Abstract
Medical case-based reasoning (CBR) systems require the handling of vague or imprecise data. The fuzzy set theory is particularly suitable for this purpose. This paper proposes a case-base preparation framework for CBR systems, which converts the electronic health record medical data into fuzzy CBR knowledge. It generates fuzzy case-base knowledge by suggesting a standard crisp entity–relationship data model for CBR case-base. The resulting data model is fuzzified using a proposed relational data model fuzzification methodology. The performances of this methodology and its resulting fuzzy case-base structure are evaluated. Diabetes diagnosis is used as a case study. A set of 60 real diabetic cases is used in the study. A fuzzy CBR system is implemented to check the diagnoses accuracy. It combines the resulting fuzzy case-base with a proposed fuzzy similarity measure. Experimental results indicate that the proposed fuzzy CBR method is superior to traditional CBR and other machine-learning methods. Our fuzzy CBR achieves an accuracy of 95%, a precision of 96%, a recall 97.96%, an f-measure of 96.97%, a specificity of 81.82%, and good robustness for dealing with vagueness. The resulting fuzzy case-base relational database enhances the representation of case-base knowledge, the performance of retrieval algorithms, and the querying capabilities of CBR systems.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Diabetes mellitus (DM) is a chronic disease with severe complications. Early DM diagnosis plays a crucial role in its control and prevention of its complications (Varma et al. 2014). It has been shown that 80% of type 2 DM complications can be avoided or delayed by early identification of those at risk (Barakat and Barakat 2010). A physician has to be able to analyze several factors to diagnose DM correctly. An inability to comprehend large amounts of data may lead to an erroneous diagnosis. As a result, we need to select an effective AI technique to build a clinical decision support system (CDSS). Case-based reasoning (CBR) is one of the most suitable intelligent techniques for an experience-based, theory-less, or ill-formed problems, such as DM diagnosis (El-Sappagh et al. 2014). Many real-world systems require support for managing imprecise data. Radha and Rajagopalan (2007) argued that any medical diagnostic system that disallows vagueness in knowledge representation would be prone to errors.
For diagnosing diseases properly, physicians depend on their experience and the patient’s vague descriptions of their symptoms. To handle the complexity of chronic disease, the fuzzy theory needs to be incorporated into CBR to allow more robust, flexible, and accurate models. Fuzzy logic has been integrated with CBR in many fields (Portinale and Montani 2002; Xiaodong et al. 2009). As far as we know, there are no studies of fuzzy CBR in diabetes diagnosis. However, fuzzy logic has been used with other AI techniques, such as rule-based reasoning (RBR), for diabetes diagnosis (Lee and Wang 2011).
Fuzzy sets enhance CBR in many ways. First, they help in building fuzzy cases to provide a characterization of imprecise information (Portinale and Montani 2002). The cases in CBR can be built from electronic health record (EHR) database. However, the crisp data model of EHR proves inadequate for representing vague data. An extended fuzzy relational database (FRDB) needs to be derived from the EHR to represent and query vague patient data. Second, they can be used in developing case retrieval algorithms that incorporate fuzzy matching techniques, which can handle partial case matching or retrieval processes. Third, they can be used in a fuzzy query and query case description. Therefore, a CBR system can accept an imprecise description of a new case (Portinale and Montani 2002). Finally, fuzzy reasoning in the form of fuzzy rule-based systems can be combined with fuzzy CBR (FCBR) to overcome their separate limitations. However, there is no way to utilize fuzziness in CBR without having a fuzzy representation of case-base (CB) knowledge.
All existing FCBR systems concentrate on the retrieval problem and disregard the fuzzy knowledge storage of fuzzy CB. This study follows a different strategy by storing the fuzzy knowledge in a standard-based fuzzy relational database. Fuzzy knowledge representation and querying play a critical role in the success of a CBR system for chronic disease diagnosis. This paper discusses the detailed process of fuzzy CB knowledge creation from EHR by fuzzifying its conceptual and logical data models. In another study (El-Sappagh et al. 2014), we handled incomplete data and NULL values in a data preprocessing phase.
The rest of the paper is organized as follows. Section 2 reviews related work on the role of fuzzy in CBR and diabetes diagnosis. Section 3 introduces our proposed CB preparation framework. Section 4 details the proposed fuzzification framework for the CB database. Section 5 considers the implementation of our fuzzy CBR using diabetes diagnosis as a case study. Section 6 evaluates the system’s performance. Finally, Sect. 7 describes the conclusion and future work.
2 Related work
Many research efforts have been invested in diagnosing diabetes using AI techniques. Mohamudally and Khan (2011) conducted a comparison between most algorithms and listed their strengths and weaknesses. Shankaracharya et al. (2010) presented a review of some diabetes diagnosis techniques, such as ANNs, SVMs, neuro-fuzzy systems, and RBR systems. Kalaiselvi and Nasira (2014) proposed an Adaptive Neuro-fuzzy Inference System (ANFIS) for diabetes diagnosis (80% accuracy). Temurtas et al. (2009) proposed a multi-layer ANN and a probabilistic NN to diagnose diabetes by using the Pima Indian Dataset (PID),Footnote 1 which had accuracies of 79.62 and 78.05%, respectively. Ganji and Abadeh (2011) developed a fuzzy classification system for diabetes diagnosis using ant colony optimization and tested it with the PID (accuracy 84.24%). Barakat and Barakat (2010) utilized SVM to provide a diabetes diagnosis system with an accuracy of 94%. However, Yao and Li (2010) asserted that CBR is better than SVM, especially when the data contain a high level of noise. Varma et al. (2014) proposed a modified Gini-index Gaussian RBR system using fuzzy decision tree algorithm and tested it with the PID with an accuracy of 75.8%. Zadeh (2003) surveyed the recent ML techniques applied in diabetes diagnosis problem.
On the other hand, RBR requires an explicit model of the domain. It is a hard task for an expert to recall all the tacit rules comprehensively. An RBR is simply not appropriate for complex problems because of the high dimensionality of the rule space, except for trivial cases with no impact on real-life applications. For example, Barakat and Barakat (2010) generated a set of rules that are no straightforward general rules for DM diagnosis. Sometimes it is very difficult for even an experienced clinician because being dependent only on the result of the laboratory tests is not enough. In addition, the maintenance of a large rule base system is a challenging task. Its size increases the system’s computational load, and building a rule base is a time-consuming task (Ganji and Abadeh 2011). Developing a CBR system is much faster and easier than constructing RBR system. Although many studies used ANN for diabetes diagnosis, ANN has many limitations compared to CBR. For example, it has no explanations and works as a “black box.” CBR is more flexible than ANN by handling missing data and a large number of features. As a result, CBR is more suitable for complex problems than ML and RBR techniques.
By concentrating on CBR, it has been used to solve many medical and non-medical problems. Montani et al. (2000) proposed a CBR for medical education named iCBLS. This system achieved accuracies of 70% for students’ interaction, 76.4% for group learning, 72.8% for solo learning, and 74.6% for improved clinical skills. El-Sappagh et al. (2015) integrated CBR and ANN to estimate the cost of new product development. They improved CBR by using ANN to calculate attribute weights, get the optimal value of k nearest neighbors, and to estimate the cost of a potential product. Wang (1997) predicted and adapted the database management system (DBMS) performance based on CBR model, and they tested it on MySQL DBMS. Hyung et al. (1994) used CBR to determine the efforts required to build new software, by integrating particle swarm optimization (PSO) with CBR. PSO is utilized to select optimum weights, and their proposal achieved better performance by using the two datasets of Maxwell and Desharnais. Gerstenkorn and Man’ko (1991) proposed a classification model based on a hybrid CBR-ANN technique. The ANN is used to calculate feature weights. The authors adopted a cost-sensitive back-propagation neural network (BPNN) to deal with unbalanced data in the network training process. The system has been tested by seven datasets, and the system achieved an average accuracy of 83.98%. Pappis and Karacapilidis (1993)
CBR has been used for diabetes diagnosis in many studies (Marling et al. 2011; Begum et al. 2009). Marling et al. (2011) used it to improve the insulin bolus calculation process. They proposed a temporal retrieval algorithm to take into account preceding events when recommending bolus insulin doses. By enhancing bolus predictions, they reduced the blood glucose risk index by up to 27%. However, CBR has been combined with fuzzy logic to handle domain imprecisions and vagueness (Rodriguez et al. 2006; Fan et al. 2011). Rodriguez et al. (2006) proposed an individualized situation recognition system in dynamic environments by combining CBR, Situation-Operator-Modeling (SOM), and fuzzy logic. Fan et al. (2011) proposed a strategy for enhancing the operational agility of petroleum refinery plants based on CBR. They suggested a case retrieval technique based on fuzzy matching based on proposed stability number and optimization model for fuzzy membership functions. In the medical domain, physicians often describe patients using imperfect and linguistic data, and their knowledge has a significant deal of imprecision and vagueness. Petrovic et al. (2011) argued much of the knowledge that humans acquire through experience be perception-based and thus subject to imprecision and inaccuracy. When such knowledge is not treated in a suitable way that can consider and convey its inherent imprecision, it usually leads to reduced effectiveness of the used knowledge-based systems. To handle imprecision and vagueness, CBR has used fuzzy reasoning in many medical and non-medical CDSS systems. However, there is a shortage of literature showing hybrid fuzzy logic and CBR for diabetes management. On the other hand, there are many fuzzy rule-based systems for implementing diabetes diagnosis CDSS (Lee and Wang 2011).
Most existing studies have not stored fuzzy CB knowledge in a separate fuzzy database. Portinale and Montani (2002) created a crisp database for CB and used fuzzy SQL queries for case retrieval. Xiaodong et al. (2009) created a fuzzy CB database. They have represented fuzziness using a single linguistic value for each fuzzy variable, which surely affects the accuracy of the retrieval algorithms and query formulation. Medical domain requires the creation of standard and interoperable knowledge bases to support the integration between distributed CDSS systems and interoperability between CDSS and EHR. As a result, the created CB fuzzy database must be based on a unified medical data model. There are no studies in the literature that consider this issue.
Diabetes diagnosis was used as a case study in this paper. We apply the fuzzification process to a crisp HL7 RIM-based CB quantitative data. The result is a fuzzy CB relational database. To start the fuzzification process, Li and Ho (2009) created a standard relational data model for the diabetes diagnosis case-base. They populated it with real diabetic cases to make the necessary data preprocessing steps using a set of ML algorithms (El-Sappagh et al. 2014). They encode the CB unstructured contents using SNOMED CT ontology. The encoding of CB knowledge using standard ontology supports the development of semantic case retrieval algorithms (Pappis and Karacapilidis 1993; Arias-Aranda et al. 2009). Qin et al. (2018) proposed a CBR system for computer-aided tolerance specification. They modeled the CB and query cases in OWL ontologies and used ontology-based similarity measure for case retrieval. Pappis and Karacapilidis (1993) utilized CBR for analyzing the response to risks connected with urban water supply network (UWSN). They utilized OWL ontologies for the representation of disaster scenario features and UWSN risks and response strategies knowledge, and for case retrieval process.
3 The CB preparation framework
EHR data are the primary source of CB knowledge. This section discusses the conversion of both the structure and content of an EHR database to a derived CB structure and content. The structures of EHR data and CBR cases could be matched. In our domain, a case is composed of a description of the problem and its solution as shown in Eq. 1.
where P is the description of the problem that is represented by several dimensions, such as patient symptoms and laboratory test results. This part is described by 58 features, all of which are derived from the EHR as shown in Eq. 2.
where LFT Liver Function Tests, LP Lipid Profile, GS Global Symptoms, A Age, B Body Mass Index, R Residence, G Gender, O Occupation, KFT Kidney Function Tests, LT Lab Tests, US Urination Symptoms, and HP Hematological Profile. The solution part S(P) is described by six features; it is the diagnosis of diabetes including diabetic, pre-diabetic, gestational-diabetic, and pre-diabetic-gestational. Moreover, S(P) includes the potential to produce other complications as presented in Eq. 3.
where DD diabetes diagnosis, L liver problem, N nephropathy problem, H hypercholesterolemia problem, G glomerulonephritis, and SP splenomegaly. However, EHRs are distributed systems in most cases. As a result, their data structures will vary. To facilitate the collection of patient medical data from distributed EHR environments, we stick to standards that include a standard medical data model, standard diabetes diagnosis data elements, and standard contents.
Figure 1 shows our previously proposed CB preparation framework to map an EHR database into high-quality CBR’s CB knowledge (El-Sappagh et al. 2014), which improves the CB semantic. The data-fuzzification phase is the final phase, which is the focus of our study. It will be discussed in details in the next sections.
4 The proposed CB database fuzzification framework
In this section, we propose a methodology to extend the CB relational model to incorporate fuzziness. In addition, an FRDB CB is created for diabetes diagnosis. In Fig. 2, we have created a crisp physical, relational database of the diabetes case-base. Then, we populated it with a dataset that is prepared in a previous study (Li and Ho 2009). Using reverse engineering, we create the crisp CB ER model from this physical database. Next, we will discuss the detailed steps of a proposed CB database fuzzification methodology, which has seven steps as follows.
Step 1 Build the crisp CB ER model. Our domain experts and the diabetes diagnosis CPGs determined a list of DM diagnoses and their complication features. From the EHR database, all of these features are collected and organized in a CB structure. We have previously proposed a standard CB relational data model (Li and Ho 2009). In this paper, we customize this model according to our dataset. Patient cases are described by a set of entities as shown in Fig. 2. The Case problem is defined by the entities, which are Patient_Case, Kidney_Function_Test, Liver_Function_Test, Lipid_Profile, Diabetes_Lab_Test, Global_Symptom, Hematological_profile, and Urination_Symptom. The Diagnosis entity represents case solution.
Step 2 Determine the fuzzy entities and fuzzy attributes. In a fuzzy database FDB, crisp entities exist side by side with fuzzy entities: \(FDB=\left\{ {CE,FE} \right\} \). For space restrictions, we concentrate only on fuzzy relations (e.g., Patient_Case in Fig. 2):
-
a.
Determine the fuzzy entities, which are entities with one or more fuzzy attributes, such as Diabetes_Lab_Test and Patient_case. For \(FE=\left\{ {E_1 ,E_2 ,\ldots ,E_n } \right\} \), \(n=\left| {FE} \right| \) and \(FE=FDB-\left\{ {\hbox {Global}\_\hbox {Symptom},\hbox {Diagnosis}} \right\} \).
-
b.
In each fuzzy entity \(E_i =\left\{ A_1^i ,\ldots ,A_q^i ,A_{q+1}^i ,\ldots , A_b^i ,A_{b+1}^i ,\ldots ,A_c^i \right\} \), determine the quantitative attributes \(A_q^i\) (not binary and categorical) to be fuzzified that may contain imprecise data. For example, in table Patient_Case, fuzzy features are BMI, Age, etc.
-
c.
For each selected fuzzy attribute \(A_q^i\):
-
1.
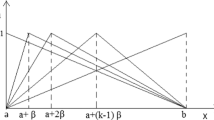
Determine the range of values for its crisp domain, \(A_q^i \in \left[ {\alpha ,\beta } \right] \).
-
2.
Determine the fuzzy linguistic terms \(A_q^i \in \left\{ A_{q1}^i,A_{q2}^i,\ldots , A_{qk}^i \right\} \) for k terms. For each term \(A_{qk}^i\) determine its MF \(\mu _{qk}^i\). For each MF, determine its shape, formula, and parameters. We select triangular and trapezoidal MFs, see Table 1. These two types of membership functions are suitable to represent the fuzzy semantics of many problems based on previous experiences (Arias-Aranda et al. 2009), and in the same time they are simple, flexible, and computationally cheaper that other nonlinear functions (Khanum et al. 2009). It is difficult to calculate the arithmetic operations in case of Bell, Sigmoidal, or Gaussian. In addition, the medical meaning of such membership functions has been deeply discussed and validated with our domain experts. Based on experience, the overlap of triangle-to-triangle and trapezoid-to-triangle fuzzy regions results in somewhere between 25 and 50% of the fuzzy set base being averaged. Our domain experts recommended fixing the normal ranges of all features and making the left and right terms overlap by 50%.
-
1.
Table 1 shows examples of fuzzy linguistic variables that are created by using MATLAB from some fuzzy tables. The attached supplementary file contains the membership functions of the 35 features.
Step 3 Extend the ER model into a fuzzy ER model by concentrating on the third fuzzification type.
An entity is said to be fuzzy if one or more of its attributes are fuzzy. All entities in Fig. 2 are fuzzy entities except Diagnosis and Global_Symptom relations, which have crisp categorical (i.e., symbolic) and ordinal features. An attribute is said to be fuzzy if its domain is a set of possibility distributions. In this step, all fuzzy entities and attributes in the crisp model are signed by the construct “F,” such as \(\hbox {Patient}\_\hbox {Case}^{\mathrm{F}}\) and \(\hbox {age}^{\mathrm{F}}\) as modeled.
Step 4 Select a strategy for the FRDB design. Three main strategies exist for FRDB design. Each method has its advantages and disadvantages. These methods concentrate on the fuzzy SQL query processing. As a result, they save a large volume of metadata in the database dictionary to describe data precisely. However, in our case, we depend on a retrieval algorithm to fetch the knowledge from the case-base. The resulting database will be used to populate the CB fuzzy ontology; we customize these models. This paper concentrates on the fuzzification of quantitative numerical data using only possibility distribution.
Step 5 Transform the fuzzy ER model to case-base fuzzy relational model. This step is performed by transforming crisp entities and relationships into tables and relationships, as done for traditional databases. For example, the Diagnosis entity is mapped to the Diagnosis relation. Fuzzification of the data is achieved by converting the crisp values into linguistic values, each with a degree of membership in every fuzzy set. The fuzzification is done by mapping each instance of a crisp attribute into the fuzzy sets of its corresponding fuzzy attributes.
More formally, let E be an entity with attributes \(<{K,A_1 ,\ldots ,A_l ,B_0 }>\), with key K, non-key attributes \(A_{i}\) for \(i=1,\ldots ,l\), and let \(B_0\) be the non-key attribute to be fuzzified. For any tuple \(z=<{k,a_1 ,\ldots ,a_l ,b_0 } >\in E\), with \(k\in K, a_{i}\in A_{i}\) for \(i=1,\ldots ,l\), and \(b_{0} \in B_{0}\), \(E^{F}\) is the fuzzy entity, and it has the tuple \(z^{F}=<{k,a_1 ,\ldots ,a_l ,\left\{ {\mu \left( {b_1 } \right) /b_1 ,\ldots ,\mu \left( {b_n } \right) /b_n } \right\} }>\), where \({b}_0 =\left\{ {\mu \left( {b_1} \right) /b_1 ,\ldots ,\mu \left( {b_n } \right) /b_n } \right\}>\) for \(i=1,\ldots ,n\). The resulting fuzzy entities are thus possibilistic relations (since one of its attributes is fuzzy), and it is not in the first normal form, as shown in Table 2 for the crisp Age=52.
Step 6 Normalize relations by using functional dependencies, multi-valued dependencies, and restricted fuzzy functional dependencies. For fuzzy attributes, the domains are sets of possibility distributions (i.e., fuzzy sets) that are non-atomic. A domain is atomic if its elements are indivisible units. For a normalized relation, all attributes are required to have atomic domains. Therefore, a possibilistic relation is not in the first normal form (1NF) unless the DBMS allows possibility distributions as a data type. In the absence of database systems that provide possibility distributions as a data types, a strategy is required to normalize possibilistic entities. In our case, each fuzzy attribute is modeled as a separate fuzzy table with key= base table key + fuzzy table’s attributes, where each attribute stores atomic value.
Consider a relation R with attributes \(< {K,A_1 ,\ldots ,A_l}>\), with a non-fuzzy key K, and non-key (fuzzy or non-fuzzy) attributes \(A_t\) for \({t}=1,\ldots ,l\). For each fuzzy attribute \(A_m\) with \(1\le m\le n\ni n\le l\) (i.e., its domain is a fuzzy set), create a new relation \(A_m =< {K_m ,\hbox {value},{\mu }}>\), and modify R to \(\acute{R}\) by replacing \(A_m\) in R with \(K_m\) in \(\acute{R}\). The terms Value and \({\mu }\) model the elements and associated grades, respectively, to the fuzzy attribute \(A_m.K_m\) is an atomic identifier in \(\acute{R}\) for \(A_m\) as shown in Eq. 4.
add the collection of tuples \(a_{m_p}^F =< {k_m ,a_{m_p } ,{\mu } \left( {a_{m_p } } \right) }>\) to the new relation \(A_m\) for \({p}=1,\ldots ,n\). Furthermore, add the instance \(< {k,a_1 ,\ldots ,k_m ,\ldots ,a_l}>\) to \(\acute{R}\), where n is the number of fuzzy sets defined for the fuzzy variable \(A_m\).
For illustration, assume that we have an entity-type set E, and type e\(\in \)E is an instance entity type, e.g., the fuzzy table \(\hbox {Patient}\_\hbox {case}^{\mathrm{F}}\). Moreover, assume that we have attribute set A for e and that a\(\in \)A is an instance attribute, e.g., \(\hbox {Patient}\_\hbox {case}^{\mathrm{F}}\) (CaseID, \(\hbox {Age}^{\mathrm{F}}\), \(\hbox {BMI}^{\mathrm{F}}\), Residence, Occupation ...), where A as the set of crisp and fuzzy attributes = {Age, BMI, Residence, and Occupation}, and a = {Age, BMI}.
If a quantitative and non-key attribute a is decided to be a fuzzy attribute, it becomes a composite or multi-valued attribute. Therefore, it can be expressed by a possibility distribution. If the attribute a has the fuzzy values (fuzzy sets): \(a_{1}, a_{2}{\ldots } a_{n}\), then its values can be represented as shown in Eq. 5.
where V(a) is the value of the attribute a, \(\mu \,(a_{i})\) is the membership function of the fuzzy value \(a_i\), and \(0<\mu \) (ai) \(\le 1, 1\le i \le n\). For example, if Age \(=\) 70 and its fuzzy values are defined as \(<{\text {young, middle}}\_{\text {aged, old}}>\), then according to the membership functions in Table 1, the fuzzification process is presented in Eq. 6.
Fuzzy attributes are modeled as separate entity-attribute-value (EAV) tables, as seen in Fig. 3. The Value attribute stores the fuzzy set name, and the \(\mu \) attribute stores its membership degree. According to the membership functions of Age and BMI, each value in the crisp table is converted to a set of tuples in the derived EAV tables.
Fuzzy attributes can be modeled as regular tables, i.e., not in EAV format. In these tables, a column is created for each fuzzy value. This design can simplify querying, but it wastes memory for fuzzy values with \(\mu = 0.0\). Moreover, the normalization can be done in the main table (e.g., Patient_case). The main table containing the crisp data can be altered by replacing the crisp column by its fuzzified columns.
Step 7 Populate the resulting fuzzy relational database. Crisp data types are represented according to the hosting RDBMS. For fuzzy relations, they are filled according to the membership function formulas of the associated attributes. In Fig. 4, data are extracted from the crisp relational database, fuzzified according to the domain expert knowledge, and stored in a fuzzy relational database. There is no need to store metadata physically about the fuzzy membership functions. We built a Java project to populate a CB FRDB from previously filled crisp CB RDB. After executing step 7, the fuzzy CB is available in a fuzzy relational database. The next stage is to build a complete CBR system that utilizes the resulting fuzzy knowledge base.
5 System implementation
Implementation issues can be grouped into three stages: (1) representation of the case features and their measures, (2) database modeling, and (3) CDSS system. The representation process involves the selection of the diagnostic features and their operationalization. Our study of the medical literature and the cooperation with our domain experts have determined the features, which are used for the diagnosis of diabetes along with their types (fuzzy or crisp), ranges, and preparation. Moreover, a fuzzy relational database for CB storage has been proposed based on our proposed methodology using the Oracle 11g DBMS.
We have implemented a fuzzy CBR prototype using our prepared dataset and fuzzification methodology results. The JCOLIBRI2 APIFootnote 2 is used as the Java platform, which supports the implementation of crisp CBR systems. On the other hand, it is not related to fuzzy logic aspects in any way. To test the results of our proposed CB fuzzification methodology, we implemented an FCBR using a fuzzy CB and added the fuzzy functionalities of the fuzzy similarity and the fuzzy query to JCOLIBRI2. Figure 5 shows a customized JCOLIBRI2 architecture according to our added and needed functionalities. To save space, we concentrate on the fuzzy CB and fuzzy retrieval steps that are the most critical steps in CBR.
-
1.
Fuzzy case-base It has been built in the previous sections. This CB is connected to JCOLIBRI2 using the Database Connector object. Our fuzzy CB does not contain any linguistic terms as feature values.
-
2.
Feature weights We calculated weights in another study for medical data preparation (El-Sappagh et al. 2014). Table 2 shows a sample of feature weights with values \(\in \) [0, 1].
-
3.
Fuzzy case retrieval algorithm This algorithm was implemented and connected to the JCOLIBRI2 environment. Very serious consideration must be given to the nature of the data that dictate the selection of a suitable similarity measurement. A single similarity measure cannot fit all situations. Our similarity measure depends on the type of feature values. The global similarity function uses a different local similarity function for each feature type. The global similarity \(\hbox {SIM}\left( {C,Q} \right) \) between two cases C and Q is calculated by Eq. 7, and sim is the local similarity function between two values.
$$\begin{aligned} \mathrm{SIM}\left( {C_i ,C_q } \right) =\sum _{k=1}^n w_k *{\text {sim}}\left( {f_{ik} ,f_{qk} } \right) \end{aligned}$$(7)
The local similarity function sim is calculated according to feature types as follows:
-
Case 1For each ordinal feature (e.g., urination frequency, urobilinogen, bilirubin, and protein), our domain experts proposed a similarity matrix. For example, the similarity matrix of urination frequency is shown in Table 3.
-
Case 2For all symbolic features (e.g., residence and gender), we use the exact-match similarity function in Eq. 8.
$$\begin{aligned} \hbox {sim}\left( {c_i ,q_i } \right) =\left\{ {{\begin{array}{ll} 0,&{}\quad \text {if}\,q_i \ne c_i \\ 1,&{}\quad \text {if}\,q_i =c_i \\ \end{array}}} \right. \end{aligned}$$(8)
-
Case 3For fuzzy features, most of the existing fuzzy case retrieval algorithms use one value to represent a fuzzy variable (the largest membership value), and they use these values to calculate the similarity. In our algorithm, we utilize all of the membership degrees of a fuzzy variable. As shown in Fig. 5, the same membership functions that are used to fuzzify the CB are used to fuzzify all numerical features in a query case (e.g., FPG, age, HbA1c, BMI, Serum Creatinine, Total cholesterol, Direct Bilirubin, Alk Phosphatase, etc.). We propose a similarity measure based on membership functions of the fuzzy sets associated with these features. The similarity is based on the degree of similarity between the fuzzy sets in the query and stored cases. A comparison is performed between the stored and fuzzy query values. The normalized Euclidean distances between the fuzzy sets of a feature are used to calculate the similarity as shown in Eq. 9.
$$\begin{aligned} \hbox {Dist}\left( {c_i ,q_i } \right) =\frac{\sqrt{\sum _{k=1}^n \left( {\mu _{ci_k } -\mu _{qi_k } } \right) ^{2}}}{\sqrt{n}} \end{aligned}$$(9)where \(q_i\) the crisp value of the query’s feature; \(c_i\) the crisp value of the stored case’s feature; n the number of fuzzy sets; \(\mu _{ci_{k}}\) the fuzzy degree of k’s fuzzy value, and it is read from the fuzzy database; \(\mu _{{qi_{k}}}\) is the corresponding fuzzy degree of \(\mu _{{ci_k}} \), and it is calculated in the application.
For example, let the fuzzy variable Age be fuzzified as the young, middle_aged and old fuzzy sets in Table 1. The value of n is 3. The current case’s age in CB is age \(=\) 30, and it is fuzzified as \(\mu _{\mathrm{Age}} \left( {30} \right) \)\(=\) {1.0 /young, 0.0/middle_age, 0.0/old}. Let the query case has age \(=\) 38, and it is fuzzified as \(\mu _{\mathrm{Age}}\left( {38} \right) \)= {0.2/young, 0.8/middle_age, 0.0/old}. Figure 6 shows the distance between the young membership values in the query and case-base cases. The same process is carried out for middle_age and old for ages 30 and 38, respectively. The normalized Euclidean distance between the two ages (i.e., 30, 38) is calculated using Eq. 9 as follows.
$$\begin{aligned}&\hbox {Dist}\left( {30,38} \right) \nonumber \\&\quad =\frac{\sqrt{\left( {1.0-v0.2} \right) ^{2}+\left( {0.8-0.0} \right) ^{2}+\left( {0.0-0.0} \right) ^{2}}}{\sqrt{3}}=0.65 \end{aligned}$$The similarity is calculated using Eq. 10.
$$\begin{aligned} \hbox {sim}\left( {c_i ,q_i } \right) =1-\hbox { Dist }\left( {c_i ,q_i } \right) \end{aligned}$$(10)The similarity level between 30 and 38 is \(\hbox {sim}\left( {30,38} \right) =0.35\). As we will discuss later, this similarity measure satisfies the defined fuzzy similarity measure properties (Sushmita and Chaudhury 2007). Moreover, in a query description, a patient can now be described using vague terms for numerical features (e.g., Age \(=\) young, BMI \(=\) obese, FPG \(=\) low) without any numerical values. In this case, the full possibility distribution of the fuzzy term is created, and Eq. 9 can calculate the distance.
Finally, fuzzy hedges such as “very” or “extremely” are possible in query case description. Hedges are fuzzy qualifiers that modify a membership value in a fuzzy set. For example, if \(\mu _{\mathrm{young}} \left( x \right) \) is the membership function, then \(\mu _{\mathrm{veryYoung}} \left( x \right) =\left( {\mu _{\mathrm{young}} \left( x \right) } \right) ^{2}\).
-
Case 4For numerical features, the similarity is calculated according to Eq. 11, where \(\max _i\), and \(\min _i\) are the feature’s maximum and minimum values, respectively.
$$\begin{aligned} \hbox {sim}\left( {c_i ,q_i } \right) =1-\frac{\left| {c_i -q_i } \right| }{\max _i -\min _i } \end{aligned}$$(11)
6 Performance evaluation
The performance assessment has four strategies as discussed in the following subsections.
6.1 Evaluation of the fuzzy similarity metric
First, we discuss the justification for the similarity measures that we proposed. We support many types of features, including numerical, nominal, ordinal, and fuzzy types. We select a fuzzy similarity measure based on the well-known Euclidean distance function. This fuzzy distance metric satisfies the following properties: nonnegative\(:d\left( {x,y} \right) \ge 0\); identity\(:d\left( {x,y} \right) =0\,\mathrm{iff}\,x=y\); symmetry\(:\,d\left( {x,y} \right) =d\left( {y,x} \right) \); and triangle inequality\(:d\left( {x,z} \right) \le d\left( {x,y} \right) +d\left( {y,z} \right) \). The defined fuzzy similarity measure satisfies the defined similarity properties as follows in Sushmita and Chaudhury (2007). Let \(x_i ,x_j \in X\) be two elements in the universe X, where \(x_i ,x_j\) are defined by fuzzy sets \(A_k \in \mathcal{F}\) and \(\mathcal{F}\) is the class of all fuzzy sets of \(X,k=1,\ldots ,N\).
-
1.
\(S\left( {x_i ,x_j } \right) =S\left( {x_j ,x_i } \right) ,x_i ,x_j \in X,\) for all \(A_k\),
-
2.
\(S\left( {x_i ,x_i } \right) =1,\)
-
3.
\(0\le S\left( {x_i ,x_j } \right) \le 1,\)
-
4.
\(S\left( {x_i ,x_j } \right) =1\) iff \(x_i =x_j\),
-
5.
If \(\mu _{A_{h}} \left( {x_i } \right) \le \mu _{A_{h}} \left( {x_j } \right) \le \mu _{A_h } \left( {x_k } \right) \) for all \(x_i ,x_j ,x_k \in X\), where \(A_h ,h=1,2,\ldots ,N\in \mathcal{F},\mathcal{F}\) is the class of all fuzzy sets of X, then \(S\left( {x_i ,x_j}\right) \ge S\left( {x_i ,x_k } \right) \) and \(S\left( {x_j ,x_k}\right) \ge S\left( {x_i ,x_k } \right) \).
-
6.
\(\max _{\forall x_i ,x_j \in X} S\left( {x_i,x_j} \right) =1,\) which means it is a normal similarity measure.
We compare the performance of this similarity measure with some of the existing measures as shown in Eq. 12 (Xiong 2011):
Moreover, the correlation coefficient of \(x_i\) and \(x_j\) is defined in Eq. 13 (Godo et al. 2009).
where \(T(x_i )=\sum _{k=1}^N \left[ {\mu _{A_k }^2 \left( {x_i } \right) +v_{A_k }^2 \left( {x_i } \right) } \right] , \quad v_{A_k } (x_i )=1-\mu _{A_k } (x_i )\), and \(C\left( {x_i ,x_j } \right) =\mathop \sum \nolimits _{k=1}^N \mu _{A_k } (x_i ).\mu _{A_k } (x_j )+v_{A_k } (x_i ).v_{A_k } (x_j )\).
Moreover, Jin et al. (2010) proposed three types of similarity measures are described by Eqs. 14, 15, and 16.
Let us now illustrate several examples that compare the present measure with the previously listed measures. Consider that we have four ages defined according to three fuzzy sets young, middle_age, and old, as shown in Table 4.
From Table 5, we see that our similarity measure satisfies the previously listed properties. However, it is difficult to say which similarity measure is the best. Nevertheless, we can make the following observations.
-
The similarity measure in Eq. 12 uses \(\max _i\) to obtain the similarity. If \(x_i =x_j\) and \(\max _i <1\), then \(S\left( {x_i ,x_j } \right) <1\), which violates the fourth property.
-
If there exists at least one (but not all) fuzzy set A such that \(\mu _{A_{k}} (x_i)=\mu _{A_{k}} (x_j)=1\), then \(S\left( {x_i ,x_j} \right) =1\), by Eq. 12. However, \(x_i \ne x_j\), here, e.g., S(Age 2, Age 3), where \(\mu _{\mathrm{young}}\) and \(\mu _{\mathrm{old}}\) are not equal in both Age 2 and Age 3.
-
From Eq. 15, if there exists a fuzzy set A such that \(\mu _{A_{k}} (x_i)\) and \(\mu _{A_{k}}(x_j)\) are significantly different, then \(L_{x_i ,x_j } \) is relatively small even if the other elements are relatively close. For example, S(Age 1, Age 3) \(=\) 0.0 for Eq. 15, which is not reliable since \(\mu _{\mathrm{old}} \left( {\mathrm{Age}\,1}\right) \) and \(\mu _{\mathrm{old}}\left( {\mathrm{Age}\,3} \right) \) are ignored.
-
Finally, our proposed measure has the property of significance of the average property. We mean by this that each element in the compared sets plays an equal role in the similarity computation. This point is not seen in the other measures.
6.2 Evaluation by comparison with domain expert decisions
In this section, we have evaluated the accuracy decisions made by the proposed system decision compared to those of our domain expert to verify its feasibility and applicability. A CBR system is characterized as a lazy learner technique. We have queried the system using all cases in the knowledge base (i.e., 60 cases) by using “leave one out” cross-validation (LOOCV). Each test query is fed into the system, and the corresponding response is recorded. The decisions of the proposed system are compared with those of our domain experts.
The effectiveness of the system is determined by the number of correct answers that it gives, i.e., answers that concur with those of the expert. In other words, the accuracy is inversely proportional to the number of times that the system fails. We have used the weight vector computed using a set of ML algorithms (El-Sappagh et al. 2014) in our previous work. The diagnoses suggested by the proposed system include a diabetes diagnosis (i.e., diabetic, pre-diabetic, normal, etc.) and the possibility of developing other chronic problems, such as nephropathy, cancer, and kidney malfunction. “Appendix A” shows a small sample of cases that were tested. The accuracy of our system is 95%; that is, in 57 out of 60 cases, the system retrieved a case with the same diagnoses suggested by the domain experts. Moreover, besides diabetes diagnoses, our system has predicted the future complications of all cases like the domain expert predictions. We have selected \(k=3\) to assert the consistency of decisions of our proposed system. For example, Case 5 decided that the patient has diabetes in all of the retrieved cases. As a result, the accuracy of the CBR system decisions would give a physician the confidence to follow them because it is accurate. Moreover, if the system diagnosis is incorrect for the first \(k=1\), it can be correct for the second or third decision (i.e., \(k=2, 3)\). The predicted complications, other than diabetes, need to be checked by a physician to prevent their occurrence or to begin their treatments.
6.3 Evaluation by comparison with other CBR systems
We have executed the same experiments with a traditional CBR system and then compared its performance with that of our proposed system. A traditional system means one that is not fuzzy; instead, it has a crisp CB representation, crisp similarity algorithms, and crisp querying capabilities. “Appendix B” presents a sample of the tested cases. The comparison includes three aspects. First, the accuracy of the traditional CBR is lower than our accuracy. The reasons for the inaccuracy of the traditional systems are as follows: (1) they do not handle the similarity of ordinal features correctly, e.g., vision, thirst, etc.; (2) the proposed fuzzy similarity measures are more accurate than the crisp similarity of traditional CBR; (3) the fuzzified version of the previously prepared CB has an accurate design that handles the fuzziness aspect perfectly. Second, the cases diagnosed correctly by traditional CBR have lower similarity levels (i.e., confidence) compared to ours. For example, case 5 was diagnosed with 100% confidence in our system, compared to 80% in the traditional system. Finally, the most critical property of our system is its ability to describe a patient with vague terms and hedges in query cases. For example, if a physician did not know the exact value of a particular test, he could use a linguistic term to describe this feature (e.g., HbA1c \(=\) high, or FPG \(=\) low). Moreover, hedges can be used to modify these linguistic terms (e.g., Age \(=\) very old, or FPG \(=\) slightly high).
The classification accuracy may not always be the most significant performance criterion in medical diagnosis; other measures such as sensitivity and specificity might outweigh it. Therefore, in our evaluation, we calculate and compare these metrics. We use a 2 \(\times \) 2 confusion matrix to calculate the precision (confidence), recall (sensitivity), accuracy, and specificity of our FCBR and the traditional system as shown in Table 6. Moreover, we compare the harmonic mean of the precision and sensitivity (i.e., F-measure) of both systems. We calculate the system performance for diabetic decisions, and the terms TP, FP, FN, and TN are interpreted as:
-
TP \(=\) the CBR system decides the diabetic case, and domain expert decides a diabetic case.
-
FP \(=\) the CBR system decides a diabetic case, but the domain expert does not.
-
FN \(=\) the CBR system decides not a diabetic case, but the domain expert decide it be diabetic.
-
TN \(=\) the CBR system decides not a diabetic case, and the expert decides not a diabetic case.
The above parameters can be evaluated for pre-diabetic and normal as well. We calculate P, R, A, E, and S for both systems using Eqs. 17, 18, 19, 20, and 21:
The effectiveness E is a single number indicator of system performance. It is the harmonic mean of precision and recall.
The values of P, R, A, E, and S for both systems regarding the diagnoses of diabetes are as follows:
-
For the proposed FCBR system
$$\begin{aligned}&P=\frac{48}{48+2}=96\% , \quad R=\frac{48}{48+1}=97.96\% , \quad \\&A=\frac{48+9}{48+9+2+1}=95\% , \quad E=\frac{1}{\frac{1}{2*0.96}+\frac{1}{2*0.9796}}\nonumber \\&\quad =96.97\% , \quad \\&S=\frac{9}{9+2}=81.82\%. \end{aligned}$$ -
For the traditional CBR system
$$\begin{aligned}&P=\frac{19}{19+6}=76\% ,\quad R=\frac{19}{19+21}=47.5\%,\\&A=\frac{19+14}{19+14+6+21}=55\% ,\\&E=\frac{1}{\frac{1}{2*0.76}+\frac{1}{2*0.475}}=58.46\% , \quad S=\frac{14}{14+20}=70\% . \end{aligned}$$
The overall performance of our proposed system is better the other systems because its similarity measures take into account the nature of all features, the cases are carefully preprocessed and cleaned, and an accurate weight vector has been defined. Moreover, the pre-calculated features weights enhance the accuracy of the calculation.
Table 7 shows a comparison between 18 CBR systems for both medical and non-medical applications. As seen in Fig. 7, our FCBR system has improved accuracy.
6.4 Evaluation by comparison with ML classifiers
In this section, we perform two types of comparison. First, Table 8 shows a comparison between classification accuracy of the proposed FCBR system and existing ML studies for diabetes diagnosis. All these studies have lower performances than that of ours. However, these systems mostly depend on the PID dataset. As a result, it is better to apply these machine-learning algorithms to our preprocessed dataset (El-Sappagh et al. 2014; Kalpana and Kumar 2011b).
Second, we run a set of ML algorithms as black boxes on our case-base data using the WEKA APIs. We compare the proposed FCBR system with the techniques of C4.5, k-NN with \(k=3\) (IBK in WEKA), SVM (SMO in WEKA), ANN, and Naive Bayes. We used the default settings of these algorithms recommended by WEKA. The k-fold cross-validation (KFCV) is the selected evaluation technique for \(k=2,{\ldots },10\). When \(k=N\) then KFCV is equivalent to LOOCV used in CBR for \(N=\) case-base size. “Appendix C” shows the results of the tested algorithms.
Many studies compare algorithms accuracy to measure the performance (Temurtas et al. 2009), but we used accuracy and other metrics. As previously demonstrated, our technique can predict diabetes with high accuracy, sensitivity, and specificity, which outperforms other machine-learning classification techniques. Moreover, to the best of our knowledge, there is no machine-learning classification algorithm with multi-attributes for classes. Figure 8 compares the performance metrics between the k-folds, and fivefold cross-validation has the best performance. However, our system achieves better classification. The 60-fold cross-validation, which is equivalent to LOOCV, did not achieve the best performance compared to fivefold cross-validation.
In Fig. 9, it is easily seen that our technique produced a better performance than the tested algorithms for diagnosing diabetes. This figure compares the proposed system against a traditional CBR, C4.5, naïve Bayes, k-NN, SVM, and ANN. Although the proposed FCBR system performs better compared to traditional one, it still has some problems dealing with semantic retrieval. When the CB is represented as a fuzzy ontology and the domain background knowledge is represented as an ontology, semantic retrieval plays a critical role in evaluating the semantic and clinical similarity. The current work has prepared the fuzzy CB database to be used in the future in building CBR systems based on fuzzy ontology.
7 Conclusion
In this paper, we proposed a CB preparation framework, which is dependent on EHR data. We proposed a CB database fuzzification methodology. The methodology has been applied to a diabetes diagnosis database and produced a fuzzy CB relational database. We implemented a case study to measure the accuracy of the fuzzification process, and a fuzzy CBR system was implemented. The accuracy and performance evaluation of our system is 95%, and it proved its applicability for diagnosing diabetes. However, the main limitation of our study is the semantic understanding of the relationships between medical concepts. This restriction will be handled in the third phase of our proposed framework (i.e., coding phase). Moreover, combining fuzzy and ontology semantics can improve the CBR performance. In future work, we will study the applicability of fuzzy ontology as a CB, and standard medical ontology, such as SNOMED CT, as domain background knowledge in CBR. We expect that this integrated architecture will enhance the intelligence of CBR systems and make them more acceptable in medical environments.
References
Abbasbandy S, Hajjari T (2010) Weighted trapezoidal approximation-preserving cores of a fuzzy number. Comput Math Appl 59(9):3066–3077
Adekunle Y (2015) The prediction, diagnosis, and treatment of diabetes mellitus using an intelligent decision support system framework. Int J Adv Res Comput Sci Softw Eng 5(3):1285–1292
Aibinu A, Salami M, Shafie A (2010) Application of modeling techniques to diabetes diagnosis. In: IEEE EMBS conference on biomedical engineering and sciences (IECBES), pp 194–198
Ali M, Han S, Bilal H, Lee S, Kang M, Kang B, Razzaq M, Amin M (2018a) iCBLS: an interactive case-based learning system for medical education. Int J Med Inform 109:55–69
Ali F, Islam SR, Kwak D, Khan P, Ullah N, Yoo SJ, Kwak KS (2018b) Type-2 fuzzy ontology-aided recommendation systems for IoT-based healthcare. Comput Commun
Arias-Aranda D, Castro J, Navarro M, Zurita J (2009) A CBR system for knowing the relationship between flexibility and operations strategy. Found Intell Syst 5722:463–472
Balakrishnan V, Shakouri M, Hoodeh H, Loo H (2012) Predictions using data mining and case-based reasoning: a case study for retinopathy. World Acad Sci Eng Technol 63:573–576
Barakat N, Barakat M (2010) Intelligible support vector machines for diagnosis of diabetes mellitus. IEEE Trans Inf Technol Biomed 14(4):1114: 1120
Begum S, Ahmed M, Funk P, Xiong N, Schéele V (2009) A case-based decision support system for individual stress diagnosis using fuzzy similarity matching. Int J Comput Intell 25(3):180–195
Bellazzi R, Montani S, Portinale L (1998) Retrieval in a prototype-based case library: a case study in diabetes therapy revision. Adv Case Based Reason 1488:64–75
Biswas S, Chakraborty M, Singh H et al (2017) Hybrid case-based reasoning system by cost-sensitive neural network for classification. Soft Comput 21(24):7579–7596
Brown D, Aldea A, Harrison R, Martin C, Bayley I (2018) Temporal case-based reasoning for type 1 diabetes mellitus bolus insulin decision support. Artif Intell Med 85:28–42
Çalisir D, Dogantekin E (2011) An automatic diabetes diagnosis system based on LDA-wavelet support vector machine classifier. Expert Syst Appl 38(7):8311–8315
Chang X, Lilly J (2004) Evolutionary design of a fuzzy classifier from data. IEEE Trans Syst Man Cybern B Cybern 34(4):1894–1906
Dogantekin E, Dogantekin A, Avci D, Avci L (2010) An intelligent diagnosis system for diabetes on linear discriminant analysis and adaptive network based fuzzy inference system: LDA-ANFIS. Digit Signal Proc 20(4):1248–1255
El-Sappagh SH, Elmogy M, Riad A, Badria F, Zaghlol H (2014) EHR data preparation for case based reasoning construction. Adv Mach Learn Technol Appl 488:483–497
El-Sappagh SH, Elmogy M, Riad A (2015) A CBR system for diabetes mellitus diagnosis: CB standard data model. Int J Med Eng Inform 7(3):191–208
Fan C, Chang P, Lin J, Hsieh J (2011) A hybrid model combining case-based reasoning and Fuzzy decision tree for medical data classification. Appl Soft Comput 11:632–644
Ganji M, Abadeh A (2011) A Fuzzy classification system based on ant colony optimization for diabetes disease diagnosis. Exp Syst Appl 38:14650–9
Gerstenkorn T, Man’ko J (1991) Correlation of intuitionistic fuzzy sets. Fuzzy Sets Syst 44:39–43
Godo L, Sandri S, Dutra L, Freitas C, Carvalho O, Guimarães R, Amaral R (2009) Classification of schistosomiasis prevalence using fuzzy case-based reasoning. Bio Inspired Syst Comput Ambient Intell 5517:1053–1060
Goncalves L, Vellasco M, Pacheco M, de Souza F (2006) Inverted hierarchical neuro-fuzzy BSP system: a novel neuro-fuzzy model for pattern classification and rule extraction in databases. IEEE Trans Syst Man Cybern C Appl Rev 36(2):236–248
Hyung L, Song Y, Lee K (1994) Similarity measure between fuzzy sets and between elements. Fuzzy Sets Syst 62:291–293
Jin Q, Jie H, Ying-hong P, Wei-ming W, Zhen-fei Z (2010) New weighted fuzzy case retrieval method for customer-driven product design. J Shanghai Jiaotong Univ (Sci) 15(6):641–650
Kahramanli H, Allahverdi N (2008) Design of a hybrid system for the diabetes and heart diseases. Expert Syst Appl 35(1–2):82–89
Kalaiselvi C, Nasira G (2014) A new approach for diagnosis of diabetes and prediction of cancer using ANFIS. In: IEEE conference of world congress on computing and communication technologies, pp 188–190
Kalpana M, Kumar A (2011a) Fuzzy expert system for diabetes using fuzzy verdict mechanism. Int J Adv Netw Appl 03(02):1128–1134
Kalpana M, Kumar A (2011b) Fuzzy expert system for diagnosis of diabetes using fuzzy determination mechanism. Int J Comput Sci Emerg Technol 2(6):354–361
Karatsiolis S, Schizas C (2012) Region based support vector machine algorithm for medical diagnosis on Pima Indian Diabetes dataset. In: IEEE 12th international conference on bioinformatics and bioengineering (BIBE), pp 139–144
Khanum A, Mufti M, Javed M, Shafiq M (2009) Fuzzy case-based reasoning for facial expression recognition. Fuzzy Sets Syst 160:231–250
Kumari S, Singh A (2013) A data mining approach for the diagnosis of diabetes mellitus. In: 7th International conference on intelligent systems and control (ISCO), pp 373–375
Lee C, Wang M (2011) A fuzzy expert system for diabetes decision support application. IEEE Trans Syst Man Cybern Part B Cybern 41(1):139–153
Li S, Ho H (2009) Predicting financial activity with evolutionary Fuzzy case-based reasoning. Expert Syst Appl 36:411–422
Li H, Sun J (2011) On performance of case-based reasoning in Chinese business failure prediction from sensitivity, specificity, positive and negative values. Appl Soft Comput 11(1):460–7
Marling C, Shubrook J, Schwartz F (2008) Case-based decision support for patients with type 1 diabetes on insulin pump therapy. In: Advances in case-based reasoning: 9th European Conference (ECCBR), vol 5239, pp 325–339
Marling C, Wiley M, Cooper T, Bunescu R, Shubrook J, Schwartz F (2011) The 4 diabetes support system: a case study in CBR research and development. Case Based Reason Res Dev 6880:137–150
Mohamudally N, Khan D (2011) Application of a unified medical data miner (UMDM) for prediction, classification, interpretation and visualization on medical datasets: the diabetes dataset case. In: Advances in data mining: applications and theoretical aspects, vol 6870, pp 78–95
Montani S, Bellazzi R, Portinale L, d’Annunzio G, Fiocchi S, Stefanelli M (2000) Diabetic patients management exploiting case-based reasoning techniques. Comput Methods Programs Biomed 62(3):205–218
Pappis C, Karacapilidis N (1993) A comparative assessment of measures of similarity of fuzzy values. Fuzzy Sets Syst 56:171–174
Patil B, Joshi R, Toshniwal D (2010) Hybrid prediction model for Type-2 diabetic patients. Expert Syst Appl 37(12):8102–8108
Petrovic S, Mishra N, Sundar S (2011) A novel case based reasoning approach to radiotherapy planning. Expert Syst Appl 38:10759–10769
Polat K, Güneş S (2007) An expert system approach based on principal component analysis and adaptive Neuro-fuzzy inference system to diagnosis of diabetes disease. Digit Signal Proc 17(4):702–710
Polat K, Gunes S, Aslan A (2008) A cascade learning system for classification of diabetes disease: generalized discriminant analysis and least square support vector machine. Expert Syst Appl 34(1):214–221
Portinale L, Montani S (2002) A fuzzy case retrieval approach based on SQL for implementing electronic catalogs. Adv Case Based Reason 2416:321–335
Qin Y, Lu W, Qi Q, Liu X, Huang M, Scott P, Jiang X (2018) Towards an ontology-supported case-base d reasoning approach for computer-aided tolerance specification. Knowl Based Syst 141:129–147
Radha R, Rajagopalan S (2007) Fuzzy logic approach for diagnosis of diabetics. Inf Technol J 61:96–102
Raza B, Kumar Y, Malik A, Anjum A, Faheem M (2018) Performance prediction and adaptation for database management system workload using case-based reasoning approach. Inf Syst 76:46–58
Relich M, Pawlewski P (2018) A case-based reasoning approach to cost estimation of new product development. Neurocomputing 272:40–45
Rodriguez Y, Garcia M, Baets B, Morell C, Bello R (2006) A connectionist fuzzy case-based reasoning model. In: MICAI: advances in artificial intelligence. Springer, Berlin, pp 176–185
Samant P, Agarwal R (2018) Machine learning techniques for medical diagnosis of diabetes using iris images. Comput Methods Programs Biomed 157:121–128
Sarkheyli-Hägele A, Söffker D (2017) Fuzzy SOM-based case-based reasoning for individualized situation recognition applied to supervision of human operators. Knowl Based Syst 137:42–53
Shankaracharya OD, Samanta S, Vidyarthi A (2010) Computational intelligence in early diabetes diagnosis: a review. Rev Diabet Stud 7(4):252–62
Sushmita S, Chaudhury S (2007) Hierarchical fuzzy case based reasoning with multi-criteria decision making for financial applications. Pattern Recognit Mach Intell 4815:226–234
Temurtas H, Yumusak N, Temurtas F (2009) A comparative study on diabetes disease diagnosis using neural networks. Expert Syst Appl 36(4):8610–8615
Varma K, Rao A, Lakshmi T, Rao P (2014) A computational intelligence approach for a better diagnosis of diabetic patients. Comput Electr Eng 40:1758–1765
Wang W (1997) New similarity measures on fuzzy sets and on elements. Fuzzy Sets Syst 85:305–309
Wu D, Li J, Bao C (2017) Case-based reasoning with optimized weight derived by particle swarm optimization for software effort estimation. Soft Comput 1–12
Xiaodong H, Jianwu W, Fuqian S, Haiyan C (2009) Apply fuzzy case-based reasoning to knowledge acquisition of product style. In: IEEE 10th international conference on computer-aided industrial design and conceptual design (CAID & CD), pp 383–386
Xiong N (2011) Learning fuzzy rules for similarity assessment in case-based reasoning. Expert Syst Appl 38:10780–10786
Yao B, Li S (2010) ANMM4CBR: a case-based reasoning method for gene expression data classification. Algorithms Mol Biol (AMB) 5:14
Yu F, Li X, Han X (2018) Risk response for urban water supply network using case-based reasoning during a natural disaster. Saf Sci 106:121–139
Zadeh LA (2003) From search engines to question-answering systems the need for new tools. In: The 12th IEEE international conference fuzzy systems, vol 2, pp 1107–1109
Zhang Z, Chen D, Feng Y, Yuan Z, Chen B, Qin W, Zou S, Qin S, Han J (2018) A strategy for enhancing the operational agility of petroleum refinery plant using case based fuzzy reasoning method. Comput Chem Eng 111:27–36
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) Grant funded by the Korea government (MSIT)-NRF-2017R1A2B2012337. The authors would like to thank Dr. Farid Badria, Prof. of Pharmacognosy, Department, and head of Liver Research Lab, Mansoura University, Egypt; and Dr. Hosam Zaghloul, Prof. at Clinical Pathology Department, Faculty of Medicine, Mansoura University, Egypt, for their efforts in this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Communicated by V. Loia.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendices
Appendix A
The performance evaluation of the proposed fuzzy CBR system
\(K=3\) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
Query cases | Retrieved cases | FKI-CBR diagnose | Domain expert diagnose | |||||||
Diabetes diagnosis | Nephropathy | Hypercholesterolemia | Cancer | Liver | Glomerulonephritis | Splenomegaly | Confidence (%) | |||
Case 1 | Case 1-1 | D | N | N | N | N | N | N | 90 | Diabetes Diagnosis: D |
Case 1-2 | P | N | N | N | N | N | N | 88.9 | Percentage of similarity for another diagnosis: 9/10 | |
Case 1-3 | D | N | H | N | N | N | N | 88.8 | ||
Case 2 | Case 2-1 | N | N | N | N | N | N | N | 95 | Diabetes Diagnosis: N |
Case 2-2 | N | N | N | N | N | N | N | 91 | Percentage of similarity for another diagnosis: 10/10 | |
Case 2-3 | N | N | N | N | N | N | N | 90 | ||
Case 3 | Case 3-1 | P | SH | N | N | N | G | N | 88 | Diabetes Diagnosis: D |
Case 3-2 | DG | SH | N | N | F | G | N | 85 | Percentage of similarity for another diagnosis: 10/10 | |
Case 3-3 | D | N | N | N | N | N | N | 84 | ||
Case 4 | Case 4-1 | D | N | H | N | N | G | N | 94 | Diabetes Diagnosis: D |
Case 4-2 | D | N | H | N | N | G | N | 93.2 | Percentage of similarity for another diagnosis: 10/10 | |
Case 4-3 | N | NP | N | N | N | G | N | 86.5 | ||
Case 5 | Case 5-1 | D | N | N | N | N | N | N | 100 | Diabetes Diagnosis: D |
Case 5-2 | D | N | N | O | N | N | N | 88.8 | Percentage of similarity for another diagnosis: 10/10 | |
Case 5-3 | D | N | N | N | N | N | N | 86.9 | ||
Case 6 | Case 6-1 | P | SH | N | N | N | G | N | 81 | Diabetes Diagnosis: P |
Case 6-2 | D | N | N | N | N | N | N | 79 | Percentage of similarity for another diagnosis: 8/10 | |
Case 6-3 | D | N | N | HCC | HCC | N | N | 79 | ||
Case 7 | Case 7-1 | N | N | N | N | N | N | N | 92.4 | Diabetes Diagnosis: N |
Case 7-2 | D | N | N | N | N | N | N | 89.6 | Percentage of similarity for another diagnosis: 10/10 | |
Case 7-3 | N | N | N | N | N | N | N | 89.4 | ||
Case 8 | Case 8-1 | D | N | N | N | HCC | N | N | 99 | Diabetes Diagnosis: D |
Case 8-2 | N | N | H | N | N | N | N | 82.5 | Percentage of similarity for another diagnosis: 9/10 | |
Case 8-3 | P | N | N | N | N | N | N | 80 | ||
Case 9 | Case 9-1 | D | N | H | N | N | N | N | 93 | Diabetes Diagnosis: D |
Case 9-2 | DG | SH | N | N | N | G | N | 91 | Percentage of similarity for another diagnosis: 10/10 | |
Case 9-3 | D | N | N | N | HCC | N | SG | 90 | ||
Case 10 | Case 10-1 | D | N | N | N | N | N | N | 91 | Diabetes Diagnosis: D |
Case 10-2 | DG | N | N | N | N | N | N | 89.9 | Percentage of similarity for another diagnosis: 10/10 | |
Case 10-3 | P | N | H | N | N | N | N | 89.2 | ||
N normal, A abnormal, D diabetic, P pre-diabetic, H hypercholesterolemia, O ovarian cancer, DG gestational diabetes, SH Shrunken Kidney, G glomerulonephritis, NP nephropathy, SG splenomegaly, HCC hepatocellular carcinoma, F fatty liver, LC liver cirrhosis
Appendix B
A comparison of the proposed fuzzy CBR system and traditional CBR system
Query case | Domain expert decision | Proposed system decision | Traditional system decision |
|---|---|---|---|
Case1 | Diabetes diagnosis: D | D (90%) | P (78%) |
Case 2 | Diabetes diagnosis: N | N (95%) | N (83%) |
Case 3 | Diabetes diagnosis: D | P (88%) | D (70%) |
Case 4 | Diabetes diagnosis: D | D (94%) | P (64%) |
Case 5 | Diabetes diagnosis: D | D (100%) | D (80%) |
Case 6 | Diabetes diagnosis: P | P (81%) | D (64%) |
Case 7 | Diabetes diagnosis: N | N (92.4%) | N (85%) |
Case 8 | Diabetes diagnosis: D | D (99%) | P (79%) |
Case 9 | Diabetes diagnosis: D | D (93%) | P (74%) |
Case 10 | Diabetes diagnosis: D | D (91%) | P (75%) |
Appendix C
A comparison of our FCBR and ML classifiers using our case-base data
Fold | Algorithm | Precision (%) | TPR-Recall (%) | Accuracy (%) | F-Measure (%) |
|---|---|---|---|---|---|
Machine-learning algorithms | |||||
Twofold | C4.5 | 90 | 93.1 | 90 | 91.5 |
k-NN (\(k=3\)) | 80 | 69 | 66.66 | 74.1 | |
SVM | 75.8 | 86.2 | 68.33 | 80.6 | |
Naive Bayes | 86.2 | 86.2 | 75 | 86.2 | |
ANN | 70.6 | 82.8 | 65 | 76.2 | |
Threefold | C4.5 | 89.7 | 89.7 | 88.33 | 89.7 |
k-NN (\(k=3\)) | 74.1 | 69 | 60 | 71.4 | |
SVM | 78.8 | 89.7 | 71.66 | 83.9 | |
Naive Bayes | 83.9 | 89.7 | 75 | 86.7 | |
ANN | 73.5 | 86.2 | 65 | 79.4 | |
Fourfold | C4.5 | 89.7 | 89.7 | 88.33 | 89.7 |
k-NN (\(k=3\)) | 76.9 | 69 | 65 | 72.7 | |
SVM | 77.4 | 82.8 | 71.66 | 80 | |
Naive Bayes | 79.3 | 79.3 | 71.66 | 79.3 | |
ANN | 76.7 | 79.3 | 66.66 | 78 | |
Fivefold | C4.5 | 93.1 | 93.1 | 91.67 | 93.1 |
k-NN (\(k=3\)) | 73.1 | 65.5 | 63.33 | 69.1 | |
SVM | 75 | 82.8 | 66.66 | 78.7 | |
Naive Bayes | 83.3 | 69 | 66.66 | 75.5 | |
ANN | 75.8 | 86.2 | 68.33 | 80.6 | |
Sixfold | C4.5 | 89.7 | 89.7 | 88.33 | 89.7 |
k-NN (\(k=3\)) | 74.1 | 69 | 63.33 | 71.4 | |
SVM | 81.3 | 89.7 | 75 | 85.2 | |
Naive Bayes | 86.2 | 86.2 | 73.33 | 86.2 | |
ANN | 76.5 | 89.7 | 70 | 82.5 | |
Sevenfold | C4.5 | 90 | 93.1 | 90 | 91.5 |
k-NN (\(k=3\)) | 63.3 | 65.5 | 58.33 | 64.4 | |
SVM | 76.7 | 79.3 | 71.66 | 78 | |
Naive Bayes | 75.9 | 75.9 | 70 | 75.9 | |
ANN | 76.7 | 79.3 | 65 | 78 | |
Eightfold | C4.5 | 89.3 | 86.2 | 86.66 | 87.7 |
k-NN (\(k=3\)) | 78.6 | 75.9 | 66.66 | 77.2 | |
SVM | 82.1 | 79.3 | 71.66 | 80.7 | |
Naive Bayes | 78.1 | 86.2 | 73.33 | 82 | |
ANN | 77.4 | 82.8 | 71.66 | 80 | |
Ninefold | C4.5 | 89.7 | 89.7 | 88.33 | 89.7 |
k-NN (\(k=3\)) | 83.3 | 69 | 66.66 | 75.5 | |
SVM | 80.6 | 86.2 | 76.66 | 83.3 | |
Naive Bayes | 82.1 | 79.3 | 73.33 | 80.7 | |
ANN | 83.9 | 89.7 | 75 | 86.7 | |
Tenfold | C4.5 | 89.7 | 89.7 | 88.33 | 89.7 |
k-NN (\(k=3\)) | 73.1 | 65.5 | 56.67 | 69.1 | |
SVM | 80.6 | 86.2 | 75 | 83.3 | |
Naive Bayes | 83.3 | 86.2 | 75 | 84.7 | |
ANN | 76.5 | 89.7 | 70 | 82.5 | |
k-fold (\(k=60\)) \(\equiv \) LOOCV | C4.5 | 89.7 | 89.7 | 88.33 | 89.7 |
k-NN (\(k=3\)) | 73.1 | 65.5 | 58.33 | 69.1 | |
SVM | 80 | 82.8 | 75 | 81.4 | |
Naive Bayes | 85.2 | 79.3 | 73.33 | 82.1 | |
ANN | 78.8 | 89.7 | 75 | 83.9 | |
Average (%) | 80.568 | 82.086 | 73.0972 | 81.164 | |
Maximum (%) | 93.1 | 93.1 | 91.67 | 93.1 | |
Conventional CBR system | 76 | 47.5 | 55 | 58.46 | |
Proposed FCBR system | 96 | 97.96 | 95 | 96.97 | |
Rights and permissions
About this article
Cite this article
El-Sappagh, S., Elmogy, M., Ali, F. et al. A case-base fuzzification process: diabetes diagnosis case study. Soft Comput 23, 5815–5834 (2019). https://doi.org/10.1007/s00500-018-3245-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00500-018-3245-3