Abstract
Rapamycin is a high-value product finding immense use as a drug, in organ transplantation, and as a potential immunosuppressant. Optimization of fermentation parameters of rapamycin production by Streptomyces hygroscopicus NRRL 5491 has been carried out. The low titer value of rapamycin in the original producer strain limits its applicability at industrial level. This study aims at improving the production of rapamycin by optimizing the nutrient requirements. Addition of l-lysine increased the production of rapamycin up to a significant level which supports the fact that it acts as precursor for rapamycin production, as found in previous studies. Effect of optimized medium on the Streptomyces growth rate as well as rapamycin production has been studied. The optimization study incorporates one at a time parameter optimization studies followed by tool-based hybrid methodology. This methodology includes the Plackett–Burman design (PBD) method, artificial neural networks (ANN), and genetic algorithms (GA). PBD screened mannose, soyabean meal, and l-lysine concentrations as significant factors for rapamycin production. ANN was used to construct rapamycin production model. This strategy has led to a significant increase of rapamycin production up to 320.89 mg/L at GA optimized concentrations of 25.47, 15.39, and 17.48 g/L for mannose, soyabean meal, and l-lysine, respectively. The present study must find its application in scale-up study for industrial level production of rapamycin.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rapamycin (also called sirolimus), an immunosuppressant, is a peptide that was isolated from actinomycetes strain Streptomyces hygroscopicus [1, 2]. Rapamycin showed a good activity against mammary, colon, and brain tumor model systems [3]. Rapamycin acts via a mechanism that is completely different from that of cyclosporine A, and it has an extraordinary advantage of having greater activity which is 150 times as that of cyclosporine A with lower toxicity [4]. Rapamycin has shown inhibitory effect on vascular smooth muscle for the treatment of coronary artery disease [5]. Nephrotoxicity has long been the vulnerable point of treatment with calcineurin inhibitors (CNIs). It has been reported that switching to rapamycin is a feasible method of treating patients with calcineurin inhibitor toxicity [6]. Up to 40 % of renal allografts have showed delayed graft function due to slowly recovering acute tubular necrosis [7].
Rapamycin binds to the same cytosolic receptor as tacrolimus, namely the immunophilin, FK-binding protein-12 [8]. This complex then binds to the mammalian target of rapamycin (mTOR)—a key regulatory kinase. Disruption of mTOR interferes with cytokine signaling, which results in the inhibition of T-cell growth and differentiation.
Natural products such as isoprenoids, flavonoids or polyketides represent structurally complex compounds which are made in nature, but require long and elaborate synthesis routes when classical chemical methods are employed. As the structural complexity makes the chemical synthesis of natural products quite difficult, fermentation is considered to be an economically feasible alternative to produce pharmaceutically useful compounds for commercial purposes [9]. Streptomyces sp. and the related filamentous bacteria produce macrocyclic polyketides, by the action of so-called type I modular polyketide synthases. It is multienzyme in which different sets (modules) of enzymatic activities catalyze each successive round of elongation [10, 11].
Rapamycin is an atypical triene macrolide containing nitrogen, and the immediate precursor of the nitrogen-containing ring has been shown to be pipecolic acid [12]. l-Pipecolate is a cyclization product of l-lysine so addition of l-lysine had shown to increase the production of rapamycin [13].
Different strains of S. hygroscopicus have different abilities for rapamycin production, which are compared in Table 1. The composition of the culture medium, closely connected with the metabolic capacities of the producing organism, greatly influences the biosynthesis of antibiotics [22, 23]. The fermentation process needs to be optimized to obtain maximum production of rapamycin. Parameter selections are based on early studies [15]. The production parameters are first optimized by varying one factor at a time, while keeping the other constituents of the medium constant. The parameters include carbon sources, nitrogen sources, agitation rate, and precursor concentration. This one factor at a time does not consider the effect due to interactive behavior of all the significant factors since other factors are maintained arbitrarily at a constant level. Therefore, the development of a multivariate non-linear process model for optimization has been considered as a superior alternative. For this approach, a hybrid of Plackett–Burman (PB) design, artificial neural networks (ANN), and genetic algorithms (GA) methodologies has been used. The PB design is used to select the significant parameters for the process [24]. For each parameter, two levels are considered, namely, “high” and “low”. Dummy variables are incorporated which give the idea of experimental error of an effect. In the PB design, various combinations of high and low values of the process parameters are used for performing the experiments and the results of these experiments are analyzed to determine the significance of process parameters [25].
The ANN tool is used to construct process model using significant parameters as inputs and rapamycin concentration as output. The ANN paradigm is the multi-layered perceptron (MLP) that approximates non-linear relationships existing between the input and the output of the model [26].
Finally to maximize the rapamycin production, the input space of ANN model has been optimized using GA tool. Multivariable non-linear problems can be solved using GA paradigm [27]. Specifically, GAs are based on the principles of “survival-of-the-fittest” and “random exchange of memory during genetic propagation”, which are followed by the biologically evolving species [28]. Thus, in the present study, rapamycin production process optimization has been done using ANN–GA hybrid methodology. The results obtained through computational techniques were experimentally verified.
Materials and methods
Chemicals
The media ingredients were from HiMedia (Mumbai, India). HPLC grade solvents were from Merck (Mumbai, India). Rapamycin (EmTOR) from Emcure Pharmaceuticals (Pune, India) was used as HPLC standard reference.
Strain
The strain S. hygroscopicus ATCC 29253 was obtained from National Centre for Agricultural Utilization Research (NRRL), USA.
Culture conditions
Streptomyces hygroscopicus was routinely cultivated at 28 °C and 200 rpm in culture broth medium containing (g/L): malt extract 3, glucose 10, and peptone 3 at pH 6.4–6.8. Fermentation was carried out in triplicate in 250-mL unbaffled Erlenmeyer flasks, each containing 50 mL production medium containing (g/L): soyabean meal 20, d(+) mannose 20, potassium dihydrogen phosphate 5, and pH 6.5 and incubated at 25 °C ± 2 for 4 days at 200 rpm [15]. Phosphate is a major factor in the synthesis of a wide range of antibiotics [29]. The data obtained throughout the present investigation were the average mean of triplicate observation.
Optimization of carbon sources for rapamycin production
Carbon source optimization studies were done using different carbon sources in the production medium. S. hygroscopicus growth was carried out in 250-mL unbaffled shake flask culture containing 50 mL of fermentation medium, using the different carbon sources which included fructose, glucose, maltose, mannose, and soluble starch, one at a time and keeping the other contents of the medium constant, at 200 rpm, pH 6.5, and temperature 28 °C. The carbon sources at a concentration of 20 g/L were sterilized and added separately to the production medium. The experiment was performed in triplicates and mean value was taken. Samples were withdrawn on the fourth day of fermentation and analyzed for production of rapamycin.
Optimization of nitrogen sources for rapamycin production
To the production medium containing d(+) mannose, different nitrogen equivalent sources were added, one at a time, as replacement of soyabean meal. The growth of S. hygroscopicus was carried out in 250-mL shake flask taking different nitrogen sources such as soyabean meal, casein, beef extract, and peptone at a concentration of 20 g/L, one at a time keeping other contents of medium constant. The experiment was carried out in triplicate to minimize the error. The batch study was performed to determine the effect of different nitrogen sources on rapamycin production at 28 °C and shaking speed of 200 rpm. Samples were collected after 96 h of fermentation and the production of rapamycin was calculated.
Detection of rapamycin
At the end of fermentation, aliquots of 3 mL were taken where microbial growth was separated by centrifuging at 2,000×g for 5 min and extracted twice by shaking with 3 mL methanol for 30 min. Then the two extracts were pooled to be assayed for rapamycin concentration. Samples were analyzed by high-performance liquid chromatography (HPLC).
The chromatographic system includes HITACHI D-6500 model gradient pump (Japan), a stainless steel injector (5 μL loop), a UV VIS detector (Jasco, Tokyo, Japan). A chromolith RP18 column (Intersil 7 ODS—346 mm i.d., 250 mm, Merck) was used as an analytical column.
Qualitative and quantitative determination was done by HPLC apparatus having C18 column at 272-nm UV detector and 10-μL sample loop injector. A mixture of acetonitrile and methanol (20:80 v/v) was used as mobile phase. The flow rate was maintained at 1.0 mL/min.
Dry cell weight measurement
For the determination of microbial dry cell weight, 5 mL sample of whole fermentation medium was placed into a pre-weighed centrifuge tube and then centrifuged at 5,000×g for 10 min. The microbial residue which remained after decantation was dried at 50 °C for 12 h. The tubes were reweighed to determine the growth yield expressed as gram dry weight per liter fermentation medium.
Computational techniques
Plackett–Burman design was performed using commercial software Minitab 15.1.0.0, USA. ANN-based GA study was done with the help of MATLAB [version 7.6.0.324 (R2008a) Mathworks Inc., MA, USA] program was used.
Screening of significant parameters using Plackett–Burman design
The PB experimental design was applied to screen the significant medium components to maximize the production of rapamycin. A total of four parameters, viz., mannose concentration, soyabean meal concentration, l-lysine concentration, and pH have been considered for the screening experiment. Each variable is represented at two levels, i.e., high (+1) and low (−1). A set of 12 experiments was performed according to PB design to determine the critically important parameters for the production of rapamycin.
If the numbers of runs is higher than the numbers of variables (at this stage, there are 4 variables and 12 trials) the resolution is better than with a saturated design [30]. Four assigned variables and seven unassigned variables (commonly referred as dummy variables) were screened in PB design of 12 experiments. The more dummy factors there are, the better the estimate of such errors, so it is not uncommon for experimenters to use a larger PB design than is strictly necessary, thus getting higher quality information on the significance of each “real” factor [31]. Dummy variable are used to estimate experimental errors in data analysis [32]. Seven dummy variables, whose levels do not change in the design, were introduced to estimate the population standard deviation. Analysis for the PB design was carried out as follows. First, the effect of all variables including dummies was calculated as follows [33]:
where E A is the effect of the variable, and \(\bar{Y}\) is the average value of the repeatedly measured responses for each experiment. The experimental error was determined as the average square of the dummy effects (E d):
where V E is the variance of the effects and n is the total number of dummy variables. The standard error of the effects (SE) was determined as the square root of the variance [33]:
Finally, the significance (p value) of the effect of each variable on rapamycin production was measured by Student’s t test [34]:
The variable which has no effect will give a t value of 0. The larger the absolute value of the t value, the more significant is the variable. The regression model was applied to screen the significant medium components. On the analysis of the regression coefficients of the medium components, mannose concentration, soyabean meal concentration, lysine concentration, and pH have shown positive effect values. The components were screened based on their absolute value of effects (either positive or negative) and p value below 0.05.
The random error variability and test for the statistical significance of the parameter estimates can be determined using the design. The regression coefficient, p value, and confidence level were determined and the variables with confidence level greater than 95 % were considered to be more significant for rapamycin production.
Study of interactive effect of parameters with central composite design (CCD)
The effects of three independent process parameters (mannose, soyabean meal, l-lysine) on rapamycin production were analyzed using a central composite design (CCD). It was performed at five experimental levels: −α, −1, 0, +1, +α. For a three-factor design, consisting of 6 central points, a total of 20 experiments are to be performed (Table 2). The various combinations used in these 20 experiments were used as input data set for training using ANN code written in MATLAB.
Of these 20 experimental data, we have taken 12 as training set, 4 as validation set to validate that the network is generalizing and to stop training before overfitting and the remaining 4 as completely independent test of network generalization. This was done using default function for random data division.
Each experiment was performed in triplicate and the mean value reported. The regression analysis was done using software (Minitab 15) with the numerical data obtained after performing the experiments. The individual as well as the interactive effect of each parameter was determined and the process was modeled using the analysis of variance (ANOVA).
Artificial neural network (ANN)
The neural networks are adjusted, or trained, so that a particular input leads to a specific target output. The ANN works on error-back-propagation method in which input vectors and the corresponding target vectors are used to train a network until it can approximate a function. The basic ANN architecture has three layers, input layer, hidden layer, and output layer. Each of these layers consists of a number of nodes which are linked to subsequent nodes of subsequent layers with weighted connections. Before training the weights are initialized to random values. The reason to initialize weights with small values is to prevent saturation. The newff command automatically initializes the weights. This function takes a network object as input and returns a network object with all weights and biases initialized.
The normalization process for the raw inputs has great effect on preparing the data to be suitable for the training. Without this normalization, training the neural networks would have been very slow. In general, the normalization step is applied to both the input vectors and the target vectors in the data set. In this way, the network output always falls into a normalized range. The network output can then be reverse transformed back into the units of the original target data when the network is put to use in the field. The ‘mapminmax’ function maps the range of input values to the range [0 1], or normalizes the input values. Training is often faster when values are normalized.
The application randomly divides the input vectors and target vectors into three sets. One set for training, second to validate that the network is generalizing and to stop training before over fitting and last as a completely independent test of network generalization. This involves randomly extracting test set, developing a neural network model based on validation set and test set. This is followed by repeating the process with several random divisions of data. The reported results are based on the average performance of all randomly extracted test sets [35]. After training, ANN can predict the output when any input similar to the pattern that it has learnt is fed. ANN tends to inherently match the input vector (i.e., medium composition) to the output vector (rapamycin production).
For training of experimental data generated by CCD with ANN, we used feed forward neural network and, for training the network back propagation, Levenberg–Marquardt algorithm was applied. Tansig and purelin transfer functions have been used for hidden and output layer, respectively, as shown below:For hidden layer [36]:
For output layer [36]:
where a 1 and a 2 are the output of the hidden layer and output layer, respectively. IW1,1 and LW2,1 are the input weight matrix and output weight matrix, whereas b 1 and b 2 are the bias of hidden layer and output layer.
Three input variables were used, which are mannose (14.95–25.04 g/L), soyabean meal (11.59–28.408 g/L), and lysine concentration (1.59–18.40 g/L). The input data were previously normalized and then fed into ANN for training. After training, these data were fed to the fitness evaluation function to get the best solution of all three input variables through GA.
Genetic algorithm
The genetic algorithm is a method for solving both constrained and unconstrained optimization problems that is based on natural selection, the process that drives biological evolution. Once a generation of ANN-based process model with good prediction accuracy is developed, a genetic algorithm can be used to optimize its input space representing process variables, with a view of maximizing the process performance. The genetic algorithm repeatedly modifies a population of individual solutions. At each step, the genetic algorithm selects individuals at random from the current population to be parents and uses them to produce children for the next generation. This is performed by a main loop of operations consisting of: (1) selection of better (fitter) parent chromosomes to create a mating pool, (2) crossover, i.e., the production of offspring solutions by pair-wise crossing-over of the contents between pairs of fitter parent chromosomes, and (3) mutating elements of the offspring strings [37]. Over successive generations, the population “evolves” toward an optimal solution. Solutions are then evaluated to measure their fitness in fulfilling the optimization target [37].
Results and discussions
Optimization using one-factor-at-a-time method
Parameters were first optimized by varying single parameter and keeping the other constituents of medium constant, i.e., using one-factor-at-a-time method. The full fermentative process was carried out for 6 days and it was found that maximum production of rapamycin took place on the fourth day of the fermentation. Hence, for further studies, samples were collected on the fourth day of the production to determine the effect of various factors on the production.
Effect of carbon sources on rapamycin production
Figure 1 shows the effect of various carbon sources on rapamycin production with initial substrate concentration of 20.0 g/L. The production of rapamycin was found to be maximum using d(+) mannose as a carbon source which was 248.7 mg/L. Fructose, as carbon source, was found to produce lower concentration of rapamycin compared to mannose.
Effect of nitrogen sources on rapamycin production
Organic nitrogen is a complex nitrogen source composed of spectrum of peptides, free amino acids, and trace elements. Among all the organic nitrogen sources, soyabean meal, which was present in the medium used for carbon source optimization [15], showed the highest production of 248.71 mg/L (Fig. 2). This may be possibly due to high l-lysine content of soyabean meal which possibly acts as a precursor for biosynthesis of rapamycin [38].
Effect of l-lysine as precursor on rapamycin production
Streptomyces hygroscopicus was grown in 250-mL shake flask containing production medium supplemented with varying concentration of l-lysine as 0, 3, 5, 7, 10, 12, 15 g/L, keeping the other contents of the medium constant. The experiment was performed in triplicates and the batch study was performed at 28 °C, 200 rpm for 4 days. Samples were collected on fourth day to calculate the production of rapamycin. It was observed that addition of l-lysine at concentration of 10 g/L showed considerable increase in the production of rapamycin of 268.87 mg/L. Further increase in l-lysine concentration showed no significant increase in rapamycin production. Thus, 10 g/L was found to yield maximum production of rapamycin. (Table 3) The results are similar to the earlier studies of Cheng et al. [8]. l-lysine stimulation is probably due to its role as precursor of pipecolic acid, which is incorporated into the nitrogen-containing ring of rapamycin [39].
Optimization using statistical techniques
Plackett–Burman experimental design for rapamycin production
The design includes various concentrations of mannose, soyabean meal, l-lysine parameters, and pH. Design of experiments was done to find out the significant factors along with the measured response (rapamycin production).
The Pareto chart (Fig. 3) which shows the ranking of variables according to the absolute values of standardized effect, is important in the design of the experiment for optimization, and is a convenient way to view the results of a PB experimental design. It consists of bars with a length proportional to the absolute value of the estimated effects, divided by the standard error. The effects of variables, which extend past the line, were known to be significant at a particular α.
In this study, the chart includes a vertical line at the critical t value (2.365) for α of 0.50, and the effect (positive or negative) of its bar smaller than the critical t value is considered as not significant and not affecting the response variable. Based on the value of coefficient for significance (p < 0.05), three of the four factors, viz., mannose, soyabean meal, l-lysine concentration, were observed to influence the production of rapamycin very significantly. Based on the results obtained of p value for pH (0.985; p ≫ 0.05), it was found insignificant. Though pH has been found to enhance the biomass, negligible impact of initial pH on rapamycin production has been reported [40]. Further experiments of rapamycin production were carried out by taking the pH value constant at its mid-value (i.e., 6).
The regression model was applied to screen the significant medium components. Using ANOVA, the following first-order regression equation was obtained showing the effect of significant factors on production of rapamycin.
where X 1 is mannose, X 2 soyabean meal, X 3 l-lysine, and Y is the concentration of rapamycin produced (response activity).
The statistical analysis consisting of main effects, value of coefficients, standard error of coefficient, t and p values of the experimental design, generated by the software is shown in Table 4. From the table, it can be concluded that soyabean meal concentration shows the highest confidence level of 99.9 %. The coefficient of determination (R 2) was calculated as 94.72 %, which indicates good agreement between the experimental and the predicted values. R 2 value is corrected by the adjusted coefficient of determination (adj R 2) for the sample size and the number of terms in the model. The predicted coefficient of determination (pred-R 2) of 83.78 % was in reasonable agreement with adj-R 2 of 91.84 %.
The main effect of each factor can be calculated as difference between both the averages of measurement made at higher and lower levels of corresponding factor. The appropriateness of the model was determined using correlation coefficient and statistically significant effects were screened via Student’s t test for ANOVA.
The levels of significant factors were optimized using central composite experimental design to obtain increased production of rapamycin [41]. By applying the multiple regression analysis (Table 5) on the experimental data, the following second-order polynomial equation was found. The equation explains the effect of screened medium components and their interactions on the production of rapamycin from S. hygroscopicus NRRL 5491:
where X 1 is mannose, X 2 soyabean meal, X 3 l-lysine, and Y is the concentration of rapamycin produced (response activity).
Coefficients having larger magnitude of the t value and a smaller p value can be associated with more significance. Also, p value <0.05 exhibits the significance of coefficient at 5 % confidence level. The results were analyzed using the ANOVA as appropriate to the experimental design used. The statistical analysis shows that the interaction of mannose with soyabean meal (p = 0) and mannose with l-lysine (p = 0.04) was found to be very significant.
The appropriateness of the model was checked by coefficient of determination, R 2, which implies that the sample variation of 98.41 % for rapamycin production is attributed to the medium components and also only 1.59 % of the total variation is not explained by the model. This explains the significance of the model optimization process.
Optimization of rapamycin production by ANN–GA method
The MLP network has three input nodes (L = 3) for representing the three influential process variables (mannose concentration, soyabean meal concentration, l-lysine concentration) and one output node (N = 1) representing the rapamycin concentration (mg/L) at the end of a batch. The process data for MLP-based modeling was generated by carrying out a number of fermentation runs by varying the input conditions. ANN model provided accurate predictions. The data were trained in 1077 epoch with R value of 0.99984 (shown in Fig. 4) and mean square error value of 0.305 (shown in Fig. 5). The results depict that ANN-based training shows better correlation with the experimental rapamycin level compared to that using only RSM regression model.
This trained data’s fitness was evaluated by fitness evaluation function using the GA tool. The parameters taken for the GA optimization were population size of 10, mutation rate of 0.1, and uniform cross overrate of 0.8.
In order to validate the optimization results, five experiments were performed under the predicted optimal conditions. Results of GA-based optimization and their experimental outcomes are shown in Table 6. From these five experiments, the observed experimental production is 320.89 mg/L, very close to the software predicted result of 323.39 mg/L as shown in Fig. 6. We obtained higher concentration of rapamycin produced by S. hygroscopicus NRRL 5491 compared to previous study (Table 1). Thus, the optimal concentrations for the three components obtained from the model are 25.47, 15.39, and 17.48 g/L for mannose, soyabean meal, and l-lysine concentration, respectively. The optimization using ANN model linked with GA is found to be more effective for rapamycin production with a high degree of accuracy.
Batch kinetic studies for rapamycin production using optimized medium
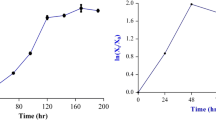
Shake flask studies were conducted in triplicate to evaluate the growth kinetics studies of S. hygroscopicus in the optimized medium. Batch fermentation was performed in flask at temperature 28 °C, pH 6.0, 200 rpm for 6 days. Samples were collected at regular intervals and analyzed for cell mass, product and substrate concentration, and the mean value was observed.
It was observed (Fig. 7) that during the growth of microorganism in production medium, the lag phase continued for 24 h, which was significantly reduced by repetitive subculturing of the organism. There was an exponential increase in the growth rate until 72 h. It was also observed that pH decreases up to third day after which it increased. Increase in pH may be due to the lysis of cells in stationary phase. Maximum specific growth rate (μ max) was observed to be 0.048 h−1.
The substrate consumption profile was evaluated for mannose using di-nitrosalicylic acid (DNS) test which was used to estimate the amount of reducing sugars in the medium [42]. Mannose utilization was rapid in the early growth stage and was completely exhausted after 144 h (Fig. 7).
With an increasing biomass, up to 70 h of fermentation, 90 % substrate was utilized. Biomass yield with respect to substrate utilized (Y x/s) was obtained as 0.410 g cells/g substrate utilized and the specific rate of substrate utilization (q s) was observed to be 0.117 h−1.
It was also observed that product formation in early stage is growth associated and starts with the growth phase and extends to the stationary phase. Hence, the maximum production occurs on the fourth day when cells have entered stationary phase (Fig. 7). The production of rapamycin in growth phase may be due to the lack of some of the nutrients in the medium. A similar result of antibiotic production has been found by Gesheva et al. [43], where maximum production of antibiotic took place at 96 h, but was also produced in the growth phase. This result is also supported by the optimization studies of Sujatha et al. [44], where the variation in the nutrients affected the growth as well as the production of antibiotics by Streptomyces sp. in a proportional manner. The specific rate of product formation (q p) was observed to be 0.1155 h−1. The rapamycin production kinetics may be termed as mixed type where the production of the metabolite continues till the late stationary phase.
Conclusion
Rapamycin, an important immunosuppressant, production was evaluated using process optimization techniques. Efforts were made to increase the microbial production of rapamycin by optimizing one parameter at a time. Production of rapamycin was found to be higher when d-mannose and soyabean meal were taken as carbon and nitrogen sources, respectively, as compared to other carbon and nitrogen sources. Production of rapamycin was found to be 248.71 mg/L on the fourth day of production by S. hygroscopicus NRRL 5491.
l-lysine acts as a precursor for rapamycin production by adding pipecolate moiety to rapamycin ring. It was observed further that l-lysine, at a concentration of 10 g/L, showed an increased production of rapamycin up to 268.87 mg/L, using the one-factor-at-a-time method of process optimization. The production medium was further optimized using computational tools. The significant factors were determined using PB design which was mannose, soyabean meal and l-lysine concentrations. After PB designing, ANN tool was used to optimize further. The input space of ANN model was optimized to obtain maximum production of rapamycin using GA and it was finally observed that 25.47, 15.39, and 17.48 g/L of mannose, soyabean meal, and l-lysine concentration, respectively, were the best suited medium composition as they increased the production up to 320.89 mg/L. Rapamycin concentration obtained by this process optimization method gave a better result, as compared to other studies.
After optimization of production parameters, the growth kinetics, substrate utilization and product formation studies were done with reference to rapamycin production. Maximum specific growth rate was found to be 0.048 h−1. The specific rate of substrate utilization (q s) and specific rate of product formation (q p) were found to be 0.117 and 0.115 h−1, respectively.
References
Vezina C, Kudelski A, Sehgal SN (1975) Rapamycin (AY-22,989), a new antifungal antibiotic. J Antibiot 28:721–726
Martel RR, Klicius J, Galet S (1977) Inhibition of the immune response by rapamycin, a new antifungal antibiotic. Can J Physiol Pharmacol 55:48–51
Durous J, Suffness M (1981) Cancer Treat Rev 8:63–69
Kojima I, Cheng YR, Mohan V, Demain AL (1995) Carbon source nutrition of rapamycin biosynthesis by Streptomyces hygroscopicus. J Ind Microbiol 14:436–439
Gallo R, Padurean A, Jayaraman T, Marx S, Roque M, Adelman S, Chesebro J, Fallon J, Fuster V, Marks A, Badimon JJ (1999) Inhibition of intimal thickening after balloon angioplasty in porcine coronary arteries by targeting regulators of the cell cycle. Circulation 27:2164–2170
Wyzgal J, Paczek L, Senatorski G, Zygier J, Rowinski W, Szmidt J, Perkowska A (2002) Sirolimus rescue treatment in calcineurin-inhibitor nephrotoxicity after kidney transplantation. Transplant Proc 34:3185–3187
Hetzel R, Klein B, Brause M, Westhoff A, Willers R, Sandmann W, Grabensee B (2002) Risk factors for delayed graft function after renal transplantation and their significance for long-term clinical outcome. Transpl Int 15:10–16
Cheng YR, Fang A, Demain AL (1995) Effect of amino acids on rapamycin biosynthesis by Streptomyces hygroscopicus. Appl Microbiol Biotechnol 43:1096–1098
Sehgal SN (1993) Immunosuppressive profile of rapamycin. Ann NY Acad Sci 696:1–8
Gonzalez R, Lakhdar B, Redon P, Potaux L, Cambar J, Aparicio M (1990) Demonstration of direct vasoconstrictive effect of cyclosporin on glomeruli isolated from human kidney. C R Acad Sci III 311:109–114
Cortes J, Haydock SF, Roberts GA, Bevitt DJ, Leadlay PF (1990) An unusually large multifunctional polypeptide in the erythromycin producing polyketide synthase of Saccharopolyspora erythraea. Nature 348:176–178
Donadio S, Staver MJ, McAlpine JB, Swanson SJ, Katz L (1991) Modular organization of genes required for complex polyketide biosynthesis. Science 252:675–679
Paiva NL, Demain AL, Roberts MF (1993) The immediate precursor of the nitrogen-containing ring of rapamycin is free pipecolic acid. Enzym Microb Technol 15:581–585
Sehgal SN, Baker H, Vezina C (1975) Rapamycin (AY-22,989), a new antifungal antibiotic II. Fermentation, isolation and characterization. J Antibiot 28:727–732
Sallam LAR, El-Refai AF, Osman ME, Hamdy AA, Ahmed EM, Mohamed MA (2010) Some Physiological factors affecting rapamycin production by Streptomyces hygroscopicus ATCC 29253. J. Am Sci 6:188–194
Lee MS, Kojima I, Demain AL (1997) Effect of nitrogen source on biosynthesis of rapamycin by Streptomyces hygroscopicus. J Ind Microbiol Biotechnol 19:83–86
Chen Y, Krol J, Sterkin V, Fan W, Yan X, Huang W, Cino J, Julien C (1999) New process control strategy used in a rapamycin fermentation. Process Biochem 34:383–389
Xu ZN, Shen WH, Chen XY, Lin JP, Cen PL (2005) A high-throughput method for screening of rapamycin-producing strains of Streptomyces hygroscopicus by cultivation in 96-well microtiter plates. Biotechnol Lett 27(15):1135–1140
Chen X, Wei P, Fan L, Yang D, Zhu X, Shen W, Xu Z, Cen P (2009) Generation of high-yield rapamycin-producing strains through protoplasts-related techniques. Appl Microbiol Biotechnol 83:507–512
Chen Y, Krol J, Huang W, Cino J, Vyas R, Mirro R, Vaillancourt B (2008) DCO2 on-line measurement used in rapamycin fed-batch fermentation process. Process Biochem 43:351–355
Zhu X, Zhang W, Chen X, Wu H, Duan Y, Xu Z (2010) Generation of high rapamycin producing strain via rational metabolic pathway-based mutagenesis and further titer improvement with fed-batch bioprocess optimization. Biotechnol Bioeng 107:506–515
Fisher S, Sonnenshein A (1991) Control of carbon and nitrogen metabolism in Bacillus subtilis. Annu Rev Microbiol 45:107–135
Vilches C, Mendez C, Hardisson C, Salas JA (1990) Biosynthesis of oleandomycin by Streptomyces antibioticus: influence of nutritional conditions and development of resistance. J Gen Microbiol 136:1447–1454
Plackett RL, Burman JP (1946) The design of optimum multifactorial experiments. Biometrika 33:305–325
Haaland PD (1989) Experimental design in biotechnology. Marcel Dekker, NY
Nandi S, Ghosh S, Tambe SS, Kulkarni BD (2001) Artificial neural-network-assisted stochastic process optimization strategies. AIChE J 47:126–141
Michalewicz Z (1994) Genetic algorithms + data structures = evolution programs, 2nd edn. Springer, New York
Desai KM, Akolkar SK, Badhe YP, Tambe SS, Lele SS (2006) Optimization of fermentation media for exopolysaccharide production from Lactobacillus plantarum using artificial intelligence-based techniques. Process Biochem 41:1842–1848
Martin JF, Demain AL (1980) Control of antibiotic biosynthesis. Microbiol Rev 44:230–251
Kalil SJ, Maugeri F, Rodrigues MI (2000) Response surface analysis and simulation as a tool for bioprocess design and optimization. Process Biochem 35:539–550
Analytical Methods Committee, AMCTB No 55 (2013) Experimental design and optimisation (4): Plackett–Burman designs. Anal Methods 5:1901–1903
Soliman NA, Mahmoud MB, Fattah A (2005) Polyglutamic acid production by Bacillus sp. SAB-26: application of Plackett–Burman experimental design to evaluate culture requirements. Appl Microbiol Biotechnol 69:259–267
Boonkerda S, Detaeverniera MR, Heydenb YV, Vindevogela J, Michottea Y (1996) Determination of the enantiomeric purity of dexfenfluramine by capillary electrophoresis: use of a Plackett–Burman design for the optimization of the separation. J Chromatogr A 736:281–289
Pardeep Kumar, Satyanarayana T (2007) Optimization of culture variables for improving glucoamylase production by alginate-entrapped Thermomucorindicae-seudaticae using statistical methods. Bioresour Technol 98:1252–1259
Smith KA, Gupta J (2002) Neural networks in business: techniques and applications. Idea Group, London
Xu M, Zeng G, Xu X, Huang G, Jiang R, Sun W (2006) Application of Bayesian regularized BP neural network model for trend analysis, acidity and chemical composition of precipitation in North Carolina. Water Air Soil Pollut 172:167–184
Freyer S, Weuster-Botz D, Wandery C (1992) Medium optimisation using genetic algorithms. J Bioeng 8:16–25
Cromwell GL (1999) Soybean meal—the “gold standard” The farmer’s pride. KPPA News 1:20
Kuscer E, Coates N, Challis I, Gregory M, Wilkinson B, Sheridan R, Petkovic H (2007) Roles of rapH and rapG in positive regulation of rapamycin biosynthesis in Streptomyces hygroscopicus. J Bacteriol 189:4756–4763
Yen HW, Hsiao PH, Chen LJ (2013) The enhancement of rapamycin production using Streptomyces hygroscopicus through a simple pH-shifted control. J Taiwan Inst Chem Eng 44:743–747
Khuri AI, Cornell JA (1987) Response surfaces: design and analysis. Marcel Decker, New York
Miller GL (1959) Use of Dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Gesheva V, Ivanova V, Gesheva R (2005) Effects of nutrients on the production of AK-111-81 macrolide antibiotic by Streptomyces hygroscopicus. Microbiol Res 160:243–248
Sujatha P, Bapi Raju KVVSN, Ramana T (2005) Studies on a new marine Streptomycete BT-408 producing polyketide antibiotic SBR-22 effective against methicillin resistant. Staphylococcus aureus 160:119–126
Acknowledgments
The authors would like to acknowledge National Centre for Agricultural Utilization Research (NRRL), USA, for providing lyophilized culture of S. hygroscopicus ATCC 29253.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sinha, R., Singh, S. & Srivastava, P. Studies on process optimization methods for rapamycin production using Streptomyces hygroscopicus ATCC 29253. Bioprocess Biosyst Eng 37, 829–840 (2014). https://doi.org/10.1007/s00449-013-1051-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-013-1051-y