Abstract
A modified Artificial Bee Colony (ABC) metaheuristics optimization technique is applied for cancer classification, that reduces the classifier’s prediction errors and allows for faster convergence by selecting informative genes. Cuckoo search (CS) algorithm was used in the onlooker bee phase (exploitation phase) of ABC to boost performance by maintaining the balance between exploration and exploitation of ABC. Tuned the modified ABC algorithm by using Naïve Bayes (NB) classifiers to improve the further accuracy of the model. Independent Component Analysis (ICA) is used for dimensionality reduction. In the first step, the reduced dataset is optimized by using Modified ABC and after that, in the second step, the optimized dataset is used to train the NB classifier. Extensive experiments were performed for comprehensive comparative analysis of the proposed algorithm with well-known metaheuristic algorithms, namely Genetic Algorithm (GA) when used with the same framework for the classification of six high-dimensional cancer datasets. The comparison results showed that the proposed model with the CS algorithm achieves the highest performance as maximum classification accuracy with less count of selected genes. This shows the effectiveness of the proposed algorithm which is validated using ANOVA for cancer classification.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The expression levels of the gene in an organism play a discriminant role in clinical studies and the management of several diseases. Microarray technology is a powerful approach for genomic research that creates many analytical challenges for data scientists. Microarray technology uses sophisticated techniques of biomarkers to identify the expressed informative gene. The experimentation with reliable cancer biomarkers plays an important in the field of clinical diagnosis (Ong 2020). Classification and analysis of various genetically linked diseases are possible through Microarray technology, which is the most widely used tool in the prognosis of different types of cancer. Accurate prediction of cancer is significant in contributing to effective treatment for the patients. The cancer classification based on gene expression profiles has provided better transparency for the possible treatment strategies. Recently, because of the increase in data generation and storage, various big data applications gain attention, which also increased interest in applying them to a wide range of biological problems (Li et al. 2017; Dwivedi 2018). The identification of genes plays an important role in detecting cancer diseases and has an essential impact for microarray cancer prediction (Selaru et al. 2002; Elek et al. 1999). Therefore dimension reduction followed by the classification acts as the major process for further analysis. However, there are difficulties in detecting cancer from microarray due to the presents of a large amount of gene expression levels in the human body (Salem et al. 2017; Garro et al. 2016). The input sets of features (genes) are the main factor that influences the quality of the performance of classification algorithms. If the features are relevant to the class labels, the classifier will be able to create a strong relationship between them. However, in most scenarios, the relevancy of features is often unknown and usually, the input data sets have issues such as irrelevancy and redundancy that are not useful during the knowledge discovery process. Thus, this can hinder the process of producing a positive classification. Machine learning techniques for data reduction require knowledge about relevant features and can substantially reduce the size of data for learning algorithms by reducing unnecessary and redundant features. In general, high-dimensional microarray data sets are difficult to interpret and data interpretation is very essential for the treatment of cancer patients. The initial studies of dimensionalities reduction problem, found that the best test error can be attained through a limited number of features (genes) that directly affect the accuracy rates (Aziz et al. 2017a; Lv et al. 2016). In a large feature space, it is common to have irrelevant and redundant genes concerning the class labels. Integrality constraints such as irrelevant and redundant features have the capacity to affect the classification performances. Therefore, this research study developed an approach for genes selection to counter all the mentioned drawbacks.
Defining an optimal decision framework for gene selection is an essential but difficult task in the field of machine learning and medical science from microarray data because the characteristic of each data is different. Recently hybrid machine learning techniques gain popularity and by using suitable ccombinations effectively obtain a few relevant and informative genes (features) (Aziz et al. 2017a). Various researches applied varieties of different data mining techniques with different combinations for the problem of identification of significant genes. Motivated by previous researchersnature-inspireded algorithms are more suitable to find optimal set of features from large and complex data of different domains. Techniques comprised of metaheuristic optimization have a broad range from the process of a local search to learning processes (Turgut et al. 2018). Nature inspired algorithm by conducting them over the search space there by bringing out its best capabilities able to obtain the best of best solutions.
Othman et al., proposed a hybrid multi-objective cuckoo search with evolutionary operators for gene selection. The evolutionary operators used in this study were double mutation and single crossover operators. Proposed approach was tested on seven publicly available, high-dimensional cancer microarray data sets. The experimental results concluded that the proposed work outperformed cuckoo search and multi-objective cuckoo search algorithms with a smaller number of selected significant genes (Othman et al. 2020).
Dash et al., proposed hybridized harmony search optimization approach for feature selection in high-dimensional data classification problem. Proposed technique select optimal minimum number of top ranked genes that provided good classification. The experental results on four well known microarray datasets showed that performance of proposed algorithms was better than other published algorithm for the same problem (Dash et al. 2021).
Hybrid approach is used to reduce the computational time and to take the benefits of the different dimension reduction method (Mafarja et al. 2020; Venkatesh and Anuradha 2019). Hybrid approaches combine different feature selection and extraction method to reduce the dimension of the data. Different researchers applied different combination of algorithm according to the requirement of different data sets.
Hameed et al., compared the performance of three well-known nature-inspired metaheuristic algorithms, namely binary particle swarm optimization (BPSO), genetic algorithm (GA) and cuckoo search algorithm (CS) with tewelve cancer data sets for gene selection and classification. In terms of accuracy, BPSO outscored GA and CS, according to the study. In comparison with GA and BPSO, CS was able to pick fewer attributable genes and was less computationally complex (Hameed 2021).
Some researchers created a classification framework and utilized it to categorise cancer gene expression patterns using various hybrid gene selection algorithms based on various nature-inspired metaheuristic methodologies, with better outcomes than single approaches. The work not only selected very few features but also reduced computational cost by using the collection of new techniques that produced good performance in classification. A comparison result expresses that the proposed hybrid approach have been successfully applied and excels with other existing methods in terms of accuracy (Baburaj 2022; Alomari 2021; Kumar and Bharti 2021). Therefore in this paper, hybrid approach based on Nature Inspired Metaheuristics technique is proposed that can produce an optimal feature space with significant genes to improve the classification performance.
Independent component analysis (ICA) method is used for finding underlying components (features) from multidimensional statistical data. The extracted components of ICA are statistically independent, this property distinguishes ICA from other methods (Hyvarinen et al. 2001). Recently, ICA feature extraction method gain attention as effective genes reduction technique of microarray data for NB classifier (Hasan et al. 2021; Li 2021). The embedded algorithm of the NB classifier based on conditional independence hypothesis, ICA technique resolved successfully this condition as the ICA extracted components (features) are statistically independent. The major problem of ICA technique is how to obtained best subsets of features from the extracted genes that enhance the classification accuracy of NB classifier. One of the author of Fan et al. (2009a) used ICA extraction method with stepwise regression for feature selection on five benchmark microarray datasets and proposed approach demonstrated in improving the classification performance of NB classifier. In Mollaee et al. (2016) proposed a three level ensemble approach for cancer prediction and classification. For gene selection firstly ensemble Fisher ratio and T test after that optimize the gene with PSO-dICA method then classified five cancer microarray data sets and found satisfactory results compare to other published results. In Mahdavi et al. (2019) selected the feature subset by using ICA that extracted essential information and at the same time separated the noise as extracted features were an independent component. The results of data sets have shown the effectiveness and improvement of the proposed approach. In Aziz et al. (2017b; c) the author used nature inspired metaheuristic techniques-based wrapper methods with ICA and found ICA increase the classification accuracy of different classification algorithm for cancer microarray profile.
Other than above nature inspired algorithm, cuckoo search algorithm is the most popular swarm intelligence algorithm, that is motivated by the egg-laying behavior of cuckoo birds. Recently, CS algorithm gain more popularity in feature selection (Pandey et al. 2020), multi-objectivetive optimization problem (Cui et al. 2019; Peng 2021), data clustering (Pandey and Rajpoot 2019), disease detection (Cristin 2020), path planning (Song et al. 2020) and soon. The literature have shown that the CS is an effective approach to solve numerous optimization problems of different domain. For different research problems CS algorithm has been widely used, but at the same time for complexity optimization problems CS still need some improvement on exploration face. On the other hand, ABC is widely used approach for finding best number of features for continuous optimization problems of microarray data (Musheer et al. 2019; Coleto-Alcudia and Vega-Rodríguez 2020; Wang 2020). ABC approach work with the help of three bees for finding best solution (food source) and gives more accurate results comparison to the other swarm-based meta-heuristic algorithm. In ABC technique, three types of bees manage the global search and local search procedure for finding best solution. The global search of ABC algorithm for finding a new solution is better in contrast to the other nature-inspired algorithms. Some authors, to maintain the balance between local search and global search procedures, some improvements with different algorithms in the onlooker bee phase (local search) of ABC are proposed that improved the performance of ABC (Alshamlan et al. 2015).
-
A.
The objective of the paper
-
•
To improved the performance of feature extraction technique (independent component analysis) by using hybrid approach.
-
•
Proposed nature inspired hybrid algorithm for solving the existing gene selection process of microarray data based on a soft computing technique.
-
•
Proposed novel framework of classification to increse the perfomance of the NB classifier algorithm by utilizing ICA with improved ABC algorithm.
1.1 Paper organization
Thus, this article would like to focus on the nature inspired metaheuristic hybrid algorithm in solving the existing gene selection process of microarray data for accurate classification of cancer. The improved ABC algorithm is used to extract the optimal features from ICA feature vectors of microarray data in this research, after that compared the obtained results of proposed framework with others recently published model of NB classifier. The remainder of this research paper is structured as follows, Section two, described the details of feature selection algorithm, Experimental setup is provided in Section three. While Section fourth discussed the experimental results and Section fifth presented the conclusion. Figure 1 shows the proposed framework.
2 Proposed algorithm
2.1 ICA gene extraction method
ICA helps in obtaining hidden features from multi-dimensional information by decomposing multivariate indications into independent nongaussian sections for the components to be statistically independent (Hyvarinen et al. 2001; Mollaee and Moattar 2016). ICA finds correlation between data by decorrelating the data by exploiting or diminishing the distinct in formation. In ICA algorithm all features X treated as a independent components S. If A signify the opposite matrix of a weighted matrix W, and columns of A characterize the source feature vectors of comment X.
ICA has been extensively utilized for biological information, recognitions and also applicable for other domain. More detailed of ICA extraction method can be found elsewhere (Aziz et al. 2016; Hsu et al. 2010).
2.2 ABC optimization method
Recently nature inspired optimization algorithm ABC is popular for genes selection problem of the microarray. The step of ABC approach is based on bees behavior for finding best food source (gene subset). Most of the researchers for the problem of different domain used ABC approach for optimization of the solution. ABC technique work with three classes of bees i.e. employed bees, onlooker bees and scouts bees by using local search and global search techniques (Garro et al. 2016). These three classes of bees convergence the problem and find the optimal solution with different effort in the different steps of algorithm. Details information about ABC can be seen in reference (Musheer et al. 2019; Coleto-Alcudia and Vega-Rodríguez 2020; Wang 2020).
2.3 Cuckoo search algorithm(CS)
CS optimization algorithms is based on the reproduction behavior of cuckoo birds. It is developed by using Levy flight rather than the isotropic random walks with infinite variance and mean which cause much longer step from its current position (Cristin 2020). Basically, the cuckoo lay one or several eggs in other birds nests which aim to ensure the continuity of their generation by directing the host birds to their natural instinct of breed, hatch, and provide food to the baby cuckoos. Three idealized steps of CS are given below (Cui et al. 2019; Peng 2021):
-
I.
A cuckoo lays one egg at a time and placed its egg randomly chosen nest of other birds.
-
II.
The best nest with the high quality of (solutions) eggs will carry over to the next generations.
-
III.
The availability number of host nests is secured and the egg laid by the cuckoo is detected with the probability, Pa ϵ [1, 0]. While, the host bird either abandon the nest or throw the egg and form a new nest. The final assumption can be estimated by the fraction Pa of n nests are exchanged with new nests with randomized results. This might increase the survival and reproductive capacity of cuckoo birds, so compare to other algorithm exploitation process of CS algorithm is more efficient (Shehab et al. 2017).
2.4 Proposed algorithm
The searching process of the optimal solution in original ABC approach based on cycle that contains three phases (Garro et al. 2016).
-
Employed bees phase: employed bees search the food sources and estimate their nectar amounts then sending the all information regarding the food sources to the onlookers.
-
Onlookers phase: the behavior of onlookers is different of employers, on the basis of received information onlooker make a decision by estimating the nectar amount of all food sources.
-
Scouts bee phase: determining the scout because the employed bee of an abandoned food source becomes a scout. Therefore, the employed and onlookers bees manage the exploitation process, on the otherhand scouts bees manage the exploration process in the searchspace.
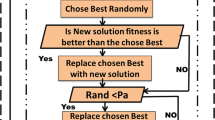
For finding the optimal subset with ABC algorithm, appropriate equilibrium amongst exploration and exploitation is essential (Garro et al. 2016). The exploration process of ABC algorithm for finding a new solution compare to the other nature expired algorithm is good, but the exploitation process required more computational time for converge to the optimal solution (Aziz et al. 2017b; Garro et al. 2015). Therefore to decrease the computational time of exploitation process of ABC, the proposed algorithm used CS (Zhu and Wang 2019). The CS algorithm has good exploitation process but it ability for searching optimal novel solution in search space is not good compare to ABC algorithm (Kıran et al. 2012; Jatoth and Rajasekhar 2010). That is why the CS obtained the local optima too quickly and suffers from the preconvergence problem which is the main issue with CS algorithm (Chen and Yu 2019). Therefore, to maintain the balanced between exploitation and exploration process, CSABC algorithm uses combination of ABC and CS algorithms. In proposed algorithm, CS algorithm is adopted in the onlooker bee phase as an exploitation process to find the best optimal solution with less computational time by improving the formation sharing between onlooker and employee bees. Furthermore, the idea of using ABC for gene selection with a CS algorithm based on researches (Ding et al. 2018). Therefore, the proposed approach uses a combination of nature-inspired metaheuristic algorithms, to reduced disadvantages of the ABC approach such as preconvergence and computational time by maintaining balanced between exploration and exploitation. Figure 2 shows how CSABC works. The code of the CSABC approach is shown in below.
Algorithm for dimensionality reduction
Feature extraction by ICA extraction method
-
1.
Firstly reduced the size of microarray dataset with ICA algorithm.
Optimizition of ICA feature vector with (CSABC) algorithm.
Next CSABC algorithm is applied for finding the best genes set from the ICA feature vector for NB classification. A main issue related with ICA is, it normally extracted number of features are equal to the sample size (m), therefore again 2 m genes sets exist (Zheng et al. 2008).

-
Return a best solution (important and relevant genes for prediction).
-
Train the NB algorithm with best obtained features.
-
Classify test set with selected features by using NB classifier.
-
Return the classification accuracy.
2.5 NB classifier
NB is famous supervised learning technique in the field of machine learning; it is widely used by many researchers to classify the objects into two or more classes by means of using Bayes theorem (Friedman et al. 1997; Hall 2007). It is used widely, when the input variable is continuous and independent then the parameters are estimated by the Bayes rule, so that the probability of output variable is exactly predicted. If E1, E2,..., En. are the selected genes from any sample of H, Naïve Bayes classifier classified the samples by using below formula with Bayes theorem as a Naïve Bayes classifier(Aziz et al. 2016; Fan et al. 2009b):
Because features of microarray are continuous so for the calculation of class-conditional probability, \(f( \, {\text{.|H}})\), probability density function with nonparametric kernel density estimation method, for each attributes is used and \(P({\text{ H}})\) is the prior probability of the particular class.
3 Experimental setup
To evaluate the performance of the proposed approach in this research used six benchmark microarray cancer datasets. In this paper, used six cancer benchmark data sets of gene expression, namely; Colon cancer (Alon et al. 1999), Acute leukemia (Golub et al. 1999), Prostate cancer (Singh et al. 2002), Lung cancer-II (Gordon et al. 2002), High-grade Glioma data (Nutt et al., 2003) of binary classification and Leukemia 2 (Armstrong et al., 2002) of multi classification. These datasets, downloaded from Kent ridge; an online repository of high-dimensional biomedical datasets (http://datam.i2r.astar.edu.sg/datasets/krbd/index.html). Table 1 shows the full detail and properties of these six datasets.
NB classifier uses either kernel density estimation or Gaussian distribution estimation for data classification according to the nature of the data. Since microarray data contain continuous feature, in this paper kernel density approximation is applied with NB classifier (Rabia et al. 2015; Campos, et al. 2011). The performance of the proposed approach was examined with two parameters classification accuracy of NB classifier and smallest number of obtained genes and the for all six datatsets. Classification accuracy of the NB classifier is evaluated by the below formula:
where, CC means correct classified samplesand N is the total number of samples in the respective class. For finiding unbiased results, In this paper implemented Leave One Out Cross-Validation (LOOCV) method. To facilitates examination of proposed approach, repeated these experiments with LOOCV at several time. Three other features selection method are also consider for comparing the results of proposed algorithm with the same parameter. The parameters of CS and ABC are adopted with the help of several research paper. For purpose of fair comparison, the same top-ranked genes were chosen for all gene selection method. Experiments were performed for ICA extracted genes in each datasets. To evaluate the performance of the proposed approach in this research, all experimental work on preprocessing the datasets has been done using MATLAB 2016b. Moreover, we used python-based tool for feature selection and classification. The experiments were conducted on a desktop computer running 64-bit Windows 10 with an Intel (R) Core (TM) i7-3770 CPU at 3.40 GHz and 8 GB of RAM. The code of ICA algorithm as the FastICA package, ABC algorithm and CS algorithm are freely available on internet.
3.1 Parameter setting and fitness function of proposed algorithm
Recently some of the researchers used Genetic Bee Colony (GBC) approach for selection of features and found better result compared to original ABC (Alshamlan et al. 2015). Genetic Bee Colony (GBC) technique is a combination ABC and Genetic Algorithm (GA) algorithm. GBC is a novel technique, purpose of GBC techniques is to find best subsets of gene for improving the accuracy of different classification algorithm. Therefore, for the sake of a fair comparison, the result of the proposed approach compared with the GBC-based hybrid algorithm called (ICA + GBC) with the same classifier. The parameters of proposed approach (ICA + CSABC) and (ICA + GBC) approach that was used in our experiments are given in below tables. For purpose of fair comparison, the same top-ranked genes were chosen for all gene selection method. Experiments were performed for ICA extracted genes in each datasets.
Parameter setting of proposed algorithm
Parameter | Value |
|---|---|
Colony size | 50 |
Max cycle | 100 |
Number of runs | 30 runs |
\(\mathrm{Levy} \left(s,\uplambda \right)\) | \({s}^{-\uplambda }\),1 < λ < \(\le 3\) |
\(\alpha \mathrm{Step size}\) | 1.0 |
\({P}_{{a}_{\mathrm{min}}}\) | 0.3 |
\({P}_{{a}_{\mathrm{max}}}\) | 0.5 |
Limit | 5 iterations |
Parameter setting of (ICA + GBC) algorithm
Parameter | Value |
|---|---|
Colony size | 50 |
Max cycle | 100 |
Number of runs | 30 runs |
\(\mathrm{Mutation Pro}bability\) | 0.02 |
\(\mathrm{Crossover Pro}bability\) | o.75 |
Limit | 5 iterations |
3.1.1 Evaluating new solutions and levy flight
CSABC-based feature selection approach find the new optimal solutions with random cuckoo by modifying the parameters using Lévy flight with below equation.
Levy walk generates some new solutions around the obtained best solutions, will accelerate the local search functionality. Here, \(C\) has been set to 0.85 from experience. Next evaluate its quality with fitness function of algorithm.
3.1.2 Fitness function
Fitness function of proposed approach is evaluated by classification accuracy of NB classifier. If the current fitness value is better than the previous one, then skip the previous result, and moves to the current; else it retains the previous solution. Finally, the fitness solution with highest value is returned for best predictive gene subset. The fitness function (fit) is defined as follows:
Where, Accuracy (fit) is Testing data (f) classifier accuracy.
3.1.3 Parameter Pa
It represents the probability of discovery of an egg. The Pa value is modified dynamically in modified cuckoo search using below equation.
\({P}_{{a}_{\mathrm{max}}}\) and \({P}_{{a}_{\mathrm{min}}}\) is set 0.5 and 0.3, respectively.
4 Experimental results and discussions
Tables 2, 3, 4, 5, 6, 7 shows the LOOCV classification accuracy of NB classifiers with different (ICA + CSABC) and (ICA + GBC) approached for above-explained six microarray datasets. The best results of all data sets (smallest selected gene size and highest classification accuracy) are highlighted using bold font. Figures 3a–d, 4, 5a–d, illustrates the output of the proposed work in the term of AUC curve on six datasets with different threshold, for best subsets genes found by (ICA + CSABC) and (ICA + GBC) approach. From Figs. 3a–d, 4, 5a–d and Tables 2, 3, 4, 5, 6, 7 give following results.
Regarding colon dataset classification accuracy of test data with the proposed approach achieved 99.13%, 92.31% and 88.07%, best mean and worst, respectively, which defeated (ICA + GBC) with best mean and worst by 2.62%, 0.46% and 1.14%, respectively. The best ROC with proposed approach was obtained 96.36 with 12 gene where as ROC value with ICA + GBC was 95.66 with 16 genes for Colon data, which shows that the proposed approach has a discrimination capability between two classes.
Based on the results of test data of Lekumia, the proposed approach obtained 95.24% on an average classification accuracy which is best compared to ICA + GBC and the other hybrid method that used in this experiment. On the other hand the proposed approach obtained 9 important and predictive gene from 72 extracted genes of ICA, which is smallest number of gene compare to obtained genes by the other competitor approaches.
The experimental Tables 4 and 8 for prostate data depicted that proposed approach provides 100% best classification accuracy that showed improvement over ICA + GBC, ICA + ABC, ICA + GA, and ICA + PSO, respectively, for the NB classifier. The AUC value of prostate data found 98.96 with proposed approach for 12 genes.
For the High-grade Glioma dataset, the best training and testing classification accuracy with all ICA features was 87.88% and 70.55%, while with the CSABC wrapper method with NB classifier increases training and testing accuracy 98.11% and 96.82%, respectively. Therefore, CSABC become a commanding optimization technique for obtaining best feature with ICA for Naïve Bayes classifier.
With respect to the test dataset of Lung cancer II dataset, proposed approach obtained best classification accuracy 93.45%, that indicates proposed approach applicable to classify all most all samples in their defined classes. Secondly, proposed approach obtained 24 features from 181 extracted features of ICA, which is low compare to other applied approaches.
In the case of multiclass classification of Lukemia 2 dataset, the proposed approach obtained slightly lower classification accuracy compare to ICA + GBC but better classification accuracy compare to other three method (Table 8). But it gives an advantage for obtaining the smallest number of informative and predictive genes, for the NB classifier its obtained 15 genes for the highest classification accuracy which is low compared with 18 obtained genes by the ICA + GBC method.
The proposed technique had effectively performance. It is proved from the experimental results, these outputs were stable and improved compared to the output acquired from the previous experiment. In summary, the proposed technique reached on average 93% for all datasets in terms of classification accuracy which highlight the strength of the proposed technique. Therefore, the experimental result proved, compare to ICA + GBC and other three hybrid methods, the proposed approach has a more significant ability for classifying different samples in their correct classes with NB classifier.
Table 9 displays the ICA + CSABC comparative results with five papular features selection approach over six cancer datasets with the same framework. In order to make fair evaluation and comparison, we applied same control parameters that have been used for our proposed algorithms when combined with ICA. Table 8 lists the parameters of several comparison algorithms as determined by a study of numerous related research papers. Such parameter setting was optimized by literatures moreover, we conducted many trials to test such parameter setting which shows the best objective value. We first extract gene by using ICA next optimized the extracted gene by using GA, PSO, ABC and GBC and evaluated its performance in terms of two evaluation criteria: classification accuracy number of selected genes Then we compared the results of algorithms named ICA + PSO, ICA + GA, ICA + CS, ICA + ABC, ICA + GBC with ICA + CSABC by using the same model for the sake of a fair comparison. The NB classifier served as a fitness function of these gene selection methods. In order to avoid selection bias, the LOOCV was used.
Experimental results revealed that the ICA + CSABC approach obtained the highest best classification accuracy (97.70 percent) of the six cancer datasets, on the other hand best classification accuracy of the other methods was 96.73 percent, 95.98 percent, 94.42 percent and 92.94 percent for ICA + GBC, ICA + ABC, ICA + GA, and ICA + PSO, respectively. This means that the ICA + CSABC algorithm has made the NB classification output more reliable and accurate. The ICA + CSABC algorithm also obtained the smallest number of optimal genes set that contain average (13.50) genes and smallest number of optimal genes set was 14.67, 15.83, 23.67 and 29.17 for ICA + GBC, ICA + ABC, ICA + GA and ICA + PSO. Such findings of the assessment indicate that ICA + CSABC is a promising technique for resolving the problems of dimension reduction and microarray data classification.
From Fig. 6, the positive outcome with the highest classification accuracy of NB classifier with features obtained by CSABC from ICA feature vectors is clearly visible. This is not surprising because the ICA technique has the ability to resolve the classification criteria of NB classifier.
For the comparision of above five feature selection techniques, a statistical hypothesis test ANOVA was applied with α = 0.05 to determine whether there exists a significant difference between them or not. The different approach shown in Fig. 7 by the name ICA + PSO, ICA + GA, ICA + GBC, ICA + ABC and ICA + CSABC, are selected as group 1 to 5, respectively, in the study. ANOVA tests the hypothesis with H0 and H1.
H0: α1 = α2 = α5 (all group means are equal).
H1: not all group means are equal.
In Fig. 7, the blue-shaded line shows the comparison interval for (ICA + CSABC) group mean. Gray line shows the comparison intervals for (ICA + GBC) and (ICA + ABC) and the red line shows the comparison interval of (ICA + PSO) and (ICA + GA). The comparison interval (ICA + GBC) and (ICA + ABC) overlap and comparison interval (ICA + GA) and (ICA + PSO) does not overlaps with the comparison interval (ICA + CSABC) group mean. Therefore, the group means (ICA + GBC) and (ICA + ABC) are not significantly different but the group means (ICA + GA) and (ICA + PSO) are significantly different from (ICA + CSABC) group mean.
For further comparisons, the proposed algorithm employed with SVM classifier, because SVM is the best and widely used classifier for data classification as it is less sensitive over the high dimension(Aziz et al. 2022a; Desai et al. 2022). Since microarray data are a type of nonlinear classification problem, therefore employed SVM with polynomial kernel, all other parameter of SVM are used on the basis of the research with LOOCV process (Aziz et al. 2022b; Xi, et al. 2016). Firstly applied ICA technique for feature extraction in the second improved ABC (CSABC) applied to optimize ICA extracted feature with SVM classifier and lastly analyze the classification accuracy.
Table 10 and Fig. 8 summarize the classification accuracy and error rate of NB and SVM with proposed approach by using LOOCV iterations for the same parameter settings (CSABC) algorithms. We can easily see from Table 9 SVM with proposed approach aslo produced good classification accuracy which is little bit less or equal to the classification accuracy of NB classifier for Colon, Acute, Prostate, High grate Glioma data and Leukemia 2 datasets. For Colon data set both the classifiers gives same classification accuracy with different number of selected genes. On the other hand for Acute, Prostate and High grate Glioma datasets SVM achived less bit more error rate compare to NB classifier (Fig. 8). While for Lung cancer II data, SVM achived mean accuracy rate 90.34% with 20 genes which is greater than NB classification rate. The error rate of the SVM and NB in classifying Lekumia 2 data was 7.67% with 12 genes and 6.33% with 15 genes, respectively, so in the term of classification accuracy NB classifier is best but the SVM classifier is the best in the term of selected genes for Lekumia 2 data.
Figure 9 shows smallest number of genes that give best performance of NB classifier; blue and red line show the best number of genes for SVM and NB classifier, respectively, with all six datasets. The number of obtained genes able to show that the number of important and relevant genes in the cancer microarray data are lesser than 50% of the ICA extracted genes. It is seen from Fig. 9, for Colon cancer data out of 62 ICA vectors proposed approach choose only 12 gene for best classification accuracy that show 19.35% of genes were relevant and important for classification. Thus, 80.65% genes are non relevant to disease and cause noise in order to make a good classification model. For other datasets 12.50% of Acute, 11.70% of Prostate, 18.40% of High grade, 13.25% of Lung-II and 20.08% of Lukemia 2 data of ICA feature vector are important for NB classification. Similar result in the term of selected genes found with SVM classifier for all datasets, for Colon and Leukemia 2 data SVM selected 10 and 15 genes which is less compare to NB classifier. For Acute, Prostat and Lung cancer II data SVM selected 12, 14 and 24 genes, respectively, which is slightly more then NB classifier. Therefore, proposed algorithm identify the important and relevant genes that constructed the best classification model for both the classifiers. The selected number of genes for Lung cancer II data both the classifier required little bit more gene compare to other datasets because every dataset has its own different characteristics.
5 Conclusion
This paper proposes, novel metahuristic hybreid approach by combining the ICA and advantages of cuckoo search and ABC-based feature optimization apprproach with NB classifier for cancer Classification. This method was successes fully reduced the misclassification errors during the classification process on six cancer microarray data. Experimental results show the superiority of the proposed approach in the term of classification accuracy with two factors, best obtained less number of genes set and best AUC score for unbiased accuracy. Therefore, metaheuristic nature-inspired algorithms act as a strong tool in solving microarray cancer data classification problems.
In future work, incorporating more than one classifier with proposed feature selection techniques to enhance the classification accuracy of the proposed work and to examine the selective classifier mode.
Data availability
All used data are benchmark high-dimensional microarray datasets of cancer and are freely available in different repositories.
References
Alomari OA, et al. (2021) Gene selection for microarray data classification based on Gray Wolf Optimizer enhanced with TRIZ-inspired operators. Knowl Based Syst 223: 107034.
Alon U et al (1999) Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc Natl Acad Sci 96(12):6745–6750
Alshamlan HM, Badr GH, Alohali YA (2015) Genetic bee colony (GBC) algorithm: a new gene selection method for microarray cancer classification. Comput Biol Chem 56:49–60
Armstrong SA et al (2002) MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet 30(1):41–47
Aziz R, Verma C, Srivastava N (2016) A fuzzy based feature selection from independent component subspace for machine learning classification of microarray data. Genom Data.
Aziz R, Verma C, Srivastava N (2017a) Dimension reduction methods for microarray data: a review. AIMS Bioeng 4(2):179–197
Aziz R et al (2017b) Artificial neural network classification of microarray data using new hybrid gene selection method. Int J Data Min Bioinform 17(1):42–65
Aziz R, Verma C, Srivastava N (2017c) A novel approach for dimension reduction of microarray. Comput Biol Chem.
Aziz RM, Hussain A, Sharma P, Kumar P (2022a) Machine learning-based soft computing regression analysis approach for crime data prediction. Karb Int J Mod Sci 8(1):1–19
Aziz RM, Baluch MF, Patel S, Ganie AH (2022b) LGBM: a machine learning approach for Ethereum fraud detection. Int J Inf Technol 13(1):1–11
Baburaj E (2022) Comparative analysis of bio-inspired optimization algorithms in neural network-based data mining classification. Int J Swarm Intell Res (IJSIR) 13(1):1–25
Chen X, Yu K (2019) Hybridizing cuckoo search algorithm with biogeography-based optimization for estimating photovoltaic model parameters. Sol Energy 180:192–206
Coleto-Alcudia V, Vega-Rodríguez MA (2020) Artificial Bee Colony algorithm based on Dominance (ABCD) for a hybrid gene selection method. Knowl Based Syst 205:106323
Cristin R et al (2020) Deep neural network based rider-cuckoo search algorithm for plant disease detection. Artif Intell Rev 2020:1–26
Cui Z et al (2019) A hybrid many-objective cuckoo search algorithm. Soft Comput 23(21):10681–10697
Dash R (2021) An adaptive harmony search approach for gene selection and classification of high dimensional medical data. J King Saud Univ Comput Inform Sci 33(2):195–207
De Campos LM, et al. (2011) Bayesian networks classifiers for gene-expression data. In: Intelligent Systems Design and Applications (ISDA), 2011 11th International Conference on 2011. IEEE.
Desai NP et al (2022) Image processing model with deep learning approach for fish species classification. Turk J Comput Math Educ 13(1):85–99
Ding Z, Lu Z, Liu J (2018) Parameters identification of chaotic systems based on artificial bee colony algorithm combined with cuckoo search strategy. Sci China Technol Sci 61(3):417–426
Dwivedi AK (2018) Artificial neural network model for effective cancer classification using microarray gene expression data. Neural Comput Appl 29(12):1545–1554
Elek J, Park K, Narayanan R (1999) Microarray-based expression profiling in prostate tumors. In Vivo (Athens Greece) 14(1):173–182
Fan L, Poh K-L, Zhou PJESWA (2009a) A sequential feature extraction approach for naïve bayes classification of microarray data 36(6): 9919–9923
Fan L, Poh K-L, Zhou P (2009b) A sequential feature extraction approach for naïve bayes classification of microarray data. Expert Syst Appl 36(6):9919–9923
Friedman N, Geiger D, Goldszmidt M (1997) Bayesian network classifiers. Mach Learn 29(2):131–163
Garro BA, Rodríguez K, Vázquez RA (2015) Classification of DNA microarrays using artificial neural networks and ABC algorithm. Appl Soft Comput.
Garro BA, Rodríguez K, Vázquez RA (2016) Classification of DNA microarrays using artificial neural networks and ABC algorithm. Appl Soft Comput 38:548–560
Golub TR et al (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286(5439):531–537
Gordon GJ et al (2002) Translation of microarray data into clinically relevant cancer diagnostic tests using gene expression ratios in lung cancer and mesothelioma. Can Res 62(17):4963–4967
Hall M (2007) A decision tree-based attribute weighting filter for naive Bayes. Knowl Based Syst 20(2):120–126
Hameed SS et al (2021) A comparative study of nature-inspired metaheuristic algorithms using a three-phase hybrid approach for gene selection and classification in high-dimensional cancer datasets. Soft Comput 2021:1–19
Hasan BMS, Abdulazeez AM (2021) A review of principal component analysis algorithm for dimensionality reduction. J Soft Comput Data Mining 2(1):20–30
Hsu C-C, Chen M-C, Chen L-S (2010) Integrating independent component analysis and support vector machine for multivariate process monitoring. Comput Ind Eng 59(1):145–156
Hyvarinen A, Karhunen J, Oja E (2001) Independent component analysis. Wiley
Jatoth RK, Rajasekhar A (2010) Speed control of pmsm by hybrid genetic artificial bee colony algorithm. In: Communication Control and Computing Technologies (ICCCCT), 2010 IEEE International Conference on IEEE
Kıran MS et al (2012) A novel hybrid approach based on particle swarm optimization and ant colony algorithm to forecast energy demand of Turkey. Energy Convers Manage 53(1):75–83
Kumar L, Bharti KKJNC (2021) A novel hybrid BPSO–SCA approach for feature selection. Natl Comput 20(1): 39–61.
Li G et al (2017) Prediction of biomarkers of oral squamous cell carcinoma using microarray technology. Sci Rep 7:42105
Li J et al (2021) Multi-source feature extraction of rolling bearing compression measurement signal based on independent component analysis. Measurement 172:108908
Lv J et al (2016) A multi-objective heuristic algorithm for gene expression microarray data classification. Expert Syst Appl 59:13–19
Mafarja M et al (2020) Efficient hybrid nature-inspired binary optimizers for feature selection. Cogn Comput 12(1):150–175
Mahdavi K, Labarta J, Gimenez J (2019) Unsupervised feature selection for noisy data. In: International Conference on Advanced Data Mining and Applications. Springer.
Mollaee M, Moattar MH (2016) A novel feature extraction approach based on ensemble feature selection and modified discriminant independent component analysis for microarray data classification. Biocybern Biomed Eng 36(3):521–529
Mollaee M, Moattar MHJB, Engineering B (2016) A novel feature extraction approach based on ensemble feature selection and modified discriminant independent component analysis for microarray data classification. Biocybern Biomed Eng 36(3):521–529
Musheer RA, Verma CK, Srivastava N (2019) Novel machine learning approach for classification of high-dimensional microarray data. Soft Comput 23(24):13409–13421
Nutt CL et al (2003) Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Can Res 63(7):1602–1607
Ong HF, et al (2020) Informative top-k class associative rule for cancer biomarker discovery on microarray data 146: 113169.
Othman MS, Kumaran SR, Yusuf LM (2020) Gene selection using hybrid multi-objective cuckoo search algorithm with evolutionary operators for cancer microarray data. IEEE Access 8:186348–186361
Pandey AC, Rajpoot DS (2019) Spam review detection using spiral cuckoo search clustering method. Evol Intel 12(2):147–164
Pandey AC, Rajpoot DS, Saraswat M (2020) Feature selection method based on hybrid data transformation and binary binomial cuckoo search. J Ambient Intell Humaniz Comput 11(2):719–738
Peng H et al (2021) Multi-strategy serial cuckoo search algorithm for global optimization. Knowl Based Syst 214:106729
Rabia A, Namita S, Chandan KV (2015) A weighted-SNR feature selection from independent component subspace for NB classification of microarray data. Int J Adv Biotechnol Res 6(2):245–255
Salem H, Attiya G, El-Fishawy N (2017) Classification of human cancer diseases by gene expression profiles. Appl Soft Comput 50:124–134
Selaru F et al (2002) Global gene expression profiling in Barrett’s esophagus and esophageal cancer: a comparative analysis using cDNA microarrays. Oncogene 21(3):475–478
Shehab M, Khader AT, Al-Betar MA (2017) A survey on applications and variants of the cuckoo search algorithm. Appl Soft Comput 61:1041–1059
Singh D et al (2002) Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 1(2):203–209
Song P-C, Pan J-S, Chu S-C (2020) A parallel compact cuckoo search algorithm for three-dimensional path planning. Appl Soft Comput 94:106443
Turgut S, Dağtekin M, Ensari T (2018) Microarray breast cancer data classification using machine learning methods. In: 2018 Electric Electronics, Computer Science, Biomedical Engineerings' Meeting (EBBT). IEEE.
Venkatesh B, Anuradha J (2019) A review of feature selection and its methods. Cybern Inform Technol 19(1):3–26
Wang X-H et al (2020) Multi-objective feature selection based on artificial bee colony: an acceleration approach with variable sample size. Appl Soft Comput 88:106041
Xi M, et al. (2016) Cancer feature selection and classification using a binary quantum-behaved particle swarm optimization and support vector machine. Comput Math Methods Med.
Zheng C-H et al (2008) Gene expression data classification using consensus independent component analysis. Genom Proteom Bioinform 6(2):74–82
Zhu X, Wang N (2019) Cuckoo search algorithm with onlooker bee search for modeling PEMFCs using T2FNN. Eng Appl Artif Intell 85:740–753
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
Material preparation, data collection analysis and all other work were performed by Dr. Rabia Musheer Aziz.
Corresponding author
Ethics declarations
Conflict of interests
The authors have no relevant financial or non-financial interests to disclose. Also the authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Aziz, R.M. Application of nature inspired soft computing techniques for gene selection: a novel frame work for classification of cancer. Soft Comput 26, 12179–12196 (2022). https://doi.org/10.1007/s00500-022-07032-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00500-022-07032-9