Abstract
Key message
Based on Casuarina equisetifolia transcriptome data, the present study developed and validated 15 transferable EST-SSR markers across Casuarina and Allocasuarina species.
Abstract
Casuarina species are valued as pioneer trees for coastal shelter forest construction. To accelerate Casuarina breeding for more robust, attractive, and cold-resistant cultivars, it is essential to introduce molecular marker-assisted breeding. Based on Casuarina equisetifolia transcriptome data, 10,291 EST-SSR loci with the types of 2–6 nucleotide repeats were identified from assembled 118,270 unigenes. An average of one EST-SSR was found every 13.54 kb, and the distribution frequency of SSRs was 8.7%. The most common repeat motifs were dinucleotide with AG/CT and trinucleotide with AAG/CTT. A total of 150 SSR markers were randomly selected for validation, among which 15 produced clear, reproducible and polymorphic bands, and thus were employed for genetic relationship analysis among 26 genotypes from four Casuarina and three Allocasuarina species. A total of 42 bands were identified, varying from 2 to 5 bands per locus with an average of 2.8. Polymorphism information content ranged from to 0.2624 to 0.6177 with a mean value of 0.4265, indicating a high level of polymorphism. The UPGMA dendrogram revealed a high genetic variation resided among species within two genera, Allocasuarina and Casuarina. Meanwhile, the 15 highly polymorphic Casuarina SSR markers were also proved to be transferable across Casuarina and Allocasuarina species. These valuable SSR markers can be applied further for population and evolutionary genetic studies in Casuarinaceae, and will be useful in selection and breeding of Casuarina species for improving cold tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Casuarina belongs to the Casuarinaceae which is member of the major group of Angiosperms. Species in the genus Casuarina, commonly known as casuarinas, beefwoods, she-oaks, Australian pines or ironwoods, are fast-growing evergreen trees native to Australia, Southeast Asia and the Pacific archipelagos (Wheeler et al. 2011). Many casuarinas are economically and ecologically important species. They are valued as pioneer trees for degraded sites and for soil improvement and no other species can replace casuarinas at the foreshores. Along the southern coasts of China, Casuarina trees are also commonly planted as windbreaks to stabilize the moving sand, used in agroforestry, and for general rehabilitation and wood production programs (Zhong et al. 2010). However, Casuarina trees suitable for constructing coastal shelter forest are few and low in quality. Low temperature is one of the main limiting factors for cultivation expansion, fast growth, and high yield of Casuarina species. So it is very important to breed high-quality Casuarina species with cold-tolerant genotypes.
Molecular marker-assisted breeding is considered a novel way to reduce the breeding and selection processes indirectly, shortening the breeding cycle, especially beneficial for resistance breeding (Du et al. 2015). Microsatellites, or simple sequence repeats (SSRs), have been applied for construction of chromosome maps and cultivar identification (Song et al. 2004; Tsukazaki et al. 2008, 2009, 2015). Compared with other molecular markers, SSR markers are more ideal due to their reproducibility, multi-allelic nature, co-dominant inheritance, relative abundance, and good genome coverage (Tóth et al. 2000). SSRs are generally categorized into two common groups based on their origins, genomic SSRs from genomic sequences, and expressed sequence tag (EST)-SSRs from transcribed RNA sequences (Varshney et al. 2005). As one of the powerful next-generation sequencing (NGS) methods, Illumina RNA-seq has proven to be robust and efficient with higher throughput and much lower cost (Garg et al. 2011; Grabherr et al. 2011; Ono et al. 2011; Yang et al. 2011; Feng et al. 2012) than other systems, and has been used in many plants to reveal gene annotation and expression under biotic and abiotic stresses (Marguerat and Bähler 2010; Oshlack et al. 2010; Mochida and Shinozaki 2011; Wang and Chen 2012; Postnikova et al. 2013). In addition to the discovery of new genes and investigations of gene expression, RNA-seq also offers many advantages, including the detection of thousands of single nucleotide polymorphisms (SNPs) and EST-SSR loci for markers development. A large number of SNPs and SSR markers will provide the foundation for future research to address questions in population and evolutionary genetic studies.
Casuarina equisetifolia L. has the largest native range in coastal regions of Northern Australia, Malaysia and Southeast Asia, and Oceania (Parrotta 1993). Presently, C. equisetifolia genome has not yet been fully sequenced. Kullan et al. (2016) validated 42 SSR markers from 86,415 ESTs of C. equisetifolia and C. junghuhniana. However, the number of SSR markers reported for Casuarina species is still far fewer than other commonly planted tree species such as Eucalyptus (Rabello et al. 2005), Live oaks (Cavender-Bares et al. 2015), Pine (Feng et al. 2014), Kandelia candel (Islam et al. 2006, 2008), etc. It is necessary to develop more novel cross-species transferable SSR markers for promoting the genetic and breeding studies in Casuarina species.
In previous work, we performed a global survey for transcriptome profiles of C. equisetifolia via the Illumina HiSeq 2500 platform. A total of 118,270 unigenes were obtained by de novo assembly, creating an initial reference transcriptome (Li et al. 2017). Hence, in the present study, our aim was to: (1) identify a large number of EST-SSR markers based on the C. equisetifolia transcriptome information obtained; (2) validate the identified EST-SSR markers for the detection of polymorphism in Casuarina and Allocasuarina species; (3) evaluate the transferability of the validated EST-SSR markers across Casuarina and Allocasuarina.
Materials and methods
Plant materials and genomic DNA extraction
A cold-resistant C. equisetifolia individual tree (Zhou 7) was used for RNA-seq. For genetic relationship analysis, fresh juvenile needles of 26 genotypes from four Casuarina species (C. equisetifolia, C. cunninghamiana, C. glauca, and C. cristata) and three Allocasuarina species (A. huegeliana, A. littoralis and Allocasuarina sp.) were collected from the Zhoushan Germplasm Bank of Institute of Tree Breeding, Zhejiang, China and dried in silica gel for DNA extraction. Detailed information for the plant materials is listed in Table 1. Genomic DNA of 26 genotypes was extracted from fresh juvenile needles using the Plant Genomic DNA Extraction kit (Bioteke, Beijing, China) following the manufacturer’s instructions. The DNA quality was evaluated by electrophoresis through a 1.5% agarose gel, and the DNA concentration was determined using a Thermo Scientific NanoDrop™ 2000C spectrophotometer (Nanodrop Technologies, South Logan, Utah, USA).
Identification of EST-SSR markers
The MicroSAtellite program (MISA, http://pgrc.ipk-gatersleben.de/misa) was used to detect and locate SSRs from unigenes (Sharma et al. 2007). The search criteria for simple motifs (di-, tri-, tetra-, penta-, and hexanucleotides) and complex (imperfect) nucleotides were a minimum of five and three repeats, respectively. Compound SSR markers are SSR loci containing multiple different repeat motifs, separated by non-repetitive sequences. In this analysis, the maximum size of interruption allowed between two different SSRs in a compound SSR was 100 bp. Mononucleotide repeats were excluded.
Development and selection of EST-SSR primers
SSR primer pairs of each unique microsatellite locus were designed using Primer 3.0 (Rozen and Skaletsky 2000), with target microsatellites containing at least five repeats and yielding PCR products of 80–500 bp size. The genomic DNA of 12 genotypes were first used for primary screening of 150 SSR primer pairs. Each 20 μL PCR reaction mixture contained 10 μL 2 × Power Taq PCR Master Mix (Bioteke, Beijing, China), 1.5 μL SSR forward and reverse primer (each 10 μM), 3 μL DNA templates (20 ng/μL), and 4 μL sterile distilled water. SSR-PCR amplification was performed in a DNA thermal cycler (Life ECO, Hangzhou Bioer) using the following parameters: 7 min at 94 °C, followed by 30 cycles of 45 s at 94 °C, 45 s at 55 °C, 2 min at 72 °C, and a final extension step was for 7 min at 72 °C. The PCR products were resolved by 6% polyacrylamide gel electrophoresis (PAGE) and silver staining in accordance with the protocol described by Creste et al. (2001).
EST-SSR markers validation and genetic relationship analysis
Fifteen polymorphic SSR primers were used as valid SSR markers for genetic relationship analysis among 26 genotypes from Casuarina and Allocasuarina species. The SSR-PCR bands obtained were scored as presence (1) or absence (0) and two binary qualitative data matrices were constructed. A pair-wise similarity matrix was constructed using the Dice similarity coefficient (DSC). The relationship between the genotypes was displayed as a dendrogram, constructed using NTSYS-pc 2.2 r software (Rohlf 1992) based on unweighed pair group method with arithmetic mean (UPGMA). Tree confidence was also performed by a bootstrap analysis with 100 replications using the DendroUPGMA tool available at http://genomes.urv.cat/UPGMA (Garcia-Vallve et al. 1999). For each locus, the number of alleles and allele frequencies were calculated using the program POPGEN version 1.32 (Yeh et al. 2000). The polymorphism information content (PIC) (Botstein et al. 1980) of each EST-SSR primer pairs was calculated using PIC-CALC 0.6 software.
Results
Distribution and frequency of the EST-SSR motifs
The EST-SSRs were mined from C. equisetifolia transcriptome data reported in the earlier study by Li et al. (2017). Based on the transcriptome data obtained in this study, a total of 10,291 SSRs were identified (8.7%) from the 118,270 assembled unigenes with 1089 unigene sequences containing more than one SSR. Among the identified SSRs, 344 (3.34%) were present in compound formation, while others were of perfect one-repeat type. Total size of examined sequences was 139,375,239 bp, and the distribution density was one SSR for every 13.54 kb. The total sequence length of SSR loci was 153,984 bp with an average of 14.96 bp. The average repeat sequence length of di-, tri-, tetra-, penta-, and hexanucleotide was 13.95, 16.12, 19.32, 27.50, 33.05 bp, respectively (Table 2). The number of SSR repeats ranged from 5 to 12, and the most frequent repeat was six (3158 or 30.69%), followed by five (2154 or 20.93%), seven (1765 or 17.15%) and nine (1067 or 10.37%). Among these repeats, dinucleotide was the most frequent type (6546 or 63.61%), followed by trinucleotide (3356 or 32.61%) and tetranucleotide (306 or 2.97%). There were far less pentanucleotide and hexanucleotide repeat motifs, only accounting for 0.23% (24) and 0.57% (59), respectively (Table 3).
Distribution and frequency of EST-SSR types
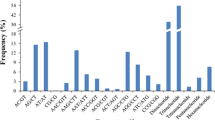
A total of 82 simple repeat motifs were identified in C. equisetifolia. The most abundant motif was AG/CT (54.24%) for dinucleotide, and AAG/CTT (13.02%) for trinucleotide motif. The motifs with low proportion of less than 1.0% were ACT/AGT (0.32%), CG/CG (0.33%), CCG/CGG (0.83%), and ACG/CGT (0.90%) (Table 4).
Validation and evaluation of polymorphic SSR markers
Of the total 10,291 EST-SSR loci identified from C. equisetifolia, 3745 were of trinucleotide, tetranucleotide, pentanucleotide, and hexanucleotide motifs with the amplicons size ranging between 100 and 300 bp. To validate the identified SSR markers, we designed 150 pairs of SSR primers among the 3745 SSR loci for PCR screening. These primers were tested for DNA amplification in 12 genotypes from Casuarina and Allocasuarina species. Subsequent to PCR, 55 SSR primers generated products in the expected size, among which, 15 (27.3%) produced clear, reproducible and polymorphic bands (Fig. 1). The 15 polymorphic SSR primers were, therefore, used as valid SSR markers for further genetic diversity study.
Polyacrylamide gel electrophoretic pattern of 26 Casuarina and Allocasuarina genotypes amplified with the primers of SSR marker CeUg19361. Lane M molecular marker; Lanes from 1 to 26 correspond to the 26 Casuarina and Allocasuarina genotypes listed in Table 1; The numbers on the left of the figure indicate the DNA size markers in kilobases (kb)
Genetic relationship analysis among 26 genotypes from four Casuarina species and three Allocasuarina species listed in Table 1 were performed with the 15 SSR primer pairs. A total of 42 polymorphic bands were detected from the 15 SSR loci with the size ranging from 131 to 280 bp. The number of bands detected by each SSR primer pairs ranged from 2 to 5, with an average of 2.8. The PIC value varied from 0.2624 to 0.6177 with a mean value of 0.4265 (Table 5). The PIC index describes diversity within accessions (intra-populational diversity) and evaluates the degree of polymorphism in each locus, a PIC value of less than 0.25 indicating low polymorphism, a value between 0.25 and 0.5 average polymorphism and a value higher than 0.5 a highly polymorphic locus (Botstein et al. 1980). Out of the 15 loci, 5 loci had PIC values exceeding 0.5 and could be considered highly polymorphic, while 10 loci had average polymorphism.
Genetic relationship analysis
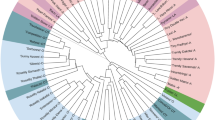
Based on 42 polymorphic bands detected from 15 SSR loci, 26 genotypes belonging to four Casuarina species and three Allocasuarina species in the UPGMA dendrogram were clearly divided into three main clades (I, II, III) with low genetic similarity coefficients (GSC) level of 0.53 with Dice, which was well supported by a 100% bootstrap value (Fig. 2). The clade I, supported by a 72% bootstrap value, consists of mixtures of species including A. huegeliana, Allocasuarina sp., A. littoralis and C. cunninghamiana. The species C. cristata formed a independent clade II, supported by a 77% bootstrap value. The species of C. equisetifolia and C. glauca representing the clade III with a 100% bootstrap support value is well separated from clade I and II, indicating that a distinct genetic relationship between the two species within clade III and the other five species within clade I–II. Meanwhile, low levels of intraspecific variation within Casuarina and Allocasuarina species were demonstrated, with high GSC level by Dice ranging from 0.68 (C. cristata) to 0.93 (A. littoralis). At the interspecific level, a low genetic variation resided between the species of C. equisetifolia and C. glauca at GSC level of 0.70, A. littoralis and C. cunninghamiana 0.73, as well as A. huegeliana and Allocasuarina sp. 0.78. In addition, the present UPGMA dendrogram did not separate the Casuarina and Allocasuarina genera and revealed a significant relationship between A. littoralis and C. cunninghamiana, as well as C. equisetifolia and C. glauca.
Discussion
Compared with time-consuming genomic SSR development, identification of SSR markers from EST sequences is less labor intensive (Varshney et al. 2005). Besides, EST-SSRs are potentially tightly linked with functional genes that can be associated with certain important genetic traits and contain high level of transferability across species and genera. In the present study, we identified a total of 10,291 SSR loci from 118,270 assembled unigene sequences of C. equisetifolia (8.7%), which is slightly lower than that observed from Casuarina transcriptome data (10.7%, Kullan et al. 2016). The distribution frequency (8.7%) is also much higher than those obtained from cereal species (3.5%) and other tree species such as Eucommia ulmoides (2.9%), Pinus massoniana (3.62%), Pinus koraiensis (4.24%), and Camellia oleifera (4.99%), and much lower than Populus trees (14.83%), also Eucalyptus trees (34.87%) (Varshney et al. 2002; Rabello et al. 2005; Wen et al. 2013; Zhang et al. 2015). The difference in distribution frequency may be mainly due to species specificity, the size of dataset, the quality of sequenced unigenes, also the database mining tools, SSR search criteria, etc. (Varshney et al. 2005; Biswas et al. 2012). Therefore, a direct comparison of abundance estimation and frequency occurrence of SSRs in different reports is difficult. In C. equisetifolia, the most abundant motif was AG/CT (54.24%) and AAG/CTT (13.02%). The same results have been observed by Kullan et al. (2016), where the types of AG/CT (62.4%) and AAG/CCT (9%) were the most prevalent motifs among dinucleotide repeats and trinucleotide repeats, respectively.
Cross-species amplification of SSR loci is considered as a cost-effective approach for developing microsatellite markers for new species. The transferability of EST-derived SSRs across different species and genera has been documented in many plant species, especially in studies involving economically important crop species (Ellis and Burke 2007). Yasodha et al. (2005; 2009) confirmed the transferability of Eucalyptus SSR markers across two species of C. equisetifolia and C. glauca. Among 42 Casuarina SSR markers, 80% were transferable across four species of C. cunninghamiana, C. junghuhniana, C. equisetifolia, and C. glauca (Kullan et al. 2016). Our present study confirmed transferability of 15 highly polymorphic Casuarina SSR markers across Casuarina and Allocasuarina species, which suggests that these SSR markers may be potentially valuable employed in the genetic diversity and genetic structure analyses in the genus of Casuarina and Allocasuarina.
The family Casuarinaceae belongs to the Gondwanic family consisting of four genera (Allocasuarina, Casuarina, Ceuthostoma and Gymnostoma) and 97 species including monoecious or dioecious shrubs and trees (Wilson and Johnson 1989; Kamalakannan et al. 2006). Allocasuarina species are considered to be the nearest relatives of the genus Casuarina since they share many common morphological features (Wilson and Johnson 1989; Pinyopusarerk and House 1993; Yasodha et al. 2004). However, our present study with SSR markers did not separate the Casuarina and Allocasuarina genera, just as reported in the previous study on morphological and molecular diversity among Casuarina and Allocasuarina species with morphometric parameters and inter simple sequence repeat (ISSR) markers (Yasodha et al. 2004; Kamalakannan et al. 2006; Chezhian et al. 2009). Therefore, further work on molecular phylogeny relationship of Casuarinaceae is needed. Thus, the 15 transferable Casuarina SSR markers can also be used for phylogenetic analysis to clarify phylogenetic relationships between the two genera. In addition, these EST-derived SSR markers can facilitate the development of gene-based markers that may increase the efficiency of marker-assisted selection and breeding of Casuarina trees.
The present study showed that all of the genotypes have low levels of genetic diversity at the species level, especially those from A. littoralis. The results are in accordance with the observation made by Kamalakannan et al. (2006) and Chezhian et al. (2009). This may be due to the less number of parent trees used for bulking of seeds and the limited genotypes used in the present study. For A. littoralis, the narrow genetic diversity may be attributed mainly to its restricted geographical distribution. In addition, the close genetic relationship between C. equisetifolia and C. glauca revealed in the present study is well supported by the results reported by Kamalakannan et al. (2006) and Chezhian et al. (2009).
Although SSR markers developed from ESTs are widely used in genetic diversity analyses, only 15 polymorphic SSR markers were validated from 150 detected SSR markers in the present study, which revealed the EST-SSR markers of C. equisetifolia have a low level of polymorphism. Compared with EST-SSR markers, the genomic SSR markers can detect more allelic diversity, thus show higher discrimination power, which were reported in a variety of plant species (Eujayl et al. 2001; Chabane et al. 2005; Hu et al. 2011; Zhang et al. 2014). This fact may be due to possible selection against alterations in the conserved coding sequences (Scott et al. 2000), which limits SSR variation in these regions. Therefore, much more valuable EST-SSR markers need to be developed based on Illumina RNA-seq or public EST databases to meet the increasing demand for genetic and breeding studies in Casuarina and Allocasuarina species.
Conclusions
Based on transcriptome data of C. equisetifolia, 10,291 EST-SSR loci with the types of 2–6 nucleotide repeats were identified from assembled 118,270 unigenes. An average of one EST-SSR was found every 13.54 kb, and the distribution frequency of SSRs was 8.7%. The most common repeat motifs were dinucleotide with AG/CT and trinucleotide with AAG/CTT. Fifteen highly polymorphic Casuarina SSR markers were developed and proved to be transferable across Casuarina and Allocasuarina species, which can be applied for population and evolutionary genetic studies in Casuarinaceae, and will be useful in selection and breeding of Casuarina trees.
Author contribution statement
NL, YQZ, HMD, HZP, BJ and HBL designed the experiments; NL and HBL performed the experiments and analyzed the data; NL and HBL wrote the manuscript.
References
Biswas MK, Chai L, Mayer C, Xu Q, Guo W, Deng X (2012) Exploiting BAC-end sequences for the mining, characterization and utility of new short sequences repeat (SSR) markers in Citrus. Mol Biol Rep 39:5373–5386
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment polymorphisms. Am J Hum Genet 32:314–331
Cavender-Bares J, González-Rodríguez A, Eaton DAR, Hipp AAL, Beulke A, Manos PS (2015) Phylogeny and biogeography of the American live oaks (Quercus subsection Virentes): a genomic and population genetics approach. Mol Ecol 24:3668–3687
Chabane K, Ablett GA, Cordeiro GM, Valkoun J, Henry RJ (2005) EST versus genomic derived microsatellite markers for genotyping wild and cultivated barley. Genet Resour Crop Evol 52:903–909 (2005)
Chezhian P, Yasodha R, Ghosh M (2009) Genetic diversity analysis in Casuarina and Allocasuarina species using ISSR markers. Madras Agric J 96:32–39
Creste S, Tulmann Neto A, Figueira A (2001) Detection of single sequence repeat polymorphisms in denaturing polyacrylamide sequencing gels by silver staining. Plant Mol Biol Rep 19:299–306
Du F, Wu Y, Zhang Z, Li X, Zhao X, Wang W, Gao Z, Xia Y (2015) De novo assembled transcriptome analysis and SSR marker development of a mixture of six tissues from Lilium Oriental Hybrid ‘Sorbonne’. Plant Mol Biol Rep 33:281–293
Ellis JR, Burke JM (2007) EST-SSRs as a resource for population genetic analyses. Heredity 99:125–132
Eujayl I, Sorrells M, Baum M, Wolters P, Powell W (2001) Assessment of genotypic variation among cultivated durum wheat based on EST-SSRS and genomic SSRS. Euphytica 119:39–43
Feng C, Chen M, Xu C, Bai L, Yin X, Li X, Allan AC, Ferguson IB, Cheng K (2012) Transcriptomic analysis of Chinese bayberry (Myrica rubra) fruit development and ripening using RNA-Seq. BMC Genom 13:1–15
Feng YH, Yang ZQ, Wang J, Luo QF, Li HG (2014) Development and characterization of SSR markers from Pinus massoniana and their transferability to P. elliottii, P. caribaea and P. yunnanensis. Genet Mol Res 13:1508–1513
Garcia-Vallve S, Palau J, Romeu A (1999) Horizontal gene transfer in glycosyl hydrolases inferred from codon usage in Escherichia coli and Bacillus subtilis. Mol Biol Evol 16(9):1125–1134
Garg R, Patel R, Jhanwar S, Priya P, Chattopadhyay D, Tyagi A, Jain M (2011) Gene discovery and tissue-specific transcriptome analysis in chickpea with massively parallel pyrosequencing and web resource development. Plant Physiol 156:1661–1678
Grabherr M, Haas B, Yassour M, Levin J, Thompson D, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol 29:644–652
Hu J, Wang L, Li J (2011) Comparison of genomic SSR and EST-SSR markers for estimating genetic diversity in cucumber. Biol Plant 55:577–580
Islam MS, Tao JM, Geng QF, Lian CL, Hogetsu T (2006) Isolation and characterization of eight compound microsatellite markers in a mangrove tree Kandelia candel (L.) Druce. Mol Ecol Notes 6:1111–1113
Islam MS, Lian C, Geng Q, Kameyama N, Hogetsu T (2008) Chloroplast microsatellite markers for the mangrove tree species Bruguiera gymnorrhiza, Kandelia candel, and Rhizophora stylosa, and cross-amplification in other mangrove species. Conserv Genet 9:989–993
Kamalakannan R, Barthwal S, Chezhian P, Balasaravanan T (2006) Morphological and molecular diversity among Casuarina and Allocasuarina species. Biotechnology 5:301–307
Kullan ARK, Kulkarni AV, Kumar RS, Rajkumar R (2016) Development of microsatellite markers and their use in genetic diversity and population structure analysis in Casuarina. Tree Genet Genomes 12:49
Li HB, Li N, Yang SZ, Peng HZ, Wang LL, Wang Y, Zhang XM, Gao ZH (2017) Transcriptomic analysis of Casuarina equisetifolia in responses to cold stress. Tree Genet Genomes 13:7
Marguerat S, Bähler J (2010) RNA-seq: from technology to biology. Cell Mol Life Sci 67:569–579
Mochida K, Shinozaki K (2011) Advances in omics and bioinformatics tools for systems analyses of plant functions. Plant Cell Physiol 12:2017–2038
Ono N, Britton M, Fass J, Nicolet C, Lin D, Tian L (2011) Exploring the transcriptome landscape of pomegranate fruit peel for natural product biosynthetic gene and SSR marker discovery. J Integr Plant Biol 53:800–813
Oshlack A, Robinson M, Young M (2010) From RNA-seq reads to differential expression results. Genome Biol 11:1–10
Parrotta JA (1993) Casuarina equisetifolia L. ex J. R. & G. Forst. Casuarina, Australian pine. Research notes. USDA Forest Service, Southern Forest Experiment Station, New Orleans, pp 11–14
Pinyopusarerk K, House APN (1993) Casuarina : An annotated bibliography of Casuarina equisetifolia, C. junghuhniana and C. oligodon. International Center for Research in Agroforestry, Nairobi
Postnikova O, Shao J, Nemchinov L (2013) Analysis of the alfalfa root transcriptome in response to salinity stress. Plant Cell Physiol 54:1041–1055
Rabello E, Souza AN, Saito D, Tsai SM (2005) In silico characterization of microsatellites in Eucalyptus spp.: abundance, length variation and transposon associations. Genet Mol Biol 28:582–588
Rohlf FJ (1992) NTSYS-pc: numerical taxonomy and multi-variate analysis system, version 1.7. Exeter Software: Setauket, NY
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Scott KD, Eggler P, Seaton G, Rossetto M, Ablett EM, Lee LS, Henry RJ (2000) Analysis of SSRs derived from grape ESTs. Theor Appl Genet 100:723–726
Sharma P, Grover A, Kahl G (2007) Mining microsatellites in eukaryotic genomes. Trends Biotechnol 25:490–498
Song YS, Suwabe K, Wako T, Ohara T, Nunome T, Kojima A (2004) Development of microsatellite markers in bunching onion (Allium fistulosum L.). Breed Sci 54:361–365
Tóth G, Gáspári Z, Jurka J (2000) Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res 10:967–981
Tsukazaki H, Yamashita K, Yaguchi S, Masuzaki S, Fukuoka H, Yonemaru J, Kanamori H, Kono I, TranThiMinhHang Shigyo M et al (2008) Construction of SSR-based chromosome map in bunching onion (Allium fistulosum). Theor Appl Genet 117:1213–1223
Tsukazaki H, Yamashita KI, Kojima A, Wako T (2009) SSR-tagged breeding scheme for allogamous crops: a trial in bunching onion (Allium fistulosum). Euphytica 169:327–334
Tsukazaki H, Yaguchi S, Sato S, Hirakawa H, Katayose Y, Kanamori H, Kurita K, Itoh T, Kumagai M, Mizuno S (2015) Development of transcriptome shotgun assembly-derived markers in bunching onion (Allium fistulosum). Mol Breed 35:1–11
Varshney RK, Thiel T, Stein N, Langridge P, Graner A (2002) In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell Mol Biol Lett 7:537–546
Varshney R, Graner A, Sorrells M (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23:48–55
Wang L, Chen F (2012) Genotypic variation of potassium uptake and use efficiency in cotton Gossypium hirsutum. J Plant Nutr Soil Sci 175:303–308
Wen Q, Xu L, Jiang X, Li J, Gu Y, Xu L, Huang M (2013) Survey and analysis of microsatellites from DNA sequences in Camellia species using 454 pyrosequencing. Sci Silv Sin 49:43–50 (in Chinese)
Wheeler G, Taylor G, Gaskin JF, Purcell M (2011) Ecology and management of Sheoak (Casuarina spp.), an invader of coastal Florida, USA. J Coastal Res 27:485–492
Wilson JL, Johnson LAS (1989) Casuarinaceae. Flora of Australia Hamamelidales to Casuarinales. Australian Government Publishing Service, Canberra, pp 100–203
Yang S, Tu Z, Cheung F, Xu W, Lamb J, Jung H, Vance C, Gronwald J, Gronwald JW (2011) Using RNA-Seq for gene identification, polymorphism detection and transcript profiling in two alfalfa genotypes with divergent cell wall composition in stems. BMC Genom 12:1–19
Yasodha R, Kathirvel M, Sumathi R, Gurumurthi K, Sunil Archak, Nagaraju J (2004) Genetic analyses of Casuarinas using ISSR and FISSR markers. Genetica 122:161–172
Yasodha R, Ghosh M, Sumathi R, Gurumurthi K (2005) Cross-species amplification of Eucalyptus SSR markers in Casuarinaceae. Acta Bot Croat 64(1):115–120
Yasodha R, Sumathi R, Modhumita G, Gurumurthi K (2009) Identification of simple sequence repeats in Casuarina equisetifolia. ICFAI Univ J Genet Evol II:46–55
Yeh FC, Yang R, Boyle TJ, Ye Z, Xiyan JM (2000) PopGene32, Microsoft Windows-based freeware for population genetic analysis, version 1.32. Molecular Biology and Biotechnology Centre, University of Alberta, Edmonton
Zhang M, Mao W, Zhang G, Wu F (2014) Development and characterization of polymorphic EST-SSR and genomic SSR markers for tibetan annual wild barley. PLoS ONE 9:e94881
Zhang Z, Zhang H, Mo C, Zhang L (2015) Transcriptome sequencing analysis and development of EST-SSR markers for Pinus koraiensis. Sci Silv Sin 51:114–120 (in Chinese)
Zhong CL, Zhang Y, Chen Y, Jiang QB, Chen Z, Liang JF, Pinyopusarerk K, Franche C, Bogusz D (2010) Casuarina research and applications in China. Symbiosis 50:107–114
Acknowledgements
The authors gratefully acknowledge funding provided by the Zhejiang Science and Technology Major Program on Agricultural New Variety Breeding (Grant no. 2016C02056-9).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by J. Carlson.
Rights and permissions
About this article
Cite this article
Li, N., Zheng, YQ., Ding, HM. et al. Development and validation of SSR markers based on transcriptome sequencing of Casuarina equisetifolia . Trees 32, 41–49 (2018). https://doi.org/10.1007/s00468-017-1607-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-017-1607-6