We found that the number of detectable contamination events of these 100 layouts ranges from 2000 to 6000. To explore the influence of sensor node layout to the performance of DLGEA, we choose another three layouts of sensor nodes under the setting of \(M=4\) and \(T=12\) for \(\mathbf{WDN }_1\), and the corresponding numbers of detectable contamination events are shown in Table 8. The layout of sensor nodes for \(\mathbf{WDN }_1\) with \(M=4\) introduced in Sect. 5 and Table 1 is indicated by layout 1. Please refer to Appendix A for the details of these four layouts of sensor nodes. In this way, we provide an evaluation of DLGEA when layouts of sensor nodes with different sensing capabilities are considered.
Table 8 shows the test accuracy of the top 1 and top 5 nodes returned by the CNN model with respect to the three layouts of sensor nodes. It can be seen that when the number of detectable events increases, the accuracy of identifying the contamination source node decreases. This can be explained by the fact that when more events can be detected it is more likely that different contamination events lead to similar sensor readings.
Table 9 shows the performance of different methods in terms of ATD, AAE, \(\hbox {TD}=0\) and \(\hbox {TD}=1\) with respect to the three layouts of sensor nodes. It can be seen that in general ATD gets worse when the number of detectable events increases for all the CSI methods. As for AAE, since it is a metric relating to contamination concentrations which may differ a lot from event to event, the results of AAE are not directly comparable across different layouts of sensor nodes. With the same layout of sensor nodes, it can be seen that the CSI methods under the DLGEA framework, i.e. CNN+NGA1 and CNN+NGA2, obtain smaller ATD, AAE and higher accuracy in terms of \(\hbox {TD}=0\), \(\hbox {TD}=1\) compared to the baselines NGA1 and NGA2. This shows that DLGEA can improve the performance of EA based CSI methods under different layouts of sensor nodes with varying sensing capabilities.
To investigate the influence of the value of K (top K nodes predicated by the DNN models) to the performance of DLGEA, we explore different values for K ranging from 1 to 8 with CNN+NGA2 under the setting of \(M=4\) and \(T=12\) for \(\mathbf{WDN }_1\) using the first layout of sensor nodes (please refer to Table 1 for the dataset size and Appendix A for the layout of the sensor nodes). Figure 5 shows the changes of prediction accuracy of the CNN model, the ATD and AAE of CNN+NGA2 along with an increasing K. It can be seen that both ATD and AAE are relatively high when K is set to 1 and then gradually decrease when K gets larger. This matches with the fact that when K is too small the source node prediction accuracy of the CNN model is relatively low, i.e. the top K nodes returned by the CNN model has a larger probability to miss the real source node. When K gets substantially large, the prediction accuracy of the CNN model will not change much. Accordingly, both ATD and AAE started to increase as there are more candidate nodes to be explored and it is more likely for the NGA to find a sub-optimal solution.
7 Conclusion
In this paper, we propose a deep learning guided evolutionary algorithm DLGEA for CSI problems which optimises the search space of EAs and thus speed up the searching efficiency. In particular, we use a DNN model to localise the set of nodes in a WDN that are most likely to be the contamination source nodes, based on which an EA can identify the contamination events more quickly. To train the DNN model, we exploit the contamination data obtained from the simulation tool EPANET. By carrying out a set of experiments, we show that the DLGEA outperforms the CSI method purely based on EAs in terms of accuracy, average topological distance as well as the accumulated concentration error. Moreover, we investigated the performance of DLGEA with both a medium-size WDN and a large-scale WDN, and showed empirically that DNN+EA outperforms EA under both settings. We also carried out a sensitivity study to prove the effectiveness of DLGEA under different layouts of sensor nodes with varying sensing capabilities and to investigate the performance of DLGEA with different values of K (top K nodes returned by the DNN models).
There are several directions for future work. First, we intend to train DNN models to provide an estimation of the other variables of the contamination event, i.e. starting time, duration, concentration, to further reduce the search space of EAs. Another important extension is to improve DLGEA to deal with multiple sources of contamination. Moreover, we will also investigate the possibility of transfer learning, i.e. applying DLGEA to an unseen WDN.
References
Bäck T, Fogel DB, Michalewicz Z (1997) Handbook of evolutionary computation. CRC Press, Boca Raton
Chung J, Gulcehre C, Cho K, Bengio Y (2014) Empirical evaluation of gated recurrent neural networks on sequence modelling. In: Proceedings of the NIPS 2014 workshop on deep learning and representation learning
Dauphin YN, Fan A, Auli M, Grangier D (2017) Language modelling with gated convolutional networks. In: Proceedings of the 34th international conference on machine learning, pp 933–941. JMLR.org
Graves A, Mohamed A, Hinton G (2013) Speech recognition with deep recurrent neural networks. In: Proceedings of the IEEE international conference on Acoustics, Speech and Signal Processing, pp 6645–6649
Guan J, Aral MM, Maslia ML, Grayman WM (2006) Identification of contaminant sources in water distribution systems using simulation-optimization method: case study. J Water Resour Plan Manag 132(4):252–262
He J, Yao X (2003) Towards an analytic framework for analysing the computation time of evolutionary algorithms. Artif Intell 145(1):59–97. https://doi.org/10.1016/S0004-3702(02)00381-8
Hochreiter S, Schmidhuber J (1997) Long short-term memory. Neural Comput 9(8):1735–1780
Holland JH (1992) Genetic algorithms. Sci Am 267(1):66–73
Hu C, Zhao J, Yan X, Zeng D, Guo S (2015) A mapreduce based parallel niche genetic algorithm for contaminant source identification in water distribution network. Ad Hoc Netw 35:116–126 (Special Issue on Big Data Inspired Data Sensing, Processing and Networking Technologies)
Huang JJ, McBean EA (2009) Data mining to identify contaminant event locations in water distribution systems. J Water Resour Plan Manag
Kong Q, Cao Y, Iqbal T, Wang Y, Wang W, Plumbley MD (2020) Panns: Large-scale pretrained audio neural networks for audio pattern recognition. IEEE/ACM Trans Audio Speech Lang Process 28:2880–2894
Krizhevsky A, Sutskever I, Hinton GE (2017) Imagenet classification with deep convolutional neural networks. Commun ACM 60(6):84–90
Laird CD, Biegler LT, van Bloemen Waanders BG (2006) Mixed-integer approach for obtaining unique solutions in source inversion of water networks. J Water Resour Plan Manag 132(4):242–251
Laird CD, Biegler LT, van Bloemen Waanders BG, Bartlett RA (2005) Contamination source determination for water networks. J Water Resour Plan Manag 131(2):125–134
Lippai I (2020) Wolf-Cordera Ranch. http://emps.exeter.ac.uk/engineering/research/cws/resources/benchmarks/expansion/wolf-cordera-ranch.php. [Online; Accessed 19 June 2020]
Liu L, Ranjithan SR, Mahinthakumar G (2010) Contamination source identification in water distribution systems using an adaptive dynamic optimization procedure. J Water Resour Plan Manag 137(2):183–192
Liu L, Zechman EM, Mahinthakumar G, Ranjithan SR (2012) Identifying contaminant sources for water distribution systems using a hybrid method. Civil Eng Environ Syst 29(2):123–136
Mahfoud SW (1995) Niching methods for genetic algorithms. Ph.D. thesis, University of Illinois at Urbana-Champaign
Perelman L, Ostfeld A (2011) Topological clustering for water distribution systems analysis. Environ Model Softw 26(7):969–972
Perelman L, Ostfeld A (2013) Bayesian networks for source intrusion detection. J Water Resour Plan Manag 139
Preis A, Ostfeld A (2006) Contamination source identification in water systems: a hybrid model trees-linear programming scheme. J Water Resour Plan Manag 132(4):263–273
Preis A, Ostfeld A (2007) A contamination source identification model for water distribution system security. Eng Optim 39(8):941–947
Qian K, Jiang J, Ding Y, Yang S (2020) Deep learning based anomaly detection in water distribution systems. In: Proceedings of the 2020 IEEE international conference on networking, sensing and control (ICNSC), pp 1–6
Quiñones-Grueiro M, Bernal-de Lázaro JM, Verde C, Prieto-Moreno A, Llanes-Santiago O (2018) Comparison of classifiers for leak location in water distribution networks. IFAC PapersOnLine 51(24):407–413
Rethage D, Pons J, Serra X (2018) A wavenet for speech denoising. In: Proceedings of the 2018 IEEE international conference on acoustics, speech and signal processing, pp 5069–5073
Rossman LA (2000) Epanet 2: Users manual
Sanctis AED, Shang F, Uber JG (2010) Real-time identification of possible contamination sources using network backtracking methods. J Water Resour Plan Manag 136(4):444–453. https://doi.org/10.1061/(ASCE)WR.1943-5452.0000050
Shang F, Uber JG, Polycarpou MM (2002) Particle backtracking algorithm for water distribution system analysis. J Environ Eng 128(5):441–450
Soldevila A, Blesa J, Tornil-Sin S, Duviella E, Fernandez-Canti RM, Puig V (2016) Leak localization in water distribution networks using a mixed model-based/data-driven approach. Control Eng Pract 55:162–173
Vankayala P, Sankarasubramanian A, Ranjithan SR, Mahinthakumar G (2009) Contaminant source identification in water distribution networks under conditions of demand uncertainty. Environ Foren 10(3):253–263
Wang H, Harrison KW (2013) Bayesian approach to contaminant source characterization in water distribution systems: adaptive sampling framework. Stoch Environ Res Risk Assess 27:1921–1928
Wang H, Harrison KW (2014) Improving efficiency of the bayesian approach to water distribution contaminant source characterization with support vector regression. Environ Model Softw 140
Yan X, Gong J, Wu Q (2020) Pollution source intelligent location algorithm in water quality sensor networks. Neural Comput Appl
Yan X, Gong W, Wu Q (2017) Contaminant source identification of water distribution networks using cultural algorithm. Concurr Comput Pract Exp 29(24):1–11
Yan X, Li T, Hu C (2019) Real-time localization of pollution source for urban water supply network in emergencies. Clust Comput 22:5941–5954
Yan X, Sun J, Hu C (2017) Research on contaminant sources identification of uncertainty water demand using genetic algorithm. Clust Comput 20(2):1007–1016
Yan X, Yang K, Hu C (2018) Pollution source positioning in a water supply network based on expensive optimization. Desalin Water Treat 110:308–318
Yan X, Zhao J, Hu C, Zeng D (2019) Multimodal optimization problem in contamination source determination of water supply networks. Swarm Evolut Comput 47:66–71
Yan X, Zhu Z, Li T (2019) Pollution source localization in an urban water supply network based on dynamic water demand. Environ Sci Pollut Res 26:17901–17910
Yu Y, Zhou ZH (2008) A new approach to estimating the expected first hitting time of evolutionary algorithms. Artif Intell 172(15):1809–1832. https://doi.org/10.1016/j.artint.2008.07.001
Acknowledgements
This research is supported by the National Natural Science Foundation of China under Grant No. 61873119, the National Key R&D Program of China under Grant No. 2019YFC0810705 and the Science and Technology Innovation Commission of Shenzhen under Grant No. KQJSCX20180322151418232.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix: Layouts of sensor nodes
Appendix: Layouts of sensor nodes
Figures 6, 7, 8 and 9 show the locations of the 4 sensor nodes in WDN\(_1\) with respect to the four layouts of sensor nodes used in this paper. The blue triangles indicate the locations of the sensor nodes and the numbers next to the blue triangles indicate the ID of the sensor nodes in \(\hbox {WDN}_1\).
Rights and permissions
About this article
Cite this article
Qian, K., Jiang, J., Ding, Y. et al. DLGEA: a deep learning guided evolutionary algorithm for water contamination source identification. Neural Comput & Applic 33, 11889–11903 (2021). https://doi.org/10.1007/s00521-021-05894-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-021-05894-y