Abstract
Promoters can direct gene expression specifically to targeted tissues or cells. Effective with both crop species and model plant systems, these tools can help researchers overcome the practical obstacles associated with transgenic protocols. Here, we identified promoters that allow one to target the manipulation of gene expression during pollen development. Utilizing published transcriptomic databases for rice, we investigated the promoter activity of selected genes in Arabidopsis. From various microarray datasets, including those for anthers and pollen grains at different developmental stages, we selected nine candidate genes that showed high levels of expression in the late stages of rice pollen development. We named these Oryza sativa late pollen-specific genes. Their promoter regions contained various cis-acting elements that could be responsible for anther-/pollen-specific expression. Promoter::GUS–GFP reporters were constructed and introduced into Arabidopsis plants. Histochemical GUS staining revealed that six of the nine rice promoters conferred strong GUS expression that was restricted to the anthers in Arabidopsis. Further analysis showed that although the GUS signals were not detected at the unicellular stage, they strengthened in the bicellular or tricellular stages, peaking at the mature pollen stage. This paralleled their transcriptomic profiles in rice. Based on our results, we proposed that these six rice promoters, which are active in the late stages of pollen formation in the dicot Arabidopsis, can aid molecular breeders in generating new varieties of a monocot plant, rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Billons of people rely on the double fertilization of flowering plants for products used in their daily life, including food and clothing. As demand for high-quality food increases with continued population growth, while the agricultural environment is altered through climate change, researchers will be challenged to develop new crop varieties. For example, the male gametophyte (pollen grain) from rice, one of the most important stable crop, presents an excellent target for molecular breeding experiments.
The most effective strategy for improving yields from fertile F1 hybrids is to create male sterile or restorer lines. This can be accomplished by either inhibiting endogenous hormone biosynthesis during pollen development or applying appropriate hormones to the reproductive tissue at the correct stage (Bae et al. 2010). Thus, identifying the best anther-/pollen-specific rice promoter is necessary so that one avoids undesirable effects on other sporophytic tissues that may alter plant growth or flowering time. Although such efforts that employ several constructs with rice tissue culture are feasible in the laboratory, field trials with transgenic plants are very labor-intensive, can usually be conducted only once in each growing season, and are constrained by safety regulations. Therefore, it would be advantageous to obtain rice promoters whose specificity for anther/pollen is equivalent to that found with the model plant Arabidopsis.
Pollen grains form within the anther during several distinct stages that involve three rounds of cell division (Borg et al. 2009). A pollen mother cell undergoes meiosis, giving rise to a tetrad of four haploid microspores enclosed in a callose wall. Each microspore is released upon the dissolution of the callose wall and undergoes pollen mitosis I (PMI). This highly asymmetric division results in a larger vegetative cell and a smaller generative cell with different fates. While the vegetative cell exits the cell cycle and differentiates for its biological role to deliver sperm cells in the vicinity of the female gametophyte via the pollen tube, the generative cell proceeds to pollen mitosis II (PMII), in which two sperm cells are produced. The majority of species, such as lily and tomato, sheds the bicellular pollen, with PMII taking place in the pollen tube. In contrast, Arabidopsis and rice shed tricellular pollen at anthesis, although their promoters did not necessarily work in bicellular plants (Ge et al. 2011).
Male gametophyte development succeeds because of the highly orchestrated expression of several genes. Numerous research groups have already identified some of the promoters that control pollen-specific expression (Honys and Twell 2004; Engel et al. 2005; Honys et al. 2006; Verelst et al. 2007; Gupta et al. 2007; Zhang et al. 2010; Khurana et al. 2013b). Both early and late pollen genes have been studied in various crops (Zimmermann et al. 2004; Twell et al. 2006; Borges et al. 2008; Zhang et al. 2010; Huang et al. 2011). Most of the late pollen genes exhibit similar patterns of expression, in which transcripts are generally accumulated beginning at PMI and then increase in abundance as the pollen grains mature (Twell et al. 1989; Xu et al. 1999).
Although several such promoters have been analyzed in crop plants, only Osg6B, OsIPP3, OsbHLH, OsIPK, OsFbox, OsIPP3, OsIPA, OsSCP1, OsSCP2, and OsSCP3 have been reported in rice (Yokoi et al. 1997; Park et al. 2006; Swapna et al. 2011; Khurana et al. 2013a, b). Among these, the OsIPK, OsIPA, OsbHLH, OsFbox and OsIPP3 promoters have conferred anther-/pollen-specific expression in the heterologous dicot system of Arabidopsis (Swapna et al. 2011; Khurana et al. 2013a, b). The most effective way to identify these promoters is to obtain genome-wide expression profiles during microsporogenesis, utilizing transcriptome analyses based on DNA microarray or RNA-seq technologies. The two most popular and reliable commercial array platforms in rice are Affymetrix and Agilent 44K. To date, 2,081 Affymetrix and 1,706 Agilent 44K array data are available from The Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (NCBI; www.ncbi.nlm.nih.gov/geo/) (Jung et al. 2011; Barrett et al. 2013). Three genome-wide perspectives have recently been published on the use of transcriptomes from developing anthers and pollen in rice: (1) transcriptome analysis of pre-meiotic (PMA), meiotic (MA), anthers at single-celled (SCP), and tri-nucleate pollen (TPA) stages (Deveshwar et al. 2011); (2) 33 laser micro-dissection (LM) microarray data and 11 microarray data covering the four stages of anther development (Aya et al. 2011); and (3) transcriptomes for developing and germinated pollen, and spern cell (Wei et al. 2010; Russell et al. 2012). Meta-expression analyses of these data are useful for identifying pollen-preferred or pollen-specific genes. Furthermore, the Rice Oligonucleotide Array Database (www.ricearray.org/) provides comprehensive expression profiles for rice genes that function in anatomy or development. These sources offer an approach for filtering out genes expressed in tissues/organs other than those being targeted (Cao et al. 2012).
Here, we used pollen-specific promoters identified in rice to evaluate their activities in Arabidopsis, a heterologous model plant. Also, for the evaluation of pollen-specific GUS expression, we performed rice transformation. Based on meta-expression analysis using microarray datasets available in rice, we selected nine genes showing high expression during the later stages of pollen grain formation. We then characterized their promoter activities using promoter::GFP–GUS reporters in Arabidopsis and rice.
Materials and methods
Selection of candidate genes showing pollen-specific expression in rice
We collected publicly available microarray datasets for rice anthers and tricellular, mature, and germinating pollen from the GEO database (Supplemental Tables 1 and 2). These data were normalized via Affy software, as described by Cao et al. (2012). Average intensities for reproductive tissues, including the anthers and grains at the trinucleate and mature stages, were compared with those for other vegetative tissues and organs. Those that showed distinct patterns of expression were chosen for further analysis.
Isolation of promoter region and construction of promoter::GFP–GUS reporters
Primers to amplify the promoter region for our nine candidate genes were designed via the oligonucleotide properties calculator (http://www.basic.northwestern.edu/biotools/oligocalc.html; Supplemental Table 3). Sizes for the selected promoter regions varied from 263 to 2,283 bp (Table 1).
We used the GATEWAY cloning system to generate promoter::GUS–GFP reporters according to the manufacturer’s instructions (Invitrogen, http://www.invitrogen.com). First, promoter fragments were amplified by two-step PCR, using genomic DNA extracted from 3-week-old japonica rice seedlings (‘Donjinbyeo’) by the cetyl trimethyl ammonium bromide (CTAB) method (Ronald and Chen 1999). For the first step, gene-specific primers with 12 bp of attB1 and attB2 sites were added, and PCR was conducted at 94 °C/5 min; then 10 cycles of 94 °C/30 s, 58 °C/30 s, and 72 °C/2 min; followed by a final extension of 72 °C/5 min. In the second step, 10 μL from the first PCR reaction was added to a 40-μL PCR reaction mixture containing the attB1 and attB2 adapter primers. Conditions included 20 cycles of 94 °C/30 s, 55 °C/30 s, and 72 °C/2 min; then a final extension of 72 °C/5 min. BP recombination reactions were conducted between the amplified promoter fragments and the pDONR201 vector, using BP clonase mix II (Invitrogen). The resulting entry clones were verified by sequencing and subjected to LR recombination reactions with the pKGWFS7 plant destination vector (Karimi et al. 2005), using LR clonase mix II (Invitrogen). The final plant expression vectors were transferred into Agrobacterium tumefaciens (GV3101).
Agrobacterium-mediated transformation of Arabidopsis and rice
Plants of Arabidopsis (‘Columbia’, or ‘Col-0’) were grown at 20–22 °C in a controlled-environment room under a 16-h light regime. Wild-type (WT) plants were transformed by the floral-dip method (Clough and Bent 1998). Transgenic plants were selected on Murashige and Skoog agar media supplemented with 50 mg L−1 kanamycin and 200 mg L−1 cefotaxime.
Mature seeds of the japonica rice cultivar Dongjinbyeo were used for Agrobacterium-mediated transformation (Hiei et al. 1997; Toki et al. 2006). We extracted genomic DNAs from the leaf tissues of T1 transgenic plants with a homogenization procedure that included TissueLyser (Qiagen, http://www.qiagen.com). PCR was performed in a 20 μL mixture containing 1 μL of genomic DNA, 1 μL of each primer (20 pmol), 0.4 μL of dNTPs (1 mM), 2 μL of 10 X PCR buffer, and 0.1 μL of e-Taq polymerase (5 units μL−1; Solgent, http://www.solgent.com). Conditions included 95 °C/2 min; then 30 cycles of 94 °C/1 min, 55 °C/1 min, and 72 °C/2 min; followed by a final extension of 72 °C/5 min.
Histochemical staining for GUS and microscopic evaluation
Histochemical GUS staining was performed with transgenic plants as described by Oh et al. (2010). Seedlings or whole inflorescences were submerged in GUS buffer [0.1 M sodium phosphate (pH 7.0), 1 mM EDTA (pH 8.0), 0.1 % Triton X-100, and 0.5 mM potassium ferricyanide] that contained 1 mM X-Gluc (5-bromo-4-chloro-3-indolyl-d-glucuronic acid). They were then vacuum-infiltrated for 10 min and incubated overnight at 37 °C. Afterward, the tissues were cleared in 70 % ethanol and viewed under a stereomicroscope (Stemi 2000C, Zeiss). Stained seedlings and inflorescences were imaged using a ProgRes C3 camera (Jenoptik, Germany). To examine the developing microspores and pollen grains at different stages, we fixed the GUS-stained inflorescences in a 3:1 (v:v) solution of ethanol and acetic acid prior to staining with DAPI (4′, 6-diamidino-2-phenylindole). Anthers in DAPI solution on the microscope slides were then opened with dissecting needles and gently squashed under a cover slip. Images were captured from either the ProgResC3 camera to show GUS signals under a bright field or else the ProgRes MFcool camera for investigating DAPI epifluorescence under a Nikon ECLIPSE 80i microscope (Nikon).
Results
Candidate genes showing pollen-specific expression
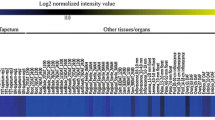
To identify pollen-specific genes in rice, we collected 65 Affymetrix arrays from five datasets. From these, we selected six slides for anthers containing tricellular and mature pollen grains, and 11 with tricellular and mature pollen grains. As controls, we used other tissue/organ types from Affymetrix meta-anatomical expression data that we had recently produced (Cao et al. 2012). In all, 261 genes showed 500-fold change in expression when compared with the other tissues/organs. Our heat map of the patterns displayed by nine randomly selected candidate genes confirmed that expression was preferential in grains at the tri-nucleate, mature, and germinating stages (Fig. 1).
Heat map analysis of expression patterns for nine genes, based on meta-anatomical expression. Blue color indicates low expression; yellow high expression. Values beneath blue–yellow bar indicate normalized average log2 intensity. Samples presented in heat map are detailed in Supplemental Tables 1 and 2 (color figure online)
Sequence analysis of nine promoter regions
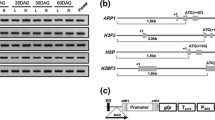
To evaluate the promoter activity for these pollen-specific genes, we used genomic DNA from ‘Dongjinbyeo’ rice as template for isolating the upstream regulatory region (URR), which comprised 263–2.3 kb, excluding the 5′-untranslated region (UTR). Amplified promoter fragments were used for constructing reporter gene fusions. The resulting binary vectors were designated as OsLPS1 through 9 (rice late pollen-specific gene 1–9) (Table 1). Sequences of the promoter fragments were compared with the ‘Nipponbare’ rice genomic sequence, using the BioEdit sequence alignment editor. Because ‘Nipponbare’ differs from ‘Dongjinbyeo’ rice, we found variations while verifying the promoter sequence of OsLPS3. Similar results have been described for the URRs of anther-/pollen-specific genes OsIPK, OsbHLH, and OsFbox (Gupta et al. 2007; Khurana et al. 2013b). By comparison, no differences between the two cultivars were found for the other eight promoters.
The cis-acting elements possess specific sequence information that is recognized by corresponding trans-acting factors. Various cis-acting elements in the promoter region are involved in anther-/pollen-specific expression (Tebbutt and Lonsdale 1995; Rogers et al. 2001; Swapna et al. 2011). We used the promoter analysis program PLACE DATABASE signal scan (Higo et al. 1999) to detect putative cis-acting elements. Regions examined here included 263 URR + 148 UTR (411 bp) for OsLPS1, 2283 URR + 122 UTR (2,405 bp) for OsLPS2, 1520 URR + 295 UTR (1,815 bp) for OsLPS3, 963 URR + 75 UTR (1,038 bp) for OsLPS4, 1792 URR (1,792 bp)) for OsLPS5, 1779 URR + 89 UTR (1,868 bp) for OsLPS6, 1784 URR + 144 UTR (1,928 bp) for OsLPS7, 1634 URR + 77 UTR (1,711 bp) for OsLPS8, and 1498 URR + 72 UTR (1,570 bp) for OsLPS9. Those elements present in all promoter regions are listed in Table 2.
Each promoter had several MYC recognition sites (CANNTG) that occur in the promoters of the dehydration gene rd22 and many other genes in Arabidopsis (Abe et al. 2003). All promoter regions had the AAAG motif, which is the recognition sequence of Dof proteins, a family of transcription factors associated with diverse promoters of plant-specific genes (Yanagisawa and Schmidt 1999). They also contained the GTGA motif, which is similar to tobacco g10 late pollen regulatory elements. The tobacco gene g10, which is homologous to pectate lyases and the tomato gene lat56, is preferentially and maximally expressed in mature pollen (Rogers et al. 2001). We also detected the DNA-binding motif for Arabidopsis ARR1 and ARR2 proteins, which are response regulators that work as transcriptional activators in plant cells (Sakai et al. 2000). All promoters, except for OsLPS1, had numerous AGAAA elements that are required for transcriptional activation of pollen-specific expression in tomato (Bate and Twell 1998). Their presence implied that all of the promoters examined here were likely to confer preferential expression during rice pollen development. Furthermore, the TGGGCY motif, which contains TCP domain-binding elements that are implicated in anther- and meristem-specific gene expression (Welchen and Gonzalez 2005), was detected in four promoters, as was the GATAA motif, which is a conserved element in many light-regulated promoters in dicots and monocots (Martínez-Hernández et al. 2002). These analytical results suggested that our nine selected promoters direct mature pollen-specific expression in a stage-specific manner, operating as late pollen genes in a heterologous system.
Analysis of promoter::GFP–GUS reporters in Arabidopsis
To assess the activity of our selected promoters, we generated promoter::GFP–GUS constructs and introduced them into WT Arabidopsis plants. PCR analysis was conducted to verify that the transformants contained each T-DNA insertion, using a forward primer specific to each promoter sequence and a reverse primer for the GFP sequence (data not shown). For each construct, we obtained 20 T1 transgenic plants and evaluated GUS activity in young buds and open flowers. Except for OsLPS5, all inflorescences accumulated GUS in the flowers and at the late stage of bud formation but showed no expression in small buds or at the early stage of pollen development. For the OsLPS5 promoter, expression was very weak, making it difficult to conclude that it could be used to drive either pollen-specific or preferential gene expression in Arabidopsis. All tested plants showed the same pattern of expression in their respective constructs. Therefore, we used the other eight promoters to examine the anther-specific expression of genes in Arabidopsis (Fig. 2).
Promoter activity was also assessed in vegetative tissues from T2 seedlings for each construct (Fig. 3). GUS activity was not detected in seedlings harboring six promoters—OsLPS1 through 4, OsLPS6, and OsLPS9—whereas plants harboring OsLPS7 and OsLPS8 promoters showed significant GUS signals in the roots and developing leaves, respectively. Because transcript levels for the first six promoters were much lower than in the open flowers and late-stage developing buds, they could be classified as pollen-preferred. Therefore, we concluded that six of the eight promoters could induce pollen-specific expression in Arabidopsis.
We produced transgenic Arabidopsis plants to monitor the expression patterns of all nine putative pollen-specific promoters fused with the GUS reporter gene. Twenty transgenic lines per construct were used for histochemical assays (Fig. 4). During pollen formation, GUS signals were detected in all promoters except for OsLPS5. At the unicellular stage (UC), no signals were detected for any of the promoters. GUS activity generally intensified over time, peaking at the mature pollen (MP) stage. This indicated that most of the promoters drove late pollen-specific or preferential expression, as supported by our microarray analysis.
Similar tissue-specific or preferential patterns of expression were observed in Arabidopsis plants harboring six vectors that expressed GUS under the control of the OsLPS1, OsLPS3, OsLPS4, OsLPS6, OsLPS8, and OsLPS9 promoters. No GUS expression occurred in the early stage of microspore development (UC) for any of the promoters. During the bicellular stage (BC), GUS activity was either absent (OsLPS3, OsLPS6, and OsLPS8) or detected only in trace amounts (OsLPS1, OsLPS4, and OsLPS9). Activity was found at the tricellular (TC) stage after PMII, peaking at the mature pollen (MP) stage (Fig. 4). The T1 transgenic plants carrying promoters for OsLPS2 and OsLPS7 showed similar patterns of GUS expression during male gametophyte development, being undetectable at the UC stage but beginning to appear at the BC stage. Activity gradually increased at the TC stage and peaked at the MP stage. In all, eight late pollen genes showed the highest expression at the MP stage. By contrast, Arabidopsis plants transformed by the OsLPS5 vector did not demonstrate strong GUS expression in the microspore and pollen development stages, thereby suggesting that its promoter is likely rice or monocot pollen-specific.
We also evaluated the activity of promoters to examine the pollen-specific expression in rice. Two late pollen-specific promoters (OsLPS1 and OsLPS5) exhibiting distinct expression patterns in Arabidopsis (Fig. 4) were analyzed in transgenic rice plants through the monitoring of the GUS gene expression pattern on developing microspore/pollen. Eight T 0 transgenic plants generated with respective constructs showed rice pollen-specific expression (Fig. 5). Rice transgenic plants carrying OsLPS1 construct exhibited similar patterns with Arabidopsis during male gametophyte development in rice. On the contrary, transgenic plants with OsLPS5 displayed the unique pattern showing strong expression at TC stage after PMII (Fig. 5). Hence, our rice data revealed that OsLPS5 is rice pollen-specific.
Discussion
Using genome-wide expression analyses and microarray datasets, we have identified nine candidate genes that are pollen-specific in rice. This approach has been proven efficient for identifying genes and/or promoters that confer spatio-temporal expression. For example, Thilmony et al. (2009) used anatomical meta-expression profiles from Genevestigator (Zimmermann et al. 2008; https://www.genevestigator.com/gv/) to determine that the promoter of Leaf Panicle 2 in rice has expression that is specific to green tissues. Similarly, Jung et al. (2008) identified light-responsive genes in rice by evaluating the data from independent NSF45K and BGI60K arrays to obtain expression profiles for light- and dark-grown seedlings. In Arabidopsis, promoters have been found that direct pollen-specific expression at the microspore stage based on transcriptomic datasets that include pollen grains sampled at different developmental stages (Honys et al. 2006; da Costa-Nunes 2013).
We used upstream regions beyond the 5′-UTR to construct promoter::GUS reporters. The presence of cis-acting elements further supported our belief that their expression is either pollen-specific or preferential. The Arabidopsis ABORTED MICROSPOROGENESIS (AMS), belonging to a MYC superfamily of basic helix-loop-helix transcription factors, is required for tapetum cell development and microspore formation (Sorensen et al. 2003; Xu et al. 2010). It has the capacity to bind in vitro to a CANNTG motif. We noted that all nine of our selected promoters had one to 11 MYC recognition sites for that motif, which also commonly occurs in the upstream regions of genes expressed in the sperm cells of rice (Sharma et al. 2011). Therefore, these findings suggest that MYC-type transcription factors are involved in the expression of our tested genes during pollen development. In our nine promoter regions, we also detected two other cis-acting elements, AAAG and GTGA, that are considered anther- or pollen-specific (Bate and Twell 1998; Yanagisawa and Schmidt 1999; Rogers et al. 2001). Another element (AGAAA) common to late pollen-specific genes was found in all selected promoter regions except for OsLPS1. For that gene, this motif was situated within its 5′-UTR region. We also located three copies of the TGGGCY motif in YY2 tapetum-specific genes, where it functions in anther- and meristem-specific gene expression for nuclear proteins PCF1 and PCF2 (Kuriakose et al. 2009). However, it remains unclear whether this motif is active in pollen-specific expression because it has been evaluated only through mutagenesis experiments and gain-of-function analysis (Gupta et al. 2007).
We performed a Blast search of genes that were homologous at the protein level for all nine rice late pollen-specific sequences tested here. Of these, five genes (OsLPS1, OsLPS2, OsLPS3, OsLPS4, and OsLPS5) had significant levels for their e-values, i.e., 4e−33, 7e−60, 2e−74, 5e−121, and 1e−13, respectively. By contrast, four other genes (OsLPS6 through OsLPS9) were not detected at significant levels in the Arabidopsis genome, even though their protein information is publicly available for that species and others. We determined that OsLPS1 (Os08g0560700) shares approximately 64–66 % identity with amino acids encoding two calmodulin-like proteins: CML28 (At3g03430) and CML29 (At5g17480). The expression profile for the former is most significantly changed during pollen germination and tube growth (Wang et al. 2008). Moreover, OsLPS2 (Os04g0638800) is 42 % identical to a DUF617 plant family protein with unknown function. The Arabidopsis gene family profiler (http://arabidopsisgfp.ueb.cas.cz/) has revealed that it is encoded by At4G39610 and shows preferential expression in mature pollen grains (Dupl’áková et al. 2007).
OsLPS3 (Os11g0683800) encodes a pectinesterase family protein with roles in pollen wall development or the production of pollen-released proteins in Arabidopsis and canola (Noir et al. 2005; Sheoran et al. 2009). It shares approximately 42 % identity with Arabidopsis pectin methylesterases that are encoded by At5g07410 and At1g69940. These are classified as pollen-specific genes because their expression is highly restricted to pollen grains and tubes (Bosch et al. 2005; Tian et al. 2006). OsLPS4 protein is 56 % identical to a peroxidase family protein encoded by At5g47000, for which expression is greatest in the late pollen stages based on NASCArrays (http://arabidopsisgfp.ueb.cas.cz/). OsLPS5 protein shows 63 % identity to DUF581 protein with unknown function encoded by At5G47060, which is expressed at a very low level in pollen grains (http://arabidopsisgfp.ueb.cas.cz/). The OsLPS6 protein encodes a Filamenting temperature-sensitive mutant Z (FtsZ) family protein involved in cell division. In Lilium longiflorum, ftsZ has roles in male gametogenesis and double fertilization (Mori and Tanaka 2000); transcripts are preferentially accumulated in the generative cells of mature pollen (Tang et al. 2009).
Both OsLPS7 and OsLPS8 encode for plant invertase/pectin methylesterase inhibitor domain proteins. The pectin methylesterase (PME) inhibitor has important roles in determining pollen development, tube growth (Bosch et al. 2005; Tian et al. 2006; Zhang et al. 2010), and the stability of cell walls at the tips of those pollen tubes (Rockel et al. 2008). A PME inhibitor gene from Brassica oleracea is preferentially expressed in the mature pollen grains and pollen tubes of Arabidopsis (Zhang et al. 2010). These findings suggest that such inhibitors have a conserved function among species.
Finally, OsLPS9 encodes Lol p 2-family proteins, which are well-characterized allergens in ryegrass pollen. These proteins are highly homologous to Zea m 3, the major Group-1 allergen from maize pollen, displaying the cell wall-loosening activity of beta expansins (Li et al. 2003). Also, OsLPS9 protein is 40 % homologous to the major Group-1 allergen from rice (Ory s 1) (Xu et al. 1995). Analysis of their regulatory regions has demonstrated that genes in this major group from Arabidopsis and tobacco show characteristic late pollen expression (Gupta et al. 2007; Swapna et al. 2011). In the rice genome, putative pollen allergens have been identified and their expression characterized using public database (Jiang et al. 2005) and the Affymetrix 57K rice GeneChip microarray (Russell et al. 2008). All of these results implicate the conserved functioning of major Group-1 allergen genes in the development of mature pollen.
Although more rice pollen-specific genes are being identified, few rice promoters have been shown to be equally specific for pollen in transgenic Arabidopsis plants (Swapna et al. 2011). However, the promoter of rice anther-specific OsIPP3 (Os05g543000), identical to OsLPS7 in our study, exhibits broad activity in anthers and roots from Arabidopsis (Khurana et al. 2013a). This is consistent with the findings reported here. Our transcriptomic data revealed that GUS activity in the Arabidopsis inflorescences and pollen grains was driven to varying degrees by the nine OsLPS promoters and followed spatio-temporal patterns. For example, at all stages, the OsLPS5 promoter led to only limited GUS signals in whole inflorescences and no signals in the grains. The other eight promoters followed similar patterns for GUS expression, starting at low levels in the young buds, but gradually increasing to a peak in the mature pollen. In particular, the earliest GUS activity was detected at the BC stage from the OsLPS1, OsLPS2, OsLPS4, OsLPS7, and OsLPS9 promoters, and at the TC stage for OsLPS3, OsLPS6, and OsLPS8. Moreover, histochemical analysis using promoter::GFP–GUS reporters indicated that six of these were late pollen-specific in our Arabidopsis model plants.
The outcome of this study provides a new set of pollen-specific promoters that can be used with both rice and Arabidopsis. This strategy will allow us to design a more efficient means for applying biotechnological approaches to rice breeding programs. For example, when combined with the expression of cytotoxic genes or the downregulation of genes essential for pollen development, constructs containing these promoters can be used to induce male sterility, a highly desirable agronomic trait (Bae et al. 2010). Therefore, future investigations should utilize a large number of transgenic Arabidopsis plants harboring the same constructs. This will greatly improve our ability to evaluate the in planta impacts of a wide variety of constructs, which is otherwise a very demanding process because of constraints related to space, time, and safety regulations that arise when working with transgenic rice.
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
Aya K, Suzuki G, Suwabe K, Hobo T, Takahashi H, Shiono K, Yano K, Tsutsumi N, Nakazono M, Nagamura Y, Matsuoka M, Watanabe M (2011) Comprehensive network analysis of anther-expressed genes in rice by the combination of 33 laser microdissection and 143 spatiotemporal microarrays. PLoS ONE 6:e26162
Bae HK, Kang HG, Kim GJ, Eu HJ, Oh SA, Song JT, Chung IK, Eun MY, Park SK (2010) Transgenic rice plants carrying RNA interference constructs of AOS (allene oxide synthase) genes show severe male sterility. Plant Breed 129:647–651
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A (2013) NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 41:D991–D995
Bate N, Twell D (1998) Functional architecture of a late pollen promoter: pollen-specific transcription is developmentally regulated by multiple stage specific and co-dependent activator elements. Plant Mol Biol 37:859–869
Borg M, Brownfield L, Twell D (2009) Male gametophyte development: a molecular perspective. J Exp Bot 60:1465–1478
Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijo JA, Becker JD (2008) Comparative transcriptomics of Arabidopsis thaliana sperm cells. Plant Physiol 148:1168–1181
Bosch M, Cheung AY, Hepler PK (2005) Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol 138:1334–1346
Cao P, Jung KH, Choi D, Hwang D, Ronald PC (2012) The rice oligonucleotide array database: an atlas of rice gene expression. Rice 5:17
Clough SJ, Bent AF (1998) Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
da Costa-Nunes JA (2013) A novel Arabidopsis marker line that strongly labels uninucleate microspores and the subsequent male gametophyte development stages. SpringerPlus 2:237
Deveshwar P, Bovill WD, Sharma R, Able JA, Kapoor S (2011) Analysis of anther transcriptomes to identify genes contributing to meiosis and male gametophyte development in rice. BMC Plant Biol 11:78
Dupl’áková N, Reňák D, Hovanec P, Honysová B, Twell D, Honys D (2007) Arabidopsis Gene Family Profiler (aGFP)—user-oriented transcriptomic database with easy-to-use graphic interface. BMC Plant Biol 7:39
Engel ML, Holmes-Davis R, McCormick S (2005) Green sperm. Identification of male gamete promoters in Arabidopsis. Plant Physiol 138:2124–2133
Ge L, Gou X, Yuan T, Strout GW, Nakashima J, Blancaflor EB, Tian HQ, Russell SD (2011) Migration of sperm cells during pollen tube elongation in Arabidopsis thaliana: behavior during transport, maturation and upon dissociation of male germ unit associations. Planta 233:325–332
Gupta V, Khurana R, Tyagi AK (2007) Promoters of two anther-specific genes confer organ-specific gene expression in a stage-specific manner in transgenic systems. Plant Cell Rep 26:1919–1931
Hiei Y, Komari T, Kubo T (1997) Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol Biol 35:205–218
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27:297–300
Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5:R85
Honys D, Oh SA, Renak D, Donders M, Solcova B, Johnson JA, Boudova R, Twell D (2006) Identification of microspore-active promoters that allow targeted manipulation of gene expression at early stages of microgametogenesis in Arabidopsis. BMC Plant Biol 6:31
Huang Z, Gan Z, He Y, Li Y, Liu X, Mu H (2011) Functional analysis of a rice late pollen-abundant UDP-glucose pyrophosphorylase (OsUgp2) promoter. Mol Biol Rep 38:4291–4302
Jiang SY, Jasmin PXH, Ting YY, Ramachandran S (2005) Genome-wide identification and molecular characterization of Ole_e_I, Allerg_1 and Allerg_2 domain-containing pollen-allergen-like genes in Oryza sativa. DNA Res 12:167–179
Jung KH, Dardick C, Bartley LE, Cao P, Phetsom J, Canlas P, Seo YS, Shultz M, Ouyang S, Yuan Q, Frank BC, Ly E, Zheng L, Jia Y, Hsia AP, An K, Chou HH, Rocke D, Lee GC, Schnable PS, An G, Buell CR, Ronald PC (2008) Refinement of light-responsive transcript lists using rice oligonucleotide arrays: evaluation of gene-redundancy. PLoS ONE 3:e3337
Jung KH, Jeon JS, An G (2011) Web tools for rice transcriptome analyses. J Plant Biol 54:65–80
Karimi M, De-Meyer B, Hilson P (2005) Modular cloning in plant cells. Trends Plant Sci 10:103–105
Khurana R, Kathuria H, Mukhopadhyay A, Kapoor S, Tyagi AK (2013a) 286 bp upstream regulatory region of a rice anther-specific gene, OSIPP3, confers pollen-specific expression in Arabidopsis. Biotechnol Lett 35:455–462
Khurana R, Kapoor S, Tyagi AK (2013b) Spatial and temporal activity of upstream regulatory regions of rice anther-specific genes in transgenic rice and Arabidopsis. Transgenic Res 22:31–46
Kuriakose B, Arun V, Gnanamanickam SS, Thomas G (2009) Tissue specific expression in transgenic rice and Arabidopsis thaliana plants of GUS gene driven by the 5′ regulatory sequences of an anther-specific rice gene YY2. Plant Sci 177:390–397
Li LC, Bedinger PA, Volk C, Jones AD, Cosgrove DJ (2003) Purification characterization of four b-expansins (Zea m 1 isoforms) from maize pollen. Plant Physiol 132:2073–2085
Martínez-Hernández A, López-Ochoa L, Argüello-Astorga G, Herrera-Estrella L (2002) Functional properties and regulatory complexity of a minimal RBCS light-responsive unit activated by phytochrome, cryptochrome, and plastid signals. Plant Physiol 128:1223–1233
Mori T, Tanaka I (2000) Isolation of the ftsZ gene from the plastid deficient generative cells in Lilium longiflorum. Protoplasma 214:57–64
Noir S, Bräutigam A, Colby T, Schmidt J, Panstruga R (2005) A reference map of the Arabidopsis thaliana mature pollen proteome. Biochem Biophys Res Commun 337:1257–1266
Oh SA, Park KS, Twell D, Park SK (2010) The SIDECAR POLLEN gene encodes a microspore-specific LOB/AS2 domain protein required for the correct timing and orientation of asymmetric cell division. Plant J 64:839–850
Park JI, Hakozaki H, Endo M, Takada Y, Ito H, Uchida M, Okabe T, Watanabe M (2006) Molecular characterization of mature pollen-specific genes encoding novel small cysteine-rich proteins in rice (Oryza sativa L.). Plant Cell Rep 25:466–474
Rockel N, Wolf S, Kost B, Rausch T, Greiner S (2008) Elaborate spatial patterning of cell-wall PME and PMEI at the pollen tube tip involves PMEI endocytosis, and reflects the distribution of esterified and de-esterified pectins. Plant J 53:133–143
Rogers HJ, Bate N, Combe J, Sullivan J, Sweetman J, Swan C, Lonsdale DM, Twell D (2001) Functional analysis of cis regulatory elements within the promoter of the tobacco late pollen gene g10. Plant Mol Biol 45:577–585
Ronald PC, Chen DH (1999) A rapid DNA mini preparation method suitable for AFLP and other PCR applications. Plant Mol Biol Rep 17:53–57
Russell SD, Bhalla PL, Singh MB (2008) Transcriptome-based examination of putative pollen allergens of rice (Oryza sativa ssp. japonica). Mol Plant 1:751–759
Russell SD, Gou X, Wong CE, Wang X, Yuan T, Wei X, Bhalla PL, Singh MB (2012) Genomic profiling of rice sperm cell transcripts reveals conserved and distinct elements in the flowering plant male germ lineage. New Phytol 195:560–573
Sakai H, Aoyama T, Oka A (2000) Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J 24:703–711
Sharma N, Russell SD, Bhalla PL, Singh MB (2011) Putative cis-regulatory elements in genes highly expressed in rice sperm cells. BMC Res Notes 4:319
Sheoran IS, Pedersen EJ, Ross AR, Sawhney VK (2009) Dynamics of protein expression during pollen germination in canola (Brassica napus). Planta 230:779–793
Sorensen AM, Krober S, Unte US, Huijser P, Dekker K, Saedler H (2003) The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J 33:413–423
Swapna L, Khurana R, Vijaya-Kumar S, Tyagi AK, Rao KV (2011) Pollen-specific expression of Oryza sativa indica pollen allergen gene (OSIPA) promoter in rice and Arabidopsis transgenic systems. Mol Biotechnol 48:49–59
Tang LY, Nagata N, Matsushima R, Chen Y, Yoshioka Y, Sakamoto W (2009) Visualization of plastids in pollen grains: involvement of FtsZ1 in pollen plastid division. Plant Cell Physiol 50:904–908
Tebbutt SJ, Lonsdale DM (1995) Deletion analysis of a tobacco pollen-specific polygalacturonase promoter. Sex Plant Reprod 8:242–246
Thilmony R, Guttman M, Thomson JG, Blechl AE (2009) The LP2 leucine-rich repeat receptor kinase gene promoter directs organ-specific, light-responsive expression in transgenic rice. Plant Biotechnol J 7:867–882
Tian GW, Chen MH, Zaltsman A, Citovsky V (2006) Pollen-specific methylesterase involved in pollen tube growth. Dev Biol 294:83–91
Toki S, Hara N, Ono K (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47:969–976
Twell D, Wing R, Yamaguchi J, McCormick S (1989) Isolation and expression of an anther-specific gene from tomato. Mol Gen Genet 217:240–245
Twell D, Oh S, Honys D (2006) Pollen development, a genetic and transcriptomic view. Plant Cell Monogr 3:15–45
Verelst W, Saedler H, Münster T (2007) MIKC* MADS-protein complexes bind motifs enriched in the proximal region of late pollen specific Arabidopsis promoters. Plant Physiol 143:447–460
Wang Y, Zhang WZ, Song LF, Zou JJ, Su Z, Wu WH (2008) Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol 148:1201–1211
Wei LQ, Xu WY, Deng ZY, Su Z, Xue Y, Wang T (2010) Genome-scale analysis and comparison of gene expression profiles in developing and germinated pollen in Oryza sativa. BMC Genom 11:338
Welchen E, Gonzalez HD (2005) Differential expression of the Arabidopsis cytochrome genes Cytc-1 and Cytc-2. Evidence for the involvement of TCP-domain protein-binding elements in anther- and meristem-specific expression of the Cytc-1 Gene 1. Plant Physiol 139:88–100
Xu H, Theerakulpisut P, Goulding N, Suphioglu C, Singh MB, Bhalla PL (1995) Cloning, expression and immunological characterization of Ory s 1, the major allergen of rice pollen. Gene 164:255–259
Xu H, Swoboba I, Bhalla PL, Singh MB (1999) Male gametic cell-specific gene expression in flowering plants. Proc Natl Acad Sci USA 96:2554–2558
Xu J, Yang C, Yuan Z, Zhang D, Gondwe MY, Ding Z, Liang W, Zhang D, Wilson Z (2010) The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 22:91–107
Yanagisawa S, Schmidt RJ (1999) Diversity and similarity among recognition sequences of Dof transcription factors. Plant J 17:209–214
Yokoi S, Tsuehiya T, Toriyama K, Hinata K (1997) Tapetum-specific expression of the Osg6B promoter-ß -glucuronidase gene in transgenic rice. Plant Cell Rep 16:363–367
Zhang GY, Feng J, Wu J, Wang XW (2010) BoPMEI1, a pollen specific pectin methylesterase inhibitor, has an essential role in pollen tube growth. Planta 231:1323–1334
Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) Genevestigator. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136:2621–2632
Zimmermann P, Laule O, Schmitz J, Hruz T, Bleuler S et al (2008) Genevestigator transcriptome meta-analysis and biomarker search using rice and barley gene expression databases. Mol Plant 1:851–857
Acknowledgments
This research was supported by the Next-Generation BioGreen21 Program, Rural Development Administration, Republic of Korea (The National Center for GM Crops, Grant No. PJ009528; Plant Molecular Breeding Center No. PJ008137, PJ009110); and in part by the Kyungpook National University Research Fund, 2012.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by Dolf Weijers.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Oo, M.M., Bae, HK., Nguyen, T.D. et al. Evaluation of rice promoters conferring pollen-specific expression in a heterologous system, Arabidopsis . Plant Reprod 27, 47–58 (2014). https://doi.org/10.1007/s00497-014-0239-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00497-014-0239-x