Abstract
The specificity of S-RNase-based self-incompatibility (SI) is controlled by two S-locus genes, the pistil S-RNase gene and the pollen S-locus-F-box gene. S-RNase is synthesized in the transmitting cell; its signal peptide is cleaved off during secretion into the transmitting tract; and the mature “S-RNase”, the subject of this study, is taken up by growing pollen tubes via an as-yet unknown mechanism. Upon uptake, S-RNase is sequestered in a vacuolar compartment in both non-self (compatible) and self (incompatible) pollen tubes, and the subsequent disruption of this compartment in incompatible pollen tubes correlates with the onset of the SI response. How the S-RNase-containing compartment is specifically disrupted in incompatible pollen tubes, however, is unknown. Here, we circumvented the uptake step of S-RNase by directly expressing S2-RNase, S3-RNase and non-glycosylated S3-RNase of Petunia inflata, with green fluorescent protein (GFP) fused at the C-terminus of each protein, in self (incompatible) and non-self (compatible) pollen of transgenic plants. We found that none of these ectopically expressed S-RNases affected the viability or the SI behavior of their self or non-self-pollen/pollen tubes. Based on GFP fluorescence of in vitro-germinated pollen tubes, all were sequestered in both self and non-self-pollen tubes. Moreover, the S-RNase-containing compartment was dynamic in living pollen tubes, with movement dependent on the actin–myosin-based molecular motor system. All these results suggest that glycosylation is not required for sequestration of S-RNase expressed in pollen tubes, and that the cytosol of pollen is the site of the cytotoxic action of S-RNase in SI.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Self-incompatibility (SI) is a reproductive strategy adopted by flowering plants to prevent inbreeding and promote outcrossing (de Nettancourt 2001). SI allows the pistil to distinguish between genetically related (self) and unrelated (non-self) pollen. As a result of this self/non-self recognition, self-pollen is rejected either on the stigmatic surface or during tube growth in the style, whereas non-self-pollen is accepted for fertilization (for a comprehensive treatise, see Frankin-Tong 2008). S-RNase-based SI, found so far in the Solanaceae, Rosaceae, and Plantaginaceae families, is of the gametophytic type, i.e., the SI phenotype of the pollen is determined by its own S-haplotype. Pollen is recognized as self-pollen only if the S-haplotype it carries is identical to one of the two S-haplotypes of the pistil.
Two separate genes, S-RNase and S-locus-F-box (SLF or SFB, SLF will be used hereafter), at the highly polymorphic S-locus, control the pollen and pistil SI specificity, respectively (for review, see Kao and Tsukamoto 2004; Hua et al. 2008; McClure 2009). S-RNase was first identified in Nicotiana alata by Anderson et al. (1986), and the function of the S-RNase gene in SI was established by both gain-of-function and loss-of-function experiments (Lee et al. 1994; Murfett et al. 1994). S-RNase shows a high degree of allelic sequence diversity (Ioerger et al. 1991), and it was so named because of its ribonuclease activity (McClure et al. 1989; Singh et al. 1991), which has been shown to be essential for its function in SI (Huang et al. 1994). Full-length cDNA sequences of S-RNases predict the existence of a signal peptide in the nascent protein, which is removed during the secretion of S-RNase from the transmitting cells to the transmitting tract of the pistil. Hereafter, the term “S-RNase” refers to the mature form (without the signal peptide), which is the form taken up by pollen tubes in the style. S-RNases are glycoproteins with various numbers of N-linked glycan chains; however, the carbohydrate moiety is not required for their recognition function, suggesting that the S-allelic specificity resides in the protein backbone (Karunanandaa et al. 1994).
S-locus-F-box was first identified at the S-locus of Antirrhinum hispanicum (Lai et al. 2002), and subsequently identified in the S-loci of Prunus mume (Entani et al. 2003) and Prunus dulcis (Ushijima et al. 2003) through sequencing of the locus. In P. inflata, sequencing of a 328-kb region that contains the S 2 -RNase gene led to the identification of P. inflataSLF (PiSLF), which is located 161 kb downstream of the S 2 -RNase gene (Wang et al. 2004). Evidence that SLF is the pollen SI determinant was obtained by a transgenic experiment, where PiSLF 2 (the S 2 -allele of PiSLF) was introduced into S 1 S 1 , S 1 S 2 and S 2 S 3 plants of P. inflata and shown to cause breakdown of SI in heteroallelic (S 1 and S 3 ) pollen, but not in homoallelic (S 2 ) pollen (Sijacic et al. 2004). These results are consistent with the prediction based on a well-documented genetic phenomenon in S-RNase-based SI, called competitive interaction, which refers to the breakdown of SI in heteroallelic pollen (containing two different S-alleles), but not in homoallelic pollen (containing duplication of the same allele). In the Rosaceae, analyses of naturally occurring pollen-part mutants of SI have attributed breakdown in SI function in pollen to defects in SLF (e.g., frameshift mutants, deletions), thus providing strong evidence for SLF being the pollen S-gene (Sonneveld et al. 2005; Tsukamoto et al. 2006). Interestingly, in the subfamily Amygdaloideae (including the genus Prunus) of the Rosaceae, SI does not break down in heteroallelic pollen, unless both pollen alleles are non-functional (Hauck et al. 2006), suggesting that the underlying biochemical mechanisms of SI in the Rosaceae and Solanaceae may not be the same.
Most F-box proteins whose functions have been characterized to date are a component of various SCF (Skp1-Cullin-F-box) E3 ubiquitin ligase complexes, which are involved in ubiquitin-26S proteasome-mediated protein degradation (Hershko and Ciechanover 1998; Moon et al. 2004; Smalle and Vierstra 2004). Qiao et al. (2004a, b) and Wang and Xue (2005) showed that AhSLF2, the S 2 -allele product of A. hispanicum SLF, might be a component of a typical SCF complex, which targets S-RNases for ubiquitination; however, the specificity of this process remains to be examined (Qiao et al. 2004a; McClure 2009). Studies from P. inflata suggest that PiSLF might be involved in a novel E3 ubiquitin ligase complex, which contains PiCUL1-G (a Cullin-1) and PiSBP1 (P. inflata S-RNase-Binding Protein; a RING-HC protein) but does not contain Skp1 or Rbx1 (Hua and Kao 2006). In vitro binding assays showed that PiSLF interacts with its non-self S-RNases more strongly than with its self S-RNase, and vice versa. This finding provides a biochemical explanation for how PiSLF and S-RNase interact inside a pollen tube to determine the specificity of the SI process (Hua et al. 2008). Results from an in vitro protein degradation assay and biochemical reconstitution of S-RNase ubiquitination suggest that ubiquitin-26S proteasome-mediated degradation of S-RNase is likely an integral part of SI (Hua and Kao 2006, 2008). Sequence and functional comparisons of PiSLF and PiSLF-like genes identified three regions that are specific to PiSLF and revealed that PiSLF is unique in its function in SI (Hua and Kao 2006; Hua et al. 2007).
S-RNase is taken up by pollen tubes in a non-S-haplotype-specific manner (Luu et al. 2000; Goldraij et al. 2006), but the mechanism of transport is unknown. Goldraij et al. (2006) observed that S-RNases are initially sequestered in compatible and incompatible pollen tubes, but later on, only the compartment in incompatible pollen tubes is disrupted, presumably releasing S-RNases to result in rejection of self-pollen tubes. This finding about the initial fate of S-RNase in the pollen tube is interesting, and it raises a question as to how the compartment is specifically disrupted in incompatible pollen tubes to trigger the SI response.
Prior to the identification of SLF as the pollen S-gene, Dodds et al. (1999) examined whether S-RNase might also control pollen specificity. They found that ectopic expression of Lycopersicon peruvianum S3-RNase in pollen of S 1 S 2 transgenic plants did not confer S 3 -haplotype specificity on the S 1 or S 2 transgenic pollen (i.e., both transgenic pollen types were accepted by S 3 -haplotype-containing pistils), nor did it affect the viability of the transgenic pollen. Interestingly, Dodds et al. (1999) showed, by immunoelectron microscopy, that S3-RNase was located in the inner part of the pollen cell wall close to the plasma membrane, and not in the cytosol or compartmentalized as reported for S-RNase taken up by pollen tubes from the transmitting tract of the pistil (Luu et al. 2000; Goldraij et al. 2006).
In this work, we ectopically expressed S2-RNase and S3-RNase of Petunia inflata in their respective self and non-self-pollen in transgenic plants and examined their effect on the SI behavior of the transgenic pollen and their localization inside transgenic pollen tubes. Moreover, we have previously shown that glycosylation is not required for the function of S-RNase in SI (Karunanandaa et al. 1994), and that deglycosylation of S3-RNase significantly enhances the extent of degradation by pollen tube extracts in vitro (Hua and Kao 2006). Thus, we also ectopically expressed the non-glycosylated form of S3-RNase in its self and non-self-pollen in transgenic plants to examine whether, if S-RNase is found to be sequestered in pollen tubes, glycosylation is required for this process.
To monitor their localization, we fused the C-terminus of each S-RNase with a GFP tag using a 13-amino-acid linker. Since this linker has previously been shown to have no effect on the normal SI function of PiSLF2 or GFP fluorescence of the PiSLF2:GFP fusion protein produced in pollen (Hua et al. 2007), the fusion of GFP will not likely affect the function of S-RNase in vivo. We found that none of these S-RNases (glycosylated or non-glycosylated) affected the SI behavior or viability of their respective self or non-self-pollen/pollen tubes, and that all the ectopically expressed S-RNases were sequestered in their self and non-self-pollen tubes. We also showed that the S-RNase-containing compartment was dynamic, and its movement inside the pollen tube was dependent on the actin–myosin, and not the microtubule, based molecular motor system.
Materials and methods
Plant material
The four S-genotypes of Petunia inflata, S 1 S 1 , S 2 S 2 , S 2 S 3 , and S 3 S 3 , used in this study were described by Ai et al. (1990).
Construction of Ti plasmids and plant transformation
The plasmids used for the transformation experiments were constructed from the Ti plasmid, pBI101 (Clontech, CA, USA), by using the coding sequences (CDS) of the mature S2-RNase, S3-RNase, and S3-RNase(N29D) of P. inflata. S3-RNase(N29D) is the non-glycosylated form of S3-RNase, with the AAC codon for the only N-linked glycosylation site, Asn-29, changed to GAC for Asp (Karunanandaa et al. 1994). The CDS for each S-RNase, with an NcoI site at the 5′ end and an NotI site at the 3′ end, was subcloned into pGEM-T Easy vector (Promega, WI, USA). The NcoI–NotI fragment containing the CDS of each S-RNase was released from pGEM-T Easy vector and used to replace the 0.72-kb NcoI–NotI fragment (containing the CDS for GFP) in pLAT52-GFP (Dowd et al. 2006) to result in a pLAT52-S-RNase construct. The CDS for GFP, plus a 39-bp linker encoding a 13-amino-acid (Ala)3(Gly)10 peptide linker, was amplified by polymerase chain reaction (PCR) from pLAT52-PiSLF 2 :GFP (Hua et al. 2007) and inserted into each pLAT52-S-RNase construct to produce a pLAT52-S-RNase:GFP construct. The Sal1–EcoRI fragment, containing pLAT52, CDS for S-RNase, linker, CDS for GFP, and the Nos terminator, was released from each pLAT52-S-RNase:GFP construct and used to replace the Sal1–EcoRI fragment (containing the CDS for GUS and the Nos terminator) in pBI101 to yield a pBI LAT52-S-RNase:GFP construct. The schematics for pBI LAT52-S 2 -RNase:GFP, pBI LAT52-S 3 -RNase:GFP, and pBI LAT52-S 3 -RNase(N29D):GFP constructs are shown in Supplementary Fig. 1 online. All of the constructs were introduced into Agrobacterium tumefaciens strain LBA4404 and used for Agrobacterium-mediated transformation as described by Lee et al. (1994).
Genomic DNA isolation and gel blot analysis
Genomic DNA was purified from 0.5 g of young leaf tissue of each plant using Plant DNAzol reagent (Invitrogen, CA, USA) according to the manufacturer’s procedure. Genomic DNA (15 μg) isolated from each plant was digested overnight with HindIII (for S 2 -RNase:GFP transgenic plants) or EcoRI (for S 3 -RNase:GFP and S 3 -RNase(N29D):GFP transgenic plants). The digests were separated on 0.7% (w/v) agarose gels and transferred to charged nylon membranes, Biodyne B (Pall, NY, USA). The S 2 -RNase and S 3 -RNase probes, obtained from PCR using each CDS as template, were labeled with 32P using the Ready-To-Go DNA Labeling kit (GE Healthcare, NJ, USA). Prehybridization, hybridization, and washing of the membranes were performed as described by Skirpan et al. (2001). The membrane was exposed to X-ray film at −80°C for 48 h with an intensifying screen.
PCR genotyping and analysis of transgenes
Genomic DNA was extracted as described under “Genomic DNA isolation and gel blot analysis” except that 0.2 g of young leaf tissue was used for each plant. For each plant, 100 ng of genomic DNA was used for PCR with one of the following primer pairs: S2-RNase_FOR (5′-TTTGACTACTTCCAACTCG-3′) and GFP_REV (5′-AGGTGGTCACGAGGGTG-3′) for S 2 -RNase:GFP; S3-RNase_FOR (5′-GCGAATTTTGACTACATC-3′) and GFP_REV (5′-AGGTGGTCACGAGGGTG-3′) for S 3 -RNase:GFP and S 3 -RNase(N29D):GFP; S2-RNase_FOR and S2-RNase_REV (5′-TCATCTCCGAAACAGAGTCT-3′) for S 2 -RNase; SLF3- _FOR (5′-AATGGTGTTTTAAAGAAATTGC-3′) and SLF3- _REV (5′-AAAGTTTTGTACTTTTGTACTGTACTC-3′) for PiSLF 3 . PCR was performed with 95°C denaturation for 2 min followed by 30 cycles of 95°C (30 s), 55°C (30 s), and 72°C (60 s). A 10 min extending reaction was further performed after the completion of the cycles. For each reaction, 15 μl of the amplified product was subjected to electrophoresis on 1% (w/v) agarose gels and visualized under ultra-violet lamp after staining with ethidium bromide.
Reverse transcription (RT)-PCR analysis
Total RNA was isolated from in vitro-germinated pollen tubes of each transgenic plant or a wild-type S 2 S 3 plant as described by Hua and Kao (2006), except that the TURBO DNA-free kit (Ambion, TX, USA) was used to remove any contaminating genomic DNA according to the manufacturer’s procedure. Reverse transcription was carried out according to Hua et al. (2007). PCR was carried out on 250 ng RNA, 250 ng cDNA, or 0.1 μg genomic DNA using one of the following primer pairs: S3-RNase_FOR and GFP_REV for S 3 -RNase:GFP and S 3 -RNase(N29D):GFP (see the “PCR genotying and analysis of transgenes” section above); S2F_FOR (5′-CCATGGCGAATGGTATTTTAAAG-3′) and GFP_REV for PiSLF 2 :GFP; Actin_FOR (5′-GGCATCACACTTCTACAATGAGC-3′) and Actin_REV (5′-GATATCCACATCACATTTCATGAT-3′) for the actin gene. PCR was performed with 95°C denaturation for 2 min followed by 25 or 30 cycles of 95°C (30 s), 56°C (30 s), and 72°C (60 s) for amplifying S 3 -RNase:GFP, S 3 -RNase(N29D):GFP and PiSLF 2 :GFP, or followed by 20 cycles of 95°C (30 s), 60°C (30 s), and 72°C (50 s) for amplifying the actin gene. A 10 min extending reaction was further performed after the completion of the cycles. For each reaction, 12 μl of the amplified product was subjected to electrophoresis on 1% (w/v) agarose gels.
Protein extraction and immunoblot analysis
Total protein was extracted from stage 5 anthers (Lee et al. 1996) from each plant by grinding the tissues in liquid nitrogen as described by Hua et al. (2007). Protein concentrations were determined using the BioRad protein assay system (Bio-Rad, CA, USA). Total protein was resolved on 10% polyacrylamide gels using a mini-protean system (Bio-Rad) and blotted onto polyvinylidene difluoride (PVDF) membranes (Millipore, MA, USA). GFP and its fusion proteins were detected by a rabbit anti-GFP antibody (1:1500 dilution; Abcam, MA, USA) and peroxidase-linked sheep anti-rabbit IgG (1:10000 dilution; GE Healthcare).
Pollen tube germination and confocal imaging
Pollen collected from open flowers of each plant was germinated in pollen germination medium [0.07% (w/v) Ca(NO3)2·4H2O, 0.02% (w/v) MgSO4·7H2O, 0.01% (w/v) KNO3, 0.01% (w/v) H3BO3, 0.2% (w/v) sucrose, 15% (w/v) polyethylene glycol 4000, and 20 mM MES, pH 6.0] for 2 h at 30°C with shaking at 200 rpm. The subcellular localization of each S-RNase:GFP protein in pollen tubes was observed under an Olympus FV1000 Laser Scanning Confocal Microscope (Olympus, PA, USA). Z-series of sections were stacked from 50 pictures, with 0.15 s per scan. GFP was imaged using 488 nm excitation and 520–560 nm emission. Data were collected and analyzed by software FV10-ASW version 1.7 (Olympus).
2,3-butanedione monoxime (BDM) and oryzalin treatment
A stock solution of 1 M 2,3-butanedione monoxime (BDM) was prepared as described by Tominaga et al. (2000). Stock solutions of 1 and 0.2 mM oryzalin were prepared by dissolving the chemical in ethanol and stored at 4°C. After pollen was germinated in pollen germination medium for 1.5 h, BDM and oryzalin were separately added in different concentrations as described in Supplementary Movies 1 and 2. The pollen tubes were cultured for 1 more hour before confocal imaging.
Results
Expression of S 2 -RNase:GFP, S 3 -RNase:GFP, and S 3 -RNase(N29D):GFP in transgenic pollen did not affect viability or SI behavior of their respective self or non-self-pollen
We used the promoter of a pollen-specific gene of tomato, LAT52 (Twell et al. 1990), to ectopically express S 2 -RNase:GFP in S 1 S 1 and S 2 S 2 plants. Table 1 summarizes numbers of transgenic plants with one or two copies of the transgene obtained in the course of this and subsequent S-RNase:GFP fusion protein experiments. For the S 1 S 1 transgenic line, nine independent transgenic plants with a single copy of the transgene (e.g., S 2 -RNase:GFP/S 1 S 1 -1 shown in lane labeled “1” in Fig. 1a) and four independent transgenic plants with two copies of the transgene were identified (e.g., S 2 -RNase:GFP/S 1 S 1 -6 shown in lane labeled “6” in Fig. 1a). For the S 2 S 2 transgenic line, two independent transgenic plants with a single copy of the transgene (one of the plants, S 2 -RNase:GFP/S 2 S 2 -2, shown in lane labeled “2” in Fig. 1b) and one transgenic plant with two copies of the transgene (S 2 -RNase:GFP/S 2 S 2 -1 shown in lane labeled “1” in Fig. 1b) were identified.
Genomic DNA gel blot analysis of the presence and copy numbers of the S 2 -RNase:GFP transgene in S 1 S 1 and S 2 S 2 transgenic plants, and the S 3 -RNase:GFP and S 3 -RNase(N29D):GFP transgenes in S 2 S 3 transgenic plants. Fifteen micrograms of genomic DNA isolated from each plant was digested with HindIII (for a, b) or EcoRI (for c, d), and separated on 0.7% (w/v) agarose gels. The blots were hybridized with a 32P-labeled cDNA probe of S 2 -RNase (a, b) or S 3 -RNase (c, d) at 65°C. a S 1 S 1 transgenic plants carrying one or two copies of the S 2 -RNase:GFP transgene. The fragment corresponding to the endogenous S 1 -RNase gene is indicated with an asterisk. b S 2 S 2 transgenic plants carrying one or two copies of the S 2 -RNase:GFP transgene. The fragment corresponding to the endogenous S 2 -RNase gene is indicated with double asterisks. c, d S 2 S 3 transgenic plants carrying one or two copies of the S 3 -RNase:GFP transgene (c), and one or two copies of the S 3 -RNase(N29D):GFP transgene (d). The fragments corresponding to the endogenous S 3 -RNase and S 2 -RNase genes are indicated with double and triple asterisks
We first examined whether expression of S2-RNase:GFP was toxic to its self (S 2 ) and/or non-self (S 1 ) pollen. Pollen from transgenic plants S 2 -RNase:GFP/S 1 S 1 -9 (Fig. 1a) and S 2 -RNase:GFP/S 2 S 2 -2 (Fig. 1a), both of which contained a single copy of the transgene, was used to pollinate wild-type plants (Table 2). The crosses S 2 S 2 × S 2 -RNase:GFP/S 1 S 1 -9, S 1 S 1 × S 2 -RNase:GFP/S 2 S 2 -2, and S 3 S 3 × S 2 -RNase:GFP/S 2 S 2 -2 were expected to be compatible, because half of the pollen produced by each transgenic plant did not inherit the transgene and should behave normally in SI. Indeed, all these pollinations resulted in large fruits with the average seed number per fruit of 130 ± 15 for S 2 S 2 × S 2 -RNase:GFP/S 1 S 1 -9, 150 ± 30 for S 1 S 1 × S 2 -RNase:GFP/S 2 S 2 -2, and 145 ± 25 for S 3 S 3 × S 2 -RNase:GFP/S 2 S 2 -2. We then used PCR to analyze the inheritance of the transgene in the resulting progenies. Approximately, half of the plants in each progeny carried the transgene (Table 2), suggesting that S2-RNase:GFP did not affect the viability of either its self (S 2 ) or non-self (S 1 ) pollen. We further examined the SI behavior of S 1 and S 2 transgenic pollen by pollinating wild-type S 1 S 1 plants with pollen from S 2 -RNase:GFP/S 1 S 1 -9 and by pollinating wild-type S 2 S 2 plants with pollen from S 2 -RNase:GFP/S 2 S 2 -2. These pollinations resulted in very small fruits with the average seed number per fruit of 30 ± 15 and 5 ± 5 (Table 2). Thus, the S 2 -RNase:GFP transgene did not affect the SI behavior of its self or non-self-pollen.
To rule out the possibility that the finding with S2-RNase:GFP is specific to this S-RNase, we next examined whether ectopic expression of S3-RNase:GFP has any effect on its non-self (S 2 ) and self (S 3 ) pollen. We identified four independent transgenic S 2 S 3 plants carrying one copy of the transgene (S 3 -RNase:GFP/S 2 S 3 -9 and -33 shown in lanes labeled “9” and “33,” respectively, in Fig. 1c) and two independent transgenic plants each carrying two copies of the transgene (S 3 -RNase:GFP/S 2 S 3 -7 and -12 shown in lanes labeled “7” and “12,” respectively, in Fig. 1c). We chose S 3 -RNase:GFP/S 2 S 3 -9 and S 3 -RNase:GFP/S 2 S 3 -33 for pollination test. Pollen of both transgenic plants were compatible with pistils of wild-type S 2 S 2 plants (with the average seed number per fruit being 155 ± 20 and 145 ± 25), but incompatible with pistils of wild-type S 2 S 3 plants (with the average seed number per fruit being 20 ± 10 and 10 ± 5) (Table 2). Thus, ectopic expression of S3-RNase:GFP in the pollen of S 2 S 3 transgenic plants did not affect their SI behavior.
We also ectopically expressed a non-glycosylated S3-RNase GFP fusion protein, S3-RNase(N29D):GFP, to study whether it had a different effect on the SI behavior or viability of self and non-self-pollen than the native glycosylated protein. We identified three independent S 2 S 3 transgenic plants with a single copy of the transgene (two, S 3 -RNase(N29D):GFP/S 2 S 3 -17 and -22, shown in lanes labeled “17” and “22,” respectively, in Fig. 1d) and four independent S 2 S 3 transgenic plants with two copies (S 3 -RNase(N29D):GFP/S 2 S 3 -12 and -112 shown in lanes labeled “12” and “112,” respectively, in Fig. 1d). We used all three single-copy transgenic plants, S 3 -RNase(N29D):GFP/S 2 S 3 -17, -22, and -23, for pollination testing. Pollinations of wild-type S 2 S 2 plants by all these transgenic plants resulted in large fruits with average seed number per fruit ranging from 135 ± 15 to 165 ± 20. Pollinations of wild-type S 1 S 1 and S 3 S 3 plants by S 3 -RNase(N29D):GFP/S 2 S 3 -22 also resulted in large fruits (Table 2). In contrast, pollinations of wild-type S 2 S 3 plants by all these three transgenic plants resulted in very small fruits (Table 2), suggesting that the S 3 -RNase(N29D):GFP transgene did not affect the SI behavior of either its self (S 3 ) or non-self (S 2 ) pollen.
To examine whether expression of S 3 -RNase(N29D):GFP was toxic to, and had any effect on the SI behavior of, its self and/or non-self-pollen, we further used PCR to determine the inheritance of the transgene in the progenies obtained from the crosses between the wild-type S 2 S 2 plants and the three single-copy transgenic plants, S 3 -RNase(N29D):GFP/S 2 S 3 -17, -22, and -23. Approximately, 50% of the plants in each progeny analyzed carried the transgene (Table 2; Fig. 2). Moreover, a total of 29 plants in the progenies from S 2 S 2 × S 3 -RNase(N29D):GFP/S 2 S 3 -22 and S 2 S 2 × S 3 -RNase(N29D):GFP/S 2 S 3 -23 (Table 2) were genotyped by PCR, using primers specific to S 2 -RNase and primers specific to PiSLF 3 , and they were all found to be S 2 S 3 (data not shown), suggesting that the expression of S 3 -RNase(N29D):GFP did not affect the viability of its self (S 3 ) pollen or the SI behavior of its non-self (S 2 ) pollen. We also examined the inheritance of the transgene in the progeny of the cross between a wild-type S 3 S 3 plant and a S 3 -RNase(N29D):GFP/S 2 S 3 -23 (Table 2). In this case, the wild-type S 3 pollen should be rejected by the S 3 S 3 pistil, and thus the observed ~50% transmission of the transgene into the progeny suggests that the expression of S 3 -RNase(N29D):GFP did not affect the viability of its non-self (S 2 ) pollen or the SI behavior of its self (S 3 ) pollen either. In the cross between a wild-type S 1 S 1 plant and a S 3 -RNase(N29D):GFP/S 2 S 3 -22, ~50% of the progeny carried the transgene (Table 2), consistent with the results described earlier that the expression of S 3 -RNase(N29D):GFP did not affect the viability of its self or non-self-pollen.
PCR analysis of the transmission of the S 3 -RNase(N29D):GFP transgene into the progenies of the crosses between three independent transgenic plants and wild-type S 2 S 2 plants. The name of each transgenic plant is indicated at the top of each panel. Genomic DNA (~100 ng) extracted from each progeny plant was used as template for PCR with a specific primer pair corresponding to the coding sequence of S 3 -RNase(N29D):GFP. The PCR products from genomic DNA of each of the three transgenic plants (indicated as T 0 in each panel) and the Ti plasmid used for transformation (indicated as P in each panel) are shown as positive controls. M indicates EcoRI and HindIII digested λ DNA used as size markers
Lastly, when in vitro-germinated pollen tubes from all the earlier mentioned single-copy and two-copy transgenic plants carrying S 2 -RNase:GFP, S 3 -RNase:GFP, or S 3 -RNase(N29D):GFP were examined under a fluorescence microscope, fluorescent pollen tubes were observed for each transgenic plant (Table 1). In the case of the single-copy transgenic plants, ~50% of the pollen tubes for each plant showed fluorescence (data not shown), consistent with the prediction that 50% of the pollen produced carried the transgene, and no morphological differences were observed among the fluorescent pollen tubes. These results also suggest that none of the S-RNase transgenes affected the viability of their self- or non-self-pollen.
Lack of cytotoxic effect of the S 2 -RNase:GFP, S 3 -RNase:GFP, and S 3 -RNase(N29D):GFP transgenes on pollen is not due to absence of their expression in transgenic pollen/pollen tubes
Pistil-expressed S-RNase is thought to act as a cytotoxin to degrade the RNAs in self-pollen tubes, thus resulting in their growth inhibition (McClure et al. 1989). However, as described earlier, none of the three ectopically expressed S-RNases affected the SI behavior or the viability of the transgenic pollen. To rule out the possibility that this lack of cytoxicity was due to an absence of the expression of the transgenes, we first performed RT-PCR on total RNA isolated from in vitro-germinated pollen tubes of five transgenic plants, S 3 -RNase:GFP/S 2 S 3 -9, -33 (Fig. 1c) and S 3 -RNase(N29D):GFP/S 2 S 3 -12, -17, -112 (Fig. 1d). As shown in Fig. 3, a DNA fragment of the expected size (~0.73 kb) was detected in the cDNA samples of all these five transgenic plants after 25 and 30 cycles of the reaction, but not in the cDNA sample of a wild-type S 2 S 3 plant (upper two panels marked with RT+), or in the RNA samples of these plants even after 30 cycles of the reaction (upper two panels marked with RT−). An RT-PCR product of the expected size was obtained using RNA isolated from in vitro-germinated pollen tubes of a previously generated transgenic plant carrying PiSLF 2 :GFP/S 2 S 3 -5 (also driven by the LAT52 promoter; Hua et al. 2007) (upper two panels marked with RT+). We had previously shown that PiSLF2:GFP produced in the pollen of this transgenic plant caused breakdown of SI in S 3 pollen, but not in S 2 pollen, similar to the effect of PiSLF2 (Hua et al. 2007). When PCR was performed on plasmid DNA containing PiSLF 2 :GFP, S 3 -RNase:GFP, or S 3 -RNase(N29D):GFP, the size of the DNA fragment produced from each plasmid DNA was similar to that of the corresponding RT-PCR product (upper two panels marked with plasmid). These results suggested that both S 3 -RNase:GFP and S 3 -RNase(N29D):GFP transgenes were transcribed in their respective transgenic pollen tubes. The same RNA samples used for RT-PCR were also amplified using a primer pair for actin, and the results show that these samples contained approximately equal amounts of RNA (lower panel marked with RT+). Finally, when the genomic DNA of the wild-type S 2 S 3 plant was amplified (lane labeled “G”) and when these RNA samples were amplified without reverse transcriptase (lower panel marked with RT−), only the genomic DNA sample yielded a fragment. This fragment, as expected, was larger in size, due to the presence of an intron, than the RT-PCR product of this wild-type plant (lane labeled “S 2 S 3 ” in the lower panel marked RT+), indicating that all the RNA samples used in RT-PCR were free of genomic DNA contamination.
RT-PCR analysis of RNA transcripts of S 3 -RNase:GFP and S 3 -RNase(N29D):GFP in in vitro-germinated pollen tubes of transgenic plants. Pollen was separately collected from five S 2 S 3 transgenic plants that carried either the S 3 -RNase:GFP or the S 3 -RNase(N29D):GFP transgene, a previously generated S 2 S 3 transgenic plant carrying the PiSLF 2 :GFP transgene (also driven by the LAT52 promoter), and a wild-type S 2 S 3 plant. Pollen was germinated in vitro for 2 h, and 5 μg of RNA isolated for each plant was used for reverse transcription; 250 ng of the resulting cDNA was used for PCR. As negative controls, 250 ng of RNA for each plant was used for PCR without reverse transcription. The upper two panels show the results of 25 and 30 cycles of amplification of the RNA samples either in the presence (RT+) or in the absence (RT−) of reverse transcriptase, as well as the results of 25 and 30 cycles of amplification of plasmid DNA containing each of the three transgenes indicated. The lower panel shows the results of 20 cycles of amplification in the presence or absence of reverse transcriptase, using a primer pair specific to the actin gene. To control for genomic DNA contamination, 0.1 μg genomic DNA of the wild-type S 2 S 3 plant was used for PCR (lane labeled G in the lower panel). Marker indicates the EcoRI and HindIII digested λ DNA used as size markers
We next used an anti-GFP antibody to examine whether the GFP fusion proteins, S2-RNase:GFP, S3-RNase:GFP, and S3-RNase(N29D):GFP, were produced in stage 5 anthers (which contain pollen in its late developmental stages; Lee et al. 1996) of their respective transgenic plants (Fig. 4). A protein band of the size expected of these fusion proteins (~50 kD) was detected in stage 5 anthers of each of the transgenic plants analyzed, but not in those of wild-type plants of S 1 S 1 , S 2 S 2 , or S 2 S 3 genotype. The PiSLF 2 :GFP/S 2 S 3 transgenic plant used as a positive control in RT-PCR was also used as a positive control for the GFP fusion protein. A PiSLF2:GFP band was detected in stage 5 anthers, and as expected, this band had a higher molecular mass than the band corresponding to the GFP fusion proteins of the S-RNases. For all the GFP fusion proteins of the S-RNases, a protein band with a molecular mass matching that of the GFP band in stage 5 anthers of a previously generated GFP/S 2 S 3 transgenic plant (with the expression of GFP driven by the LAT52 promoter; Dowd et al. 2006) was also detected. The GFP was most likely released from these fusion proteins during sample preparations by the abundant proteases in the anther extract. Indeed, different preparations produced different intensities of this protein band. The detection of the GFP fusion proteins, S2-RNase:GFP, S3-RNase:GFP, and S3-RNase(N29D):GFP, suggested that their lack of cytotoxicity was not due to absence of these proteins in pollen/pollen tubes.
Protein gel blot analysis of the presence of S2-RNase:GFP, S3-RNase:GFP and S3-RNase(N29D):GFP in stage 5 anthers of transgenic plants. The transgenic plants, as well as wild-type S 1 S 1 , S 2 S 2 and S 2 S 3 plants and two previously generated transgenic plants used as controls, are indicated at the top of each panel. The single asterisk indicates the protein band corresponding to S2-RNase:GFP, S3-RNase:GFP, or S3-RNase(N29D):GFP. The double asterisks indicate the band corresponding to the free GFP, and the arrow indicates the band corresponding to PiSLF2:GFP. Molecular mass markers are shown on the left of the figure
Pollen expressed S2-RNase:GFP, S3-RNase:GFP, and S3-RNase(N29D):GFP are sequestered in in vitro-germinated pollen tubes
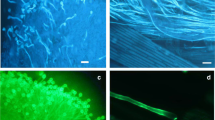
Taking advantage of the GFP tag in the fusion proteins, we used confocal microscopy to examine real-time subcellular localization of S2-RNase:GFP, S3-RNase:GFP, and S3-RNase(N29D):GFP in in vitro-germinated pollen tubes of the following transgenic plants: S 2 -RNase:GFP/S 1 S 1 -9, S 2 -RNase:GFP/S 2 S 2 -2, S 3 -RNase:GFP/S 2 S 3 -9, -33, and S 3 -RNase(N29D):GFP/S 2 S 3 -17, -22 (Fig. 1; Table 1). As controls, we also examined the two previously generated transgenic plants, PiSLF 2 :GFP-5 (Hua et al. 2007) and GFP/S 2 S 3 (Dowd et al. 2006) mentioned earlier. Pollen grains were separately collected from all the transgenic plants and germinated for 2 h. At least 50 pollen tubes were examined for each transgenic plant; representative results are shown in Fig. 5 and Supplementary Movies 1 and 2. S2-RNase:GFP was sequestered in both S 1 (Fig. 5a) and S 2 pollen tubes (Fig. 5b). In contrast, GFP fluorescence was distributed throughout the cytosol of in vitro-germinated pollen tubes of GFP/S 2 S 3 (Fig. 5c) and PiSLF 2 :GFP/S 2 S 3 -5 (Fig. 5d). The cytosolic localization of PiSLF2 is consistent with the finding by Wang and Xue (2005), based on electron-immuno imaging, that AhSLF2 of A. hispanicum is cytosolically localized. Like S2-RNase:GFP, S3-RNase:GFP was also sequestered in in vitro-germinated pollen tubes of both transgenic plants analyzed (Fig. 5e). Figure 5f shows that none of the S3-RNase:GFP-containing compartments were present in the subapical region of the pollen tube shown in Fig. 5e, as would be expected for live and actively growing pollen tubes (Hepler et al. 2001). Although native S-RNases are glycosylated, glycosylation is not required for the sequestration of S-RNase:GFP in pollen tubes, as S3-RNase(N29D):GFP was also sequestered in the in vitro-germinated pollen tubes of all three transgenic plants analyzed (Fig. 5g). Moreover, there was no discernible difference in the morphology of the compartments containing either S3-RNase(N29D):GFP or S3-RNase:GFP, and the S3-RNase(N29D):GFP:containing compartments were also absent from the subapical region of the pollen tube (results not shown). The compartments varied in size, from 2 to 7 μm, and the smaller compartments were somewhat circular, whereas the larger compartments were ellipsoid or linear in shape. To further visualize the S-RNase:GFP-containing compartments observed from single optical sections of pollen tubes by confocal microscopy, we took a Z-series of optical sections of a transgenic pollen tube expressing S3-RNase:GFP, and present in Fig. 5h two different views of the distribution of the compartments in the pollen tube. Note that the compartments are absent in the tip region.
Subcellular localization of S2-RNase:GFP, S3-RNase:GFP, S3-RNase(N29D):GFP, PiSLF2:GFP, and GFP in in vitro-germinated pollen tubes of transgenic plants. For each transgenic plant, the left panel is the GFP channel, and the right panel is the bright-field channel. The bar in each image represents 5 μM. Sequestration of S2-RNase:GFP in S 1 (a) and S 2 (b) transgenic pollen tubes. c Cytosolic localization of GFP in an S 2 or S 3 transgenic pollen tube. d Cytosolic localization of PiSLF2:GFP in an S 2 or S 3 transgenic pollen tube. e Sequestration of S3-RNase:GFP in an S 2 or S 3 transgenic pollen tube; f magnification of the subapical region of the pollen tube shown in e. g Sequestration of S3-RNase(N29D):GFP in an S 2 or S 3 transgenic pollen tube. h Two different views of a Z-series of 50 optical sections (0.15 s per scan) of S3-RNase:GFP in an S 2 or S 3 transgenic pollen tube. The blue arrowhead in each picture denotes the growing tip region
Thus, in contrast to the distribution of GFP fluorescence throughout the cytoplasm of pollen tubes expressing GFP alone (Fig. 5c) or PiSLF2:GFP fusion protein (Fig. 5d), all the ectopically expressed S-RNase:GFPs were compartmentalized (Fig. 5 a, b, e–h), and this compartmentalization is independent of the S-haplotype of the pollen tube and does not require glycosylation of S-RNase. This finding suggested that the compartmentalization blocked the cytotoxic function of S-RNase expressed in pollen tubes.
The S-RNase:GFP-containing compartments are associated with actin–myosin-based motor system
We monitored the movement of the compartments containing S3-RNase:GFP or S3-RNase(N29D):GFP in the cytoplasm of in vitro-germinated pollen tubes (Supplementary Movies 1 and 2). By tracing the movement of the compartments, we determined that both S3-RNase:GFP-containing compartments and S3-RNase(N29D):GFP-containing compartments moved at a speed of 0.5–0.7 μm/s. The mobility of sub-organelles inside a cell can be driven by either a microtubule based motor system or an actin–myosin-based motor system, with different speeds; the former is approximately 150 nm/s and the latter is approximately several micrometers per second (Romagnoli et al. 2003). The observed speed of the movement of the S-RNase:GFP-containing compartments fell in between that of the microtubule and actin–myosin-based systems. To identify which system was responsible for the movement of the S-RNase-containing compartments, we incubated in vitro-germinated pollen tubes expressing S3-RNase:GFP or S3-RNase(N29D):GFP with either 2,3-butanedione monoxime (BDM), a myosin inhibitor (Yagi et al. 1992; McKillop et al. 1994), or oryzalin, which causes depolymerization of microtubules (Morejohn et al. 1987; Cleary and Hardham 1988). Incubation of pollen tubes with 30 mM BDM for 1 h significantly slowed down the movement of both S3-RNase:GFP- and S3-RNase(N29D):GFP-containing compartments, and incubation of pollen tubes with 50 mM BDM, a moderate concentration for inhibiting the movement of the actin–myosin system (Tominaga et al. 2000), completely inhibited the movement of both compartments. However, treatment of pollen tubes with oryzalin for 1 h at 50 μM, a concentration higher than normally required to inhibit the activity of microtubules (Poulter et al. 2008), did not significantly affect the trafficking of either S3-RNase:GFP- or S3-RNase(N29D):GFP-containing compartments (Supplementary Movies 1 and 2). Thus, we concluded that both the glycosylated and the non-glycosylated forms of S-RNase were sequestered in compartments that are associated with the actin–myosin molecular motor system.
Discussion
Since S-RNase is secreted into the transmitting tract of the style, it must be taken up by self-pollen tubes in order to exert its cytotoxic function to inhibit tube growth. It was challenging to demonstrate the localization of S-RNase in the pollen tube, because the presence of very high levels of S-RNase in the transmitting tissue/transmitting tract of the pistil makes it difficult to discern the relatively small amount of S-RNase taken up by pollen tubes during their growth through the transmitting tract. Therefore, the detection of S-RNase inside pollen tubes by immunolocalization by Luu et al. (2000) and Goldraij et al. (2006) represents a major step forward. Both Luu et al. (2000) and Goldraij et al. (2006) found that S-RNase is localized in compatible and incompatible pollen tubes, suggesting that uptake of S-RNase by the pollen tube is not S-haplotype-specific. These two studies, however, differ in their findings of the subcellular localization of S-RNase. Luu et al. (2000) showed that S-RNase is localized in the cytosol, whereas Goldraij et al. (2006) showed that S-RNase is initially sequestered in a vacuolar compartment in both compatible and incompatible pollen tubes, and that this compartment is later disrupted in incompatible pollen tubes but remains intact in compatible pollen tubes.
In this work, we directly expressed three GFP tagged S-RNases, S2-RNase, S3-RNase and non-glycosylated S3-RNase, in pollen of transgenic plants and used GFP as a reporter to monitor the subcellular localization of these S-RNases in in vitro-germinated pollen tubes (both compatible and incompatible) of the transgenic plants. This approach does not require fixation of pollen tubes, which may cause artifacts, and can also provide a dynamic picture of S-RNase inside the pollen tube, rather than a static one revealed by immunolocalization. Moreover, this approach bypasses the uptake step of S-RNase, an as-yet poorly understood process, and allows examination of the fate of S-RNase alone in pollen tubes and of any effect the ectopically expressed S-RNase may have on the SI behavior of self and non-self-pollen tubes.
Two groups have previously examined the effect of ectopic expression of S-RNase in pollen/pollen tubes. Kirch et al. (1995) used the LAT52 promoter to express the coding sequence of S2-RNase (including the signal peptide) of Solanum tuberosum in self-compatible Nicotiana tabacum and showed that the fertility of the pollen of the transgenic plants was not affected, even though S2-RNase was produced. However, the effect of S2-RNase on the SI behavior of pollen could not be addressed as it was expressed in pollen of a different and self-compatible species. Dodds et al. (1993, 1999) observed low levels of expression of the S-RNase gene in developing pollen of Nicotiana alata and Lycopersicon peruvianum, and they used the LAT52 promoter to express cDNA encoding mature S3-RNase (without the signal peptide) of L. peruvianum in pollen of S 1 S 2 plants to test whether S-RNase also controls the pollen specificity in SI (Dodds et al. 1999). The results showed that expression of S3-RNase did not confer on transgenic pollen S 3 -haplotype specificity, and, similar to the finding of Kirch et al. (1995), did not affect the viability of the transgenic pollen. Since S3-RNase was only expressed in its non-self (S 1 and S 2 ) pollen, the lack of inhibitory effect on its compatible pollen tubes was no different from what would be expected for pistil-expressed S3-RNase on its compatible pollen tubes. In this work, two different S-RNases of P. inflata were separately expressed in their respective self (incompatible) and non-self (compatible) pollen in transgenic plants, so that we could examine whether there is any difference between the fates of ectopically expressed S-RNase in compatible and incompatible pollen tubes.
Consistent with the results of Dodds et al. (1999), we found that the S 2 -RNase:GFP and S 3 -RNase:GFP transgenes did not affect the viability of their respective non-self-pollen tubes (S 1 pollen tubes for the former and S 2 pollen tubes for the latter). But, interestingly, neither S 2 -RNase:GFP nor S 3 -RNase:GFP affected the viability of their respective self-pollen tubes either. For the S 2 S 3 transgenic plants that carried one copy of S 3 -RNase:GFP, we further used RT-PCR and protein gel blotting to show that the lack of toxic effect on both self and non-self-pollen tubes was not due to lack of expression of the transgene. Subcellular localization of S2-RNase:GFP in living in vitro-germinated pollen tubes by confocal imaging showed that it was sequestered inside both its self (S 2 ) and non-self (S 1 ) pollen tubes, thus suggesting that compartmentalization of S2-RNase precludes the protein from gaining access to pollen tube RNA to exert its cytotoxic effect. Similarly, S3-RNase:GFP was sequestered in in vitro-germinated pollen tubes of S 2 S 3 transgenic plants. Although we cannot distinguish between S 2 and S 3 transgenic pollen, the fact that all the ~100 fluorescent pollen tubes we examined from two independent transgenic plants showed the same localization would suggest that, like S2-RNase:GFP, S3-RNase:GFP was sequestered in both its non-self (S 2 ) and self (S 3 ) pollen tubes.
Both Kirch et al. (1995) and Dodds et al. (1999) used immunoelectron microscopy to examine the subcellular localization of the S-RNase expressed in pollen of the transgenic plants of N. tabacum and L. peruvianum, respectively. Kirch et al. (1995) showed that the ectopically expressed S2-RNase of S. tuberosum was localized in the cytosol of mature pollen of transgenic N. tabacum plants, whereas Dodds et al. (1999) showed that the ectopically expressed S3-RNase of L. peruvianum was located in the cell wall of mature pollen. It might be possible that the sequestration of S2-RNase:GFP and S3-RNae:GFP in the pollen tube was caused by the GFP tag, as GFP has been reported to cause aggregation of GFP fusion proteins (Hitchcock et al. 2005). However, this does not seem likely based on our examination of the subcellular localization of PiSLF2:GFP (Fig. 5d) in in vitro-germinated pollen tubes of an S 2 S 3 transgenic plant we had previously generated (Hua et al. 2007). This GFP fusion protein contains the same 13-amino-acid linker and GFP tag as was used in the S2-RNase:GFP and S3-RNase:GFP fusion proteins examined in this work, but it was localized in the cytosol, a location consistent with the electron immunolocalization results of AhSLF2 of A. hispanicum (Wang and Xue 2005).
In this study, we also examined whether glycosylation is required for the compartmentalization of S-RNase in the pollen tube. This was prompted by the following observations we had previously made. First, S3-RNase(N29D) produced in the pistil of transgenic S 1 S 2 plants was able to reject S 3 pollen (Karunanandaa et al. 1994), suggesting that glycosylation is not required for the uptake of S-RNase by the pollen tube, or the SI function of S-RNase inside the pollen tube. Second, deglycosylated native S3-RNase and recombinant S3-RNase(N29D) were much more efficiently degraded in pollen tube extracts than native S3-RNase purified from pistils (Hua and Kao 2006). These findings raised a possibility that deglycosylation of S-RNase might be necessary for S-RNase to locate to the cytosol and be subjected to degradation mediated by SLF. The results obtained in this work, however, suggest that the compartmentalization of S-RNase is independent of glycosylation.
Goldraij et al. (2006) identified the S-RNase-containing compartment in both compatible and incompatible pollen tubes as a vacuolar compartment, based on the co-localization of S-RNase and a vacuolar membrane marker, vacuolar pyrophosphatase. The identity of the S-RNase-containing compartment observed in this work remains to be determined, but it is interesting to note that this compartment is dynamic in living pollen tubes with the movement driven by the actin–myosin-based motor system. Goldraij et al. (2006) observed that the S-RNase-containing compartment is disrupted in incompatible pollen tubes, but not in compatible pollen tubes, when the morphology of incompatible pollen tubes begins to differ from that of compatible pollen tubes. However, we did not observe any disruption of the S-RNase-containing compartment in in vitro-germinated pollen tubes (compatible or incompatible), even after pollen tubes had grown for up to 16 h (data not shown).
If the S-RNase-containing compartment observed in this work is the same as the vacuolar compartment observed by Goldraij et al. (2006), it raises a question as to what the fundamental difference is between uptake of S-RNase by the pollen tube and direct expression of S-RNase in the pollen that could account for the different fates of the compartment in incompatible pollen tubes. One possibility is that one or more of the pistil-expressed proteins that are required for the SI response in the pollen tube are missing when S-RNase is directly produced in the pollen. For example, HT-B, a pistil-specific protein, has been shown to be required for SI in N. alata (McClure et al. 1999), S. chacoense (O’Brien et al. 2002), and L. peruvianum (Kondo et al. 2002), as suppression of its expression leads to breakdown of SI. Goldraij et al. (2006) showed that HT-B is taken up by both compatible and incompatible pollen tubes, is associated with the membrane of the S-RNase-containing vacuolar compartment, and is less stable in compatible pollen tubes than in incompatible pollen tubes. Thus, they proposed that rejection of self-pollen tubes during incompatible pollination is caused by the disruption of the S-RNase-containing compartment mediated by HT-B (Goldraij et al. 2006; McClure 2009). However, in P. inflata, the role of HT-B in SI is not as clear-cut because Puerta et al. (2009) showed that >1000-fold suppression of HT-B transcript in the pistil of transgenic plants only leads to partial breakdown of SI. Regardless, the absence of non-S-specific pistil proteins (HT-B and/or some other proteins) that are required for SI and have to be taken up by the pollen tube during its growth in the pistil can explain why the S-RNase-containing compartment is not disrupted in in vitro-germinated pollen tubes.
However, the lack of non-S-specific pistil proteins cannot explain why the ectopically expressed S-RNase does not reject its self-pollen tubes in a compatible pistil (i.e., in vivo). For example, when pollen of a transgenic S 2 S 3 plant ectopically expressing S3-RNase is used to pollinate a wild-type plant of S 2 S 2 genotype, the S 3 transgenic pollen tubes would take up S2-RNase, along with all the non-S-specific factors, during its initial growth in the S 2 S 2 pistil. The S3-RNase-containing compartment might then fuse with the S2-RNase-containing compartment to result in a compartment containing both S2- and S3-RNases. This compartment would be equivalent to the compartment formed when a wild-type S 3 pollen tube is growing in an incompatible S 2 S 3 pistil, and thus we would expect that S 3 transgenic pollen ectopically expressing S3-RNase:GFP to be rejected by an S 2 S 2 pistil. But, this is not what we found. Thus, it is likely that the ectopically expressed S-RNase:GFP-containing compartment, observed in this work, and the pistil-expressed S-RNase-containing compartment, observed by Goldraij et al. (2006), are different.
Considering that none of the ectopically expressed S-RNases affect pollen viability, the compartment is most likely formed as soon as S-RNases are produced. At present, it is not known whether this compartment, like the compartment observed by Goldraij et al. (2006), would be disrupted when pollen tubes are growing in an incompatible pistil. It is possible that the sequestration of ectopically expressed S-RNase in the pollen/pollen tube is a general defense mechanism to detoxify toxic molecules in pollen tubes.
The compartmentalization of S-RNase in pollen/pollen tubes is an interesting phenomenon, and since it is clear that S-RNase must be released from the compartment in order to exert its function in SI, the challenge is to determine how SLF, located in the cytosol, and/or any other pistil factors mediate this process to allow the S-specific interaction with SLF in the cytosol.
References
Ai Y, Singh A, Coleman CE, Ioerger TR, Kheyr-Pour A, Kao T-h (1990) Self-incompatibility in Petunia inflata: isolation and characterization of cDNAs encoding three S-allele-associated proteins. Sex Plant Reprod 3:130–138
Anderson MA, Cornish EC, Mau S-L et al (1986) Cloning of cDNA for a stylar glycoprotein associated with expression of self-incompatibility in Nicotiana alata. Nature 321:38–44
Cleary AL, Hardham AR (1988) Depolymerization of microtubule arrays in root tip cells by oryzalin and their recovery with modified nucleation patterns. Can J Bot 66:2353–2366
de Nettancourt D (2001) Incompatibility and incongruity in wild and cultivated plants. Springer, Berlin
Dodds PN, Bönig I, Du H, Rödin J, Anderson MA, Newbigin E, Clarke AE (1993) S-RNase gene of Nicotiana alata is expressed in developing pollen. Plant Cell 5:1771–1782
Dodds PN, Ferguson C, Clarke AE, Newbigin E (1999) Pollen-expressed S-RNases are not involved in self-incompatibility in Lycopersicon peruvianum. Sex Plant Reprod 12:76–87
Dowd PE, Coursol S, Skirpan A, Kao T-h, Gilroy S (2006) Petunia phospholipase C1 is involved in pollen tube growth. Plant Cell 18:1438–1453
Entani T, Iwano M, Shiba H, Che F-S, Isogai A, Takayama S (2003) Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: identification of a pollen-expressed F-box gene with allelic diversity. Genes Cells 8:203–213
Frankin-Tong V (2008) Self-incompatibility in flowering plants—evolution, diversity, and mechanisms. Springer, Berlin
Goldraij A, Kondo K, Lee CB, Hancock CN, Sivaguru M, Vazquez-Santana S, Kim S, Phillips TE, Cruz-Garcia F, McClure B (2006) Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature 439:805–810
Hauck NR, Yamane H, Tao R, Iezzoni AF (2006) Accumulation of non-functional S-haplotypes results in the breakdown of gametophytic self-incompatibility in tetraploid Prunus. Genetics 172:1191–1198
Hepler PK, Vidall L, Cheung AY (2001) Polarized cell growth in higher plants. Annu Rev Cell Dev Biol 17:159–187
Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67:425–479
Hitchcock AL, Kahana JA, Silver PA (2005) The uses of green fluorescent protein in yeasts. In: Chalfie M, Kain S (eds) Methods of biochemical analysis, green fluorescent proteins: properties, applications and protocols, vol 47, 2nd edn. Wiley, Hoboken, pp 179–202
Hua Z, Kao T-h (2006) Identification and characterization of components of a putative Petunia S-Locus F-Box-containing E3 ligase complex involved in S-RNase-based self-incompatibility. Plant Cell 18:2531–2553
Hua Z, Kao T-h (2008) Identification of major lysine residues of S3-RNase of Petunia inflata involved in ubiquitin-26S proteasome-mediated degradation in vitro. Plant J 54:1094–1104
Hua Z, Meng XY, Kao T-h (2007) Comparison of Petunia inflata S-locus F-box protein (Pi SLF) with Pi SLF-like proteins reveals its unique function in S-RNase-based self-incompatibility. Plant Cell 19:3593–3609
Hua Z, Fields A, Kao T-h (2008) Biochemical models for S-RNase-based self-incompatibility. Mol Plant 1:575–585
Huang S, Lee H–S, Karunanandaa B, Kao T-h (1994) Ribonuclease activity of Petunia inflata S proteins is essential for rejection of self-pollen. Plant Cell 6:1021–1028
Ioerger TR, Gohlke JR, Xu B, Kao T-h (1991) Primary structural features of the self-incompatibility protein in Solanaceae. Sex Plant Reprod 4:81–87
Kao T-h, Tsukamoto T (2004) The molecular and genetic bases of S-RNase-based self-incompatibility. Plant Cell 16:S72–S83
Karunanandaa B, Huang S, Kao T-h (1994) Carbohydrate moiety of the Petunia inflata S3 protein is not required for self-incompatibility interactions between pollen and pistil. Plant Cell 6:1933–1940
Kirch H–H, Li Y-Q, Seul U, Thompson RD (1995) The expression of a potato (Solanum tuberosum) S-RNase gene in Nicotiana tabacum pollen. Sex Plant Reprod 8:77–84
Kondo K, Yamamoto M, Matton DP, Sato T, Hirai M, Norioka S, Hattori T, Kowyama Y (2002) Cultivated tomato has defects in both S-RNase and HT genes required for stylar function of self-incompatibility. Plant J 29:627–636
Lai Z, Ma W, Han B, Liang L, Zhang Y, Hong G, Xue Y (2002) An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol Biol 50:29–42
Lee H–S, Huang S, Kao T-h (1994) S proteins control rejection of incompatible pollen in Petunia inflata. Nature 367:560–563
Lee H–S, Karunanandaa B, McCubbin A, Gilroy S, Kao T-h (1996) PRK1, a receptor-like kinase of Petunia inflata, is essential for postmeiotic development of pollen. Plant J 9:613–624
Luu D-T, Qin X, Morse D, Cappadocia M (2000) S-RNase uptake by compatible pollen tubes in gametophytic self-incompatibility. Nature 407:649–651
McClure B (2009) Darwin’s foundation for investigating self-incompatibility and the progress toward a physiological model for S-RNase-based SI. J Exp Bot 60:1069–1081
McClure B, Haring V, Ebert PR, Anderson MA, Simpson RJ, Sakiyama F, Clarke AE (1989) Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature 342:955–957
McClure B, Mou B, Canevascini S, Bernatzky R (1999) A small asparagine-rich protein required for S-allele-specific pollen rejection in Nicotiana. Proc Natl Acad Sci USA 96:13548–13553
McKillop DF, Fortune NS, Ranatunga KW, Geeves MA (1994) The influence of 2,3-butanedione 2-monoxime (BDM) on the interaction between actin and myosin in solution and skinned muscle fibres. J Muscle Res Cell Motil 15:309–318
Moon J, Parry G, Estelle M (2004) The ubiquitin–proteasome pathway and plant development. Plant Cell 16:3181–3195
Morejohn LC, Bureau TE, Mole-Bajer J, Bajer AS, Fosket DE (1987) Oryzalin, a dinitroaniline herbicide, binds to plant tubulin and inhibits microtubule polymerization in vitro. Planta 172:252–264
Murfett J, Atherton TL, Mou B, Gasser CS, McClure B (1994) S-RNase expressed in transgenic Nicotiana causes S-allele-specific pollen rejection. Nature 367:563–566
O’Brien M, Kapfer C, Major G, Laurin M, Bertrand C, Kondo K, Kowyama Y, Matton DP (2002) Molecular analysis of the stylar-expressed Solanum chacoense small asparagine-rich protein family related to the HT modifier of gametophytic self-incompatibility in Nicotiana. Plant J 32:985–996
Poulter NS, Vatovec S, Franklin-Tong VE (2008) Microtubules are a target for self-incompatibility signaling in Papaver pollen. Plant Physiol 146:1358–1367
Puerta AR, Ushijima K, Koba T, Sassa H (2009) Identification and functional analysis of pistil self-incompatibility factor HT-B of Petunia. J Exp Bot 60:1309–1318
Qiao H, Wang H, Zhao L, Zhou J, Huang J, Zhang Y, Xue Y (2004a) The F-box protein Ah SLF-S2 physically interacts with S-RNases that may be inhibited by the ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in Antirrhinum. Plant Cell 16:582–595
Qiao H, Wang F, Zhao L, Zhou J, Lai Z, Zhang Y, Robbins TP, Xue Y (2004b) The F-box protein AhSLF-S2 controls the pollen function of S-RNase-based self-incompatibility. Plant Cell 16:2307–2322
Romagnoli S, Cai G, Cresti M (2003) In vitro assays demonstrate that pollen tube organelles use kinesin-related motor proteins to move along microtubules. Plant Cell 15:251–269
Sijacic P, Wang X, Skirpan AL, Wang Y, Dowd PE, McCubbin AG, Huang S, Kao T-h (2004) Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 429:302–305
Singh A, Ai Y, Kao T-h (1991) Characterization of ribonuclease activity of three S-allele-associated proteins of Petunia inflata. Plant Physiol 96:61–68
Skirpan AL, McCubbin AG, Ishimizu T, Wang X, Hu Y, Dowd PE, Ma H, Kao T-h (2001) Isolation and characterization of kinase interacting protein 1, a pollen protein that interacts with the kinase domain of PRK1, a receptor-like kinase of Petunia. Plant Physiol 126:1480–1492
Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55:555–590
Sonneveld T, Kenneth R, Tobutt KR, Simon P, Vaughan SP, Robbins TP (2005) Loss of pollen-S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype-specific F-box gene. Plant Cell 17:37–51
Tominaga M, Yokota E, Sonobe S, Shimmen T (2000) Mechanism of inhibition of cytoplasmic streaming by a myosin inhibitor, 2, 3-butanedione monoxime. Protoplasma 213:46–54
Tsukamoto T, Hauck NR, Tao R, Jiang N, Iezzoni AF (2006) Molecular characterization of three non-functional S-haplotypes in sour cherry (Prunus cerasus). Plant Mol Biol 62:371–383
Twell D, Yamaguchi J, McCormick S (1990) Pollen-specific gene expression in transgenic plants: coordinate regulation of two different tomato gene promoters during microsporogenesis. Development 109:705–713
Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H (2003) Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15:771–781
Wang H, Xue Y (2005) Subcellular localization of the S-locus F-box protein AhSLF2 in pollen and pollen tubes of self-incompatible Antirrhinum. J Integr Plant Biol 47:76–83
Wang Y, Tsukamoto T, Yi K-w, Wang X, Huang S, McCubbin AG, Kao T-h (2004) Chromosome walking in the Petunia inflata self-incompatibility (S-) locus and gene identification in an 881-kb contig containing S 2 -RNase. Plant Mol Biol 54:727–742
Yagi N, Takemori S, Watanabe M, Horiuti K, Amemiya Y (1992) Effects of 2, 3-butanedione monoxime on contraction of frog skeletal muscle: an X-ray diffraction study. J Muscle Res Cell Motil 13:153–160
Acknowledgments
We thank Simon Gilroy and Richard Cyr for advice and suggestions on confocal experiments. This work was supported by National Science Foundation grants IOB 05-43201 and IOS 08-43195 to T.-h.K.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Celestina Mariani.
X. Meng and Z. Hua contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
497_2009_114_MOESM2_ESM.tif
Supplementary Fig. 1 Schematic drawing of the constructs, pBI LAT52-S 2 -RNase:GFP, pBI LAT52-S 3 -RNase:GFP, and pBI LAT52-S 3 -RNase(N29D):GFP, used in transformation. The region between the right border (RB) and the left border (LB) is integrated into transgenic plants. Nos, the gene encoding nopaline synthase; pro, promoter; ter, transcription terminator; P1 and P2 denote the locations of the forward and reverse primers, respectively, used in PCR for each transgene (see text for sequences); NPTII, the gene encoding neomycin phosphotransferase II (conferring kanamycin resistance). (TIFF 261 kb)
497_2009_114_MOESM3_ESM.zip
Supplementary Movies 1 Trafficking of S3-RNase:GFP-containing compartments in living in vitro-germinated pollen tubes and the effect of BDM and oryzalin on trafficking. The movies show in sequence: 1. GFP in in vitro-germinated pollen tubes of a transgenic S 2 S 3 plant carrying the GFP transgene expressed by the LAT52 promoter; 2. PiSLF2:GFP in in vitro-germinated pollen tubes of a transgenic S 2 S 3 plant carrying the PiSLF 2 :GFP transgene expressed by the LAT52 promoter; 3. Trafficking of S3-RNase:GFP-containing compartments in living in vitro-germinated pollen tubes of a transgenic S 2 S 3 plant carrying the S 3 -RNase:GFP transgene expressed by the LAT52 promoter; 4. Effect of 30 mM BDM on the trafficking of S3-RNase:GFP-containing compartments in living in vitro-germinated pollen tubes; 5. Effect of 50 mM BDM on the trafficking of S3-RNase:GFP-containing compartments in living in vitro-germinated pollen tubes; 6. Effect of 50 μM oryzalin on the trafficking of S3-RNase:GFP-containing compartments in living in vitro-germinated pollen tubes. In each movie, the left panel is the GFP channel and the right panel is the bright-field channel. (TAR 5,924 kb)
497_2009_114_MOESM4_ESM.zip
Supplementary Movies 2 Trafficking of S3-RNase(N29D):GFP-containing compartments in living in vitro-germinated pollen tubes and the effect of BDM and oryzalin on the trafficking. The movies show in sequence: 1. GFP in in vitro-germinated pollen tubes of a transgenic S 2 S 3 plant carrying the GFP transgene expressed by the LAT52 promoter; 2. PiSLF2:GFP in in vitro-germinated pollen tubes of a transgenic S 2 S 3 plant carrying the PiSLF 2 :GFP transgene expressed by the LAT52 promoter; 3. Trafficking of S3-RNase(N29D):GFP-containing compartments in living in vitro-germinated pollen tubes of a transgenic S 2 S 3 plant carrying the S 3 -RNase(N29D):GFP transgene expressed by the LAT52 promoter; 4. Effect of 30 mM BDM on the trafficking of S3-RNase(N29D):GFP-containing compartments in living in vitro-germinated pollen tubes; 5. Effect of 50 mM BDM on the trafficking of S3-RNase(N29D):GFP-containing compartments in living in vitro-germinated pollen tubes; 6. Effect of 50 μM oryzalin on the trafficking of S3-RNase(N29D):GFP-containing compartments in living in vitro-germinated pollen tubes. In each movie, the left panel is the GFP channel and the right panel is the bright-field channel. (TAR 5,720 kb)
Rights and permissions
About this article
Cite this article
Meng, X., Hua, Z., Wang, N. et al. Ectopic expression of S-RNase of Petunia inflata in pollen results in its sequestration and non-cytotoxic function. Sex Plant Reprod 22, 263–275 (2009). https://doi.org/10.1007/s00497-009-0114-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00497-009-0114-3