Abstract
We have constructed a linkage map of bunching onion (Allium fistulosum L., 2n = 16) using an F2 population of 225 plants. The map consists of 17 linkage groups with 212 bunching onion SSR markers and 42 bulb onion (A. cepa L.) SSR, InDel, CAPS or dCAPS markers, covering 2,069 cM. This is the first report of a linkage map mainly based on SSR markers in the genus Allium. With the 103 anchor markers [81 bunching onion SSRs, 11 bulb onion SSRs and 11 bulb onion non-SSRs (1 InDel, 9 CAPSs and 1 dCAPS)] whose chromosome assignments were identified in A. cepa and/or A. fistulosum, via the use of several kinds of Allium alien addition lines, 16 of the 17 linkage groups were connected to the 8 basic chromosomes of A. cepa.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bunching onion (Allium fistulosum L.), also called Japanese bunching onion or Welsh onion, is thought to originate from northwestern China, and is mainly cultivated in East Asian countries, particularly in Japan, China and Korea (Kumazawa and Katsumata 1965; Ford-Lloyd and Armstrong 1993). In Japan, bunching onion accounts for the fourth highest annual output of vegetables, following tomato, strawberry and cucumber (MAFF 2005).
The main breeding objectives for bunching onion are disease resistance, high yield, late bolting, high qualities (e.g., low pungency, high sugar content) and suitability for mechanized farming (vigorous seedling growth, etc.). These traits are also important in bulb onion (A. cepa L.) breeding. Quantitative trait loci (QTL) analysis based on a genetic linkage map would be efficient for revealing the mode of inheritance of these traits. However, despite the economic importance of bunching onion and bulb onion, its genetic characteristics are poorly studied and molecular-based approaches need to be utilized to clarify such characteristics. By revealing these genetic characteristics, it will be possible to utilize comparative genomics approaches between bunching onion and bulb onion, and molecular breeding for both crops will be conducted.
Simple sequence repeat (SSR) markers are ideal DNA markers owing to their simplicity, reproducibility and codominant inheritance (Jones et al. 1997). In the genus Allium, Fischer and Bachmann (2000) first reported the development of SSR markers from a bulb onion genomic library. From a large-scale sequencing of bulb onion expressed sequence tags (ESTs), Kuhl et al. (2004) detected 336 SSR cores among 313 EST sequences and developed 88 EST-SSR markers. In addition, Martin et al. (2005) located 35 EST-SSRs, 43 single nucleotide polymorphisms (SNPs) and 4 insertion-deletion (InDel) markers on medium-density linkage map in bulb onion. This map was simultaneously anchored to bulb onion chromosomes by use of A. fistulosum- A. cepa monosomic addition lines developed by Shigyo et al. (1996) (Martin et al. 2005). Furthermore, we isolated 1,940 SSR clones from a genomic library and 10 SSR-enriched DNA libraries of bunching onion (Wako et al. 2002a; Song et al. 2004; Tsukazaki et al. 2007).
In the present study, we screened the above-mentioned markers for polymorphisms in a bunching onion F2 population, developed additional informative markers, and constructed a linkage map of bunching onion mainly based on codominant SSR markers from bunching onion and bulb onion. To assign the detected linkage groups to the 8 basic chromosomes of A. cepa, we directly identified the chromosomes to which some anchor markers should belong to using Allium alien chromosome addition lines.
Materials and Methods
Plant materials for linkage analysis
A total of 225 individuals of the F2 progeny derived from a cross between 2 bunching onion inbred lines, D1s-15s-10s and J1s-14s-23s, were used as the mapping population. D1s-15s-10s was an S3 line from ‘Saiko’, an open-pollinated cultivar of the Senju group, and J1s-14s-23s was an S3 line from ‘Kujo Futo’, an open-pollinated cultivar of the Kujo group. The F2 mapping population was the selfed progeny of a single F1 plant between these S3 lines. Total DNA was extracted from each plant according to the method described by Song et al. (2004).
SSR markers
We screened 266 primer sets previously developed from bunching onion genomic SSR clones (Wako et al. 2002a; Song et al. 2004; Ohara et al. 2005; Tsukazaki et al. 2006, 2007), 1,456 new ones, and 30 from bulb onion (Fischer and Bachmann 2000) for markers showing polymorphisms between the parental lines.
As for bulb onion EST-derived SSR markers, we screened 11,008 bulb onion EST sequences from the Dana-Farber Cancer Institute (DFCI) A. cepa Gene Index v.1.0 (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=onion) to identify SSRs and design polymerase chain reaction (PCR) primer sequence sets for the detected SSR cores with ‘read2Marker’ (Fukuoka et al. 2005), and obtained 99 effective primer sequence sets for them. These markers were named as Allium cepa EST (ACE) and screened for polymorphisms between the parental lines. We also screened 118 other bulb onion EST-derived SSR markers (ACM) reported by Kuhl et al. (2004) and Martin et al. (2005).
PCR condition was the same as that for the cleaved amplified polymorphic sequence (CAPS) analysis by Ohara et al. (2005). PCR products were separated on 3% (w/v) agarose gel or 5% (w/v) denatured polyacrylamide gel according to the method of Song et al. (2004) or Ohara et al. (2005). In addition, some forward primers were fluorescent-labeled with 6-FAM, NED, PET or VIC dyes (Applied Biosystems, CA, USA) prior to use for PCR. PCR products were loaded on a capillary DNA sequencer (ABI3730; Applied Biosystems), and analyzed using GeneMapper ver. 3.0 software (Applied Biosystems).
InDel and SNP markers
Previously, we developed 39 EST-derived sequence-tagged site (STS) markers of bulb onion named as Allium cepa CAPS (ACC) (Wako et al. 2002b). In addition, 208 EST-derived bulb onion STS markers reported by McCallum et al. (2001), Kuhl et al. (2004), and Martin et al. (2005) were used. An STS marker, AOB272 (Gökçe et al. 2002), reported to be closely linked to the bulb onion nuclear male-fertility restoration locus (Ms), was also used for our mapping. Moreover, we developed 93 intron spanning EST-derived markers for bunching onion from rice genome sequences. Rice cDNA sequences and their annotation data were obtained from The Institute for Genomic Research Rice Genome Pseudomolecules ver. 4.0 (ftp://ftp.tigr.org/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_4.0/). We screened the bulb onion EST sequences from DFCI database for those homologous to the rice cDNA sequences (first hit score > 80) considered unique in the rice genome (the ratio of first to second hit E-values was higher than 1 × 1010) by conducting a BLASTN search. The selected bulb onion ESTs were aligned to the rice genome sequences, and primer pairs were designed from highly conserved segments interposing rice intron regions between them so that they could amplify putative intron regions during PCR with the genomic DNA template of either bulb onion or bunching onion.
In total, 341 bulb onion EST-derived marker candidates were screened for InDels and SNPs between the bunching onion parental lines D1s-15s-10s and J1s-14s-23s.
Primer sets producing single amplicons were selected and the fragment sizes were compared between the parental lines on 2% (w/v) agarose gel. When distinct allele differences (>0.1 kbp in fragment size) were identified, the primers were directly applied to F2 individuals. For identification of SNPs and small InDels, each PCR product was purified using the Wizard CV Gel and PCR clean-up system (Promega, WI, USA), and sequenced with the DNA sequencer ABI3730 (Applied Biosystems). Sequence alignment and SNP/InDel detection were conducted using ATGC ver. 3 software (Genetyx, Tokyo, Japan). The SNPs detected at restriction sites were converted to CAPS markers using GENETYX-WIN ver. 5.1 software (Genetyx). Some other SNPs were converted to the derived cleaved amplified polymorphic sequence (dCAPS) markers using dCAPS Finder 2.0 software (Neff et al. 2002) followed by Primer 3 (Rozen and Skaletsky 2000). When InDels of more than 7 bp were detected, primer sets were redesigned with Primer 3 to amplify smaller fragments with clearer banding differences.
Segregation scoring (or observation) and linkage analyses
Markers showing polymorphisms between the parental lines were applied to F2 individuals. Some markers did not segregate in a codominant manner probably due to the preferential annealing of primers or small differences in the size of restriction enzyme-digested PCR fragments, and these were treated as dominant markers. Linkage analysis was performed using the computer program MAPMAKER/EXP 3.0b (Lander et al. 1987). The Kosambi function was used to obtain cM values (Kosambi 1944). A log of odds threshold of 6.0 was used to group and order the markers.
Chromosomal assignment of linkage groups
To determine the chromosome identity of the detected bunching onion linkage groups, we used a complete set of bunching onion–shallot (the A. cepa Aggregatum group) monosomic addition lines (2n = 17; FF + 1C to +8C, where F stands for the basic chromosome set of A. fistulosum, and 1C and 8C the first and eighth chromosomes, respectively, of A. cepa) developed by Shigyo et al. (1996; note that Shigyo et al. (1996) used the symbol “A” instead of “C” for shallot chromosomes). The chromosomal locations of 30 bunching onion SSRs were directly determined using these monosomic addition lines.
We used the bulb onion chromosome map constructed by Martin et al. (2005) as a reference. Their map consisted of 222 bulb onion markers (RFLPs, SSRs, SNPs and InDels), with the linkage groups assigned to the 8 basic chromosomes of A. cepa using Shigyo et al.’s monosomic addition lines described above. We directly compared our bunching onion map with the bulb onion reference map with respect to the 13 bulb onion markers located on both maps. Some other bulb onion-derived markers were assigned to these monosomic addition lines.
On the other hand, we applied 141 markers (139 bunching onion- and 2 bulb onion-derived SSR markers) to hypo-allotriploids in which one of the alien chromosomes are missing from the A. fistulosum complement in the diploid background of shallot (2n = 23; CCF-1F, CCF-4F, CCF-6F, CCF-7F and CCF-8F, where C and F stand for the basic chromosome sets of A. cepa and A. fistulosum, respectively, and 1F, for example, stands for the first chromosome of A. fistulosum) developed by Hang et al. (2004) and Yaguchi et al. (2008). According to a suggestion in their previous report, these hypo-allotriploids are designated hereafter as single-alien deletions in this report.
Results
SSR markers
Of 1,722 bunching onion genomic SSR primer sets, 1,406 (81.6%) appeared to amplify single loci, and 398 of them detected polymorphisms between the parental lines (Table 1). The efficiency of polymorphism detection was 23.1% by the primer sets designed and 28.3% by the STS markers amplifiable with the primer sets. On the other hand, STS frequency was low (43.3%) for bulb onion genomic SSRs in bunching onion (Table 1). More than 50% of these primers could not amplify bunching onion DNA fragments, or produced multiple bands. In contrast, 75.1% of bulb onion EST-derived SSR markers were clearly amplified in bunching onion. The frequency of polymorphic markers in bulb onion EST-SSRs (11.0%) was comparable to that in bulb onion genomic SSRs (15.4%).
We thus obtained 398 bunching onion SSR markers and 20 bulb onion SSR markers (18 EST-derived and 2 genomic SSR markers) showing polymorphisms between the bunching onion parental lines, and applied 259 of the bunching onion SSR markers and all of the bulb onion ones to our F2 population (Table 1).
InDel and SNP markers
From 11,008 bulb onion EST sequences, 1,090 were selected as highly homologous to rice unigene cDNA sequences. Of them, 19 were already used for developing EST markers for bulb onion by Kuhl et al. (2004) and Martin et al. (2005). From the remaining 1,071 sequences, we designed 93 new primer sets for intron spanning EST markers for bunching onion (Table 2). These 93 original marker candidates, together with 248 previously reported ones, were examined for amplifiability in bunching onion.
From a total of 341 primer sets, 253 (74.2%) appeared to amplify single loci in bunching onion (Table 2). So far, we have sequenced 195 of the amplified loci in each parent and found 3 informative markers (ACAAJ79, ACAAX07 and TC1952) with large (>0.1 kbp) InDels between the parents (Fig. 1a). These InDels were detected in intron regions (Fig. 1b).
DNA polymorphisms of bulb onion EST-derived markers within F2 individuals of bunching onion. M 100 bp ladder; D D1s-15s-10s; J J1s-14s-23s. a A large InDel marker ACAAJ79. b DNA sequence alignment of a part of the ACAAJ79 locus; PCR products from the bunching onion parental lines, D1s-15s-10s (ACAAJ79_D) and J1s-14s-23s (ACAAJ79_J), and the bulb onion EST sequence (CF434863) which was submitted to GenBank for bulb onion cDNA clone of ACAAJ79. A large deletion (243 bp) was detected in a putative intron region in the parental line ‘J’
Sequence comparison of the 192 other loci between the parental lines detected 55 polymorphic loci: 13 small InDels (1–11 bp) at 10 loci and 134 SNPs at 54 loci, with 9 loci containing both types of polymorphisms. By including the large InDels described above, the frequency of polymorphic markers was 29.7% [(3 + 55)/195 loci]. As for the SNPs detected, most (95 SNPs, 70.9%) of them were due to transition. The frequency of SNP detection was estimated at 1.5 per 1 kbp sequence.
For a small InDel marker, TC0678 (containing 8 bp InDel), primer sets were redesigned to differentiate the parental alleles clearly (data not shown). Of the 54-containing loci, 17 were directly converted to CAPS markers, and the other 4 loci (ACAEK46, ACAER72, CF439394 and TC2026) were successfully converted to dCAPS markers (data not shown).
We thus obtained 25 polymorphic InDel, CAPS or dCAPS markers (Table 2). These markers, except for ACAAX07, AOB272 and TC1891, segregated codominantly in the F2 population.
Linkage map of bunching onion
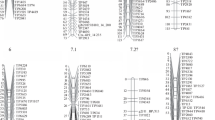
The genetic map we constructed for the F2 population consists of 17 linkage groups with 229 SSRs (212 bunching onion SSRs and 17 bulb onion SSRs) and 25 bulb onion-derived non-SSR markers (4 InDels, 17 CAPSs and 4 dCAPSs) covering 2,069 cM (Fig. 2). Of the markers located on this map, 151 bunching onion SSRs, 11 bulb onion SSRs and 5 bulb onion-derived non-SSRs were newly developed in this study (Tables 1, 2; Suppl. Tables 1, 2, 3). The other polymorphic markers were not mapped mainly due to distorted segregation or absence of significant linkages with other markers. The average marker distance was 8.1 cM. Ten linkage groups in our map corresponded to the 9 groups of a previously reported linkage map of bunching onion (Ohara et al. 2005) with 11 SSR and 2 CAPS markers in common (Fig. 2).
Bunching onion chromosome map consisting of 17 linkage groups with 229 SSRs (212 bunching onion SSRs and 17 bulb onion SSRs) and 26 bulb onion-derived non-SSR markers (4 InDels, 17 CAPSs and 4 dCAPSs) covering 2,069 cM. The genetic markers used to assign linkage groups to their relevant chromosomes are marked with [1C] to [8C] (assigned to A. cepa chromosomes) and [1F] to [8F] (to A. fistulosum chromosomes). Markers located on a previously reported bunching onion linkage map (Ohara et al. 2005) are indicated in italic with underlines
Assignment of bunching onion linkage groups to their corresponding A. cepa chromosomes
Thirty of the 212 bunching onion SSR markers located on our bunching onion map were examined for their chromosomal location using Shigyo et al.’s complete set of bunching onion–shallot monosomic addition lines. Of them, only 8 (AFAA02D08, AFB20G05, AFS015, AFS039, AFS104, AFS145, AFS149 and AFRA11F11) could be assigned to corresponding A. cepa chromosomes (Table 3). The others could not be assigned due to their poor amplifiability in shallot, monomorphism between bunching onion and shallot, or inconsistent polymorphisms (probably caused by some residual heterozygousity in the shallot and/or bunching onion lines used for developing the monosomic addition lines).
A total of 42 bulb onion-derived markers (17 SSRs, 4 InDels, 17 CAPSs and 4 dCAPSs) were mapped on 13 of the 17 linkage groups in our bunching onion map (Fig. 2). Of them, 13 markers were already mapped on a A. cepa chromosome map by Martin et al. (2005). We independently identified relevant A. cepa chromosomes for 3 of these markers, that is, ACC013, ACC033 (named as API40 and AJB64, respectively, in Martin et al.’s bulb onion map) (Wako et al. 2002b) and ACADR60 (in this study).
Of the 29 other bulb onion markers, we previously identified relevant A. cepa chromosomes for 4 markers (ACC008, ACC015, ACC043 and AMS14) (Wako et al. 2002b, Masuzaki et al. 2006). In the present study, we identified relevant A. cepa chromosomes for 3 other markers (ACE039, ACE044 and TC1010; Table 3). Besides these newly assigned markers, ACADR60, which Martin et al. (2005) assigned to the 6C chromosome, was assigned to 2C on the basis of our data (Fig. 3). Five markers could not be assigned to relevant A. cepa chromosomes due to monomorphism between bunching onion and shallot. The remaining 17 bulb onion-derived markers on our bunching onion map have not, so far, been examined for relevant chromosomes.
Twelve linkage groups in our bunching onion map were connected to 7 A. cepa chromosomes (Chrs. 1C–6C and 8C) with 8 bunching onion-derived SSR markers and 20 bulb onion-derived markers (1 genomic SSR, 8 EST-SSRs, 1 InDel, 9 CAPSs and 1 dCAPS) (Table 3).
Assignment of bunching onion linkage groups to their relevant chromosomes
Of the 212 bunching onion SSR markers on our map, 139 were examined for their chromosome assignment using the allotriploid-bunching onion single-alien deletion lines. Seventy markers were not amplified in shallot, and 39 of them were assigned to A. fistulosum chromosomes on the basis of the absence of bunching onion-specific bands in one of the deletion lines. The other 69 markers were amplifiable in shallot. Of these, 55 were polymorphic between bunching onion and shallot, and 38 of them were assigned to A. fistulosum chromosomes (Fig. 4a, b). In addition, the relevant bunching onion chromosomes for 2 bulb onion EST-derived SSR markers (ACM096 and ACE113) were determined with the deletion lines.
Eleven bunching onion linkage groups were assigned to 5 chromosomes (Chrs. 1F, 4F, 6F, 7F and 8F) with 77 bunching onion- and 2 bulb onion-derived SSR markers (Table 4).
Discussion
In our previous study, we constructed a low-density bunching onion linkage map covering 947 cM mainly based on amplified fragment length polymorphism markers (Ohara et al. 2005). In the present study, we constructed a new bunching onion linkage map covering 2,069 cM (Fig. 2). This is the first report of linkage map mainly based on SSR markers in the genus Allium. Compared with previously reported bulb onion maps (694 cM, van Heusden et al. 2000; 1,907 cM, Martin et al. 2005), our map is thought to be a standard map in bunching onion. This linkage map will be useful for genetic studies such as QTL analysis of agronomic traits in bunching onion.
Here, we also demonstrated that many bulb onion-derived markers were useful for mapping in bunching onion (Tables 1, 2). Among bulb onion EST-derived markers, the frequency of informative (i.e., polymorphic between the parental lines of bunching onion) markers were higher in those with InDels and/or SNPs (29.2%) than in those with SSRs (11.0%) (Tables 1, 2). We developed or identified 75 polymorphic markers (18 SSRs and 57 InDels and/or SNPs) from 558 bulb onion EST-derived primer sets, and 42 of them were located on the bunching onion map (Fig. 2).
From 11,008 bulb onion EST sequences (Kuhl et al. 2004), we found 1,090 sequences highly homologous to rice unigene cDNA sequences, and designed 93 primer sets for intron spanning EST-derived markers for bunching onion. We did not design primer sets for the other sequences mainly due to the large sizes (>1 kb) of introns in corresponding rice gene sequences and substantial nucleotide differences at intron-flanking sequences between bulb onion and rice. Kuhl et al. (2004) reported a lower average similarity at intron regions (39%) between bulb onion and rice than at cording (78%). We found no correlation (r = 0.07) between the PCR amplicon sizes of intron-spanning markers in bunching onion and the known sizes of corresponding rice genomic regions (data not shown). In addition, Kuhl et al. (2004) reported that the average GC content at the third codon position was very different between bulb onion (40.9%) and rice (61.8%). In this regard, it will be possible to design many additional primer sets for EST-markers for bunching onion by neglecting synonymous third codon differences between bulb onion and rice. Therefore, many more bulb onion EST-derived markers may subsequently be added to the bunching onion map.
Forty-two bulb onion-derived markers, including 1 genomic SSR, were located on 13 of the 17 linkage groups in our bunching onion map (Fig. 2). In the bulb onion map reported by Martin et al. (2005), AOB272 was located on Chr. 2C and 4 other markers (ACADR60, ACM024, ACM066 and ACM177) were located on Chr. 6C. Masuzaki et al. (2006) directly identified the relevant A. cepa chromosome (2C) for AMS14. In our bunching onion map, these markers were located on either of the linkage groups of LG2a [ACADR60, ACM024, ACM177 (ACE111 in our map) and AMS14] or LG2b [ACM066 (ACE101 in our map)]. Of them, 3 markers (ACE101, ACE111 and ACM024) were monomorphic between bunching onion and shallot (data not shown). In the present study, we directly identified the relevant A. cepa chromosome for ACADR60 and ACC008, which was tightly linked to ACE101, using Shigyo et al.’s shallot monosomic addition lines (Fig. 3). In addition, ACM024 located on LG2a and ACE111 on LG2b in our bunching onion map were linked to AOB272 in another bulb onion population (J. McCallum, personal communication). Therefore, we conclude that LGs 2a and 2b in our bunching onion map correspond to A. cepa chromosome 2 (Table 2).
Bunching onion and bulb onion belong to the section Cepa and have the same chromosome number (2n = 16), although they were classified into different alliances—Altaicum alliance and Cepa alliance (Fritsch and Friesen 2002), respectively. By using the monosomic additions and/or single-alien deletions, the direct comparisons of the relevant chromosomes for 103 markers consistently assigned 16 of the 17 linkage groups in our A. fistulosum map to the 8 basic chromosomes of A. cepa; 28 markers were assigned to A. cepa chromosomes, and 79 to A. fistulosum chromosomes, with 4 bunching onion SSR markers (AFAA02D08, AFS015, AFS039 and AFS149) assigned to both A. cepa and A. fistulosum chromosomes (Fig. 2; Tables 3, 4). This newly developed set of anchor markers comprises 81 bunching onion SSRs, 11 bulb onion SSRs and 11 bulb onion non-SSRs (1 InDel, 9 CAPSs and 1 dCAPS). We therefore conclude that a very high level of macrosynteny exists between these two species.
The remaining linkage group, temporarily named as LG X, could not be assigned to any chromosome. We consider two possible explanations for this: (1) the incompleteness of the set of allotriploid-bunching onion single-alien deletion lines suggests that LG X might belong to any of the 2F, 3F or 5F chromosome; (2) all the markers located on LG X were bunching onion-derived SSR markers, which might have resulted in the exclusive amplification of bunching onion genome regions. In another bunching onion F2 population, some markers located on bunching onion LG X were loosely linked to ACE010 (ACM133 in Martin et al. 2005, assigned to A. cepa Chr. 5) (unpublished data). The relevant chromosome of LG X would be identified by mapping new bulb onion-derived markers and/or by integrating different bunching onion linkage maps with common markers.
As compared with bulb onion, bunching onion is suitable for genetic studies because this crop is annual and has a 28% smaller genome size (Ricroch et al. 2005). Genome synteny is, as mentioned above, highly conserved between bunching onion and bulb onion. However, it was reported that a minimum of two heteromorphic bivalents were observed in meiotic analysis of interspecific hybrids between bunching onion and bulb onion (Peffley 1986), suggesting that chromosomal mutations such as translocation and inversion might be involved in the speciation between A. fistulosum and A. cepa. For detailed studies of such chromosomal mutations, microsynteny should be evaluated at the genome-wide level by plotting many more common markers onto the maps of both species.
References
Fischer D, Bachmann K (2000) Onion microsatellites for germplasm analysis and their use in assessing intra- and interspecific relatedness within the subgenus Rhizirideum. Theor Appl Genet 101:153–164
Ford-Lloyd BV, Armstrong SJ (1993) Welsh onion Allium fistulosum L. In: Kalloo G, Bergh BO (eds) Genetic improvement of vegetable crops, Pergamon Press, pp 51–58
Fritsch RM, Friesen N (2002) Evolution, domestication and taxonomy. In: Rabinowitch HD, Currah L (eds) Allium crop science: Recent advances, CABI Publishing, pp 5–30
Fukuoka H, Nunome T, Minamiyama Y, Kono I, Namiki N, Kojima A (2005) read2Marker: a data processing tool for microsatellite marker development from a large data set. BioTechniques 39:472–476
Gökçe AF, McCallum J, Sato Y, Havey MJ (2002) Molecular tagging of the Ms locus in onion. J Amer Soc Hort Sci 127:576–582
Hang TTM, Shigyo M, Yamauchi N, Tashiro Y (2004) Production and characterization of alien chromosome additions in shallot (Allium cepa L. Aggregatum group) carrying extra chromosome(s) of Japanese bunching onion (A. fistulosum L.). Genes Genet Syst 79:263–269
Jones CJ, Edwards KJ, Castaglione S, Winfield MO, Sala F, van de Wiel C, Bredemeijer G, Vosman B, Matthes M, Daly A, Brettschneider R, Bettini P, Buiatti M, Maestri E, Malcevschi A, Marmiroli N, Aert R, Volckaert G, Rueda J, Linacero R, Vazquez A, Karp A (1997) Reproducibility testing of RAPD, AFLP and SSR markers in plants by a network of European laboratories. Mol Breed 3:381–390
King JJ, Bradeen JM, Bark O, McCallum JA, Havey MJ (1998) A low-density map of onion reveals a role for tandem duplication in the evolution of an extremely large diploid genome. Theor Appl Genet 96:52–62
Kosambi D (1944) The estimation of map distance from recombination values. Ann Eugen 12:172–175
Kuhl JC, Cheung F, Yuan Q, Martin W, Zewdie Y, MaCallum J, Catanach A, Rutherford P, Sink KC, Jenderk M, Prince JP, Town CD, Havey MJ (2004) A unique set of 11,008 onion expressed sequence tags reveals expressed sequence and genomic differences between the monocot orders Asparagales and Poales. Plant Cell 16:114–125
Kumazawa S, Katsumata H (1965) Negi (Japanese bunching onion). In: Kumazawa S (ed) Sosai-engei kakuron (vegetable crops), Yokendo Press, Tokyo, pp 280–289 (in Japanese)
Lander E, Green P, Abrahamson J, Barlow A, Daly M, Lincoln S, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
MAFF (2005) Nousanbutsu-Sanshutsugaku no Jun-I (The ranking of vegetables for annual output in Japan). In: Pocket nourinsuisan toukei, 2005 (in Japanese). http://www.tdb.maff.go.jp/toukei/toukei
Martin WJ, McCallum J, Shigyo M, Jakše J, Kuhl JC, Yamane N, Pither-Joyce M, Gökçe AF, Sink KC, Town CD, Havey MJ (2005) Genetic mapping of expressed sequences in onion and in silico comparisons with rice show scant colinearity. Mol Gen Genomics 274:197–204
Masuzaki S, Araki N, Yamane N, Yamauchi N, Wako T, Kojima A, Shigyo M (2006) Chromosomal locations of microsatellites in onion. Hort Sci 41:315–318
McCallum J, Leite D, Pither-Joyce M, Havey MJ (2001) Expressed sequence markers for genetic analysis of bulb onion (Allium cepa L.). Theor Appl Genet 103:979–991
Neff MM, Turk E, Kalishman M (2002) Web-based primer design for single nucleotide polymorphism analysis. Trends Genet 18:613–615
Ohara T, Song YS, Tsukazaki H, Wako T, Nunome T, Kojima A (2005) Construction of a genetic linkage map of Japanese bunching onion (Allium fistulosum) based on AFLP and SSR markers. Euphytica 144:255–263
Peffley EB (1986) Evidence for chromosomal differentiation of Allium fistulosum and A. cepa. J Amer Soc Hort Sci 111:126–129
Ricroch A, Yockteng R, Brown SC, Nadot S (2005) Evolution of genome size across some cultivated Allium species. Genome 48:511–520
Rozen S, Skaletsky HJ (2000) Primer 3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, pp 365–386
Shigyo M, Tashiro Y, Issiki S, Miyazaki S (1996) Establishments of a series of alien monosomic addition lines of Japanese bunching onion (Allium fistulosum L.) with extra chromosomes from shallot (A. cepa L. Aggregatum group). Genes Genet Syst 71:363–371
Song YS, Suwabe K, Wako T, Ohara T, Nunome T, Kojima A (2004) Development of microsatellite markers in Japanese bunching onion (Allium fistulosum L.). Breed Sci 54:361–365
Tsukazaki H, Fukuoka H, Song YS, Yamashita K, Wako T, Kojima A (2006) Considerable heterogeneity in commercial F1 varieties of bunching onion (Allium fistulosum) and the proposal of a breeding scheme for conferring genetic traceability using SSR markers. Breed Sci 56:321–326
Tsukazaki H, Nunome T, Fukuoka H, Kanamori H, Kono I, Yamashita K, Wako T, Kojima A (2007) Isolation of 1,796 SSR clones from SSR-enriched DNA libraries of bunching onion (Allium fistulosum). Euphytica 157:83–94
Wako T, Ohara T, Song YS, Kojima A (2002a) Development of SSR markers in bunching onion. Breed Sci 4(suppl 1):83 (in Japanese)
Wako T, Shigyo M, Tashiro Y, Kataoka S, Ohara T, Kojima A (2002b) Development of chromosome-specific CAPS markers in shallot. J Jpn Soc Hort Sci 71(suppl 1):348 (in Japanese)
Yaguchi S, Hang TTM, Tsukazaki H, Vu QH, Masuzaki S, Wako T, Masamura N, Onodera S, Shiomi N, Yamauchi N, Shigyo M (2008) Molecular and biochemical identification of alien chromosome additions in shallot (Allium cepa L. Aggregatum group) carrying extra chromosome(s) of bunching onion (A. fistulosum L.). Genes Genet Syst (submitted)
Acknowledgments
We are grateful to Ms. K. Tanaka, K. Wakita and S. Negoro for the technical assistance. This work was supported by the project “Green Technology Project (DM-1604)”, Ministry of Agriculture, Forestry and Fisheries, Japan and Grant-in-Aid for Young Scientists (B) (19780010), Japan Society for the Promotion of Science. The authors wish to thank Ms. N. Matsubara, Y. Kousabara and M. Anraku (former students in Yamaguchi University) for their contributions to this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Xu.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tsukazaki, H., Yamashita, Ki., Yaguchi, S. et al. Construction of SSR-based chromosome map in bunching onion (Allium fistulosum). Theor Appl Genet 117, 1213–1223 (2008). https://doi.org/10.1007/s00122-008-0849-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-008-0849-5