Abstract
Research work was implemented to describe the kinetics of cell growth, substrate utilization and product formation using a mutant strain of Streptomyces hygroscopicus NTG-30-27 in a 3-L bioreactor under optimized condition. Various substrate inhibition mathematical models were applied and it was found that the cell growth and substrate utilization kinetic data fitted well to those models. Andrew’s kinetic model was fitted very well (R2 = 0.998) with our experimental data among different models tested for analysis whereas Luedeking–Piret model suggested that our product is mixed growth associated. The values for maximum specific growth rate (µmax), doubling time (td), saturation constant (KS), inhibition constant (KI) and yield coefficient (YX/S) were found to be 0.03985 h−1, 17.16 h, 2.076 g/l, 0.009 g/l and 0.290 g g−1. Final rapamycin yield with mutant strain was found to be 531.4 mg/l on its 5th day of fermentation which is 6.7-fold higher than the wild type (79.31 mg/l). The effect of aeration on rapamycin production was studied by batch fermentation in a stirred tank reactor (STR) using S. hygroscopicus NTG-30-27. Dynamic behaviour and aeration efficiency of the reactor, as well as rheology pattern of the fermentation broth, were determined by calculating volumetric mass transfer coefficient (KLa) of the process using “Dynamic gassing out method”. KLa was found to be 54.53 h−1 which is quite significant for rapamycin production. Further purification and structural analysis of the extracted sample were carried out by liquid chromatography–mass spectrophotometry (LC–MS) technique in positive ionization mode and molecular mass was found to be 936 D. Finally, 90.62% purified rapamycin was obtained from the study.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Immunosuppressants are finding increased use since 1970s as protective agents against the acute and chronic rejections of organ transplantation—a globally concerned problem in the medical field. Rapamycin (rap), also known as sirolimus is a promising immunosuppressant which has no side effects unlike other macrolides, i.e. cyclosporine (Cohen et al. 1984). However, rapamycin was first reported as an antifungal agent (Vezina et al. 1975) produced by soil-borne actinomycetes Streptomyces hygroscopicus; later on, it was shown to have neurotrophic (Steiner et al. 1997) and potent immunosuppressant activity (Calne et al. 1989). During clinical trials of rapamycin as an anti-transplant rejection drug, antitumor effects were also observed (Douros and Suffness 1981). Khan et al. (2006) suggested the possible assistance of rapamycin in heart attack treatment (Khan et al. 2006). Anti-parasitic effect (Barquilla et al. 2008) and anti-ageing property of rap on mice were also established (Harrison et al. 2009). Studies discovered that rapamycin has excellent anti-Candida property (High 1994). In 1999, it has been approved by the FDA under the brand name Rapamune as an anti-rejection drug in kidney transplantation.
Lung transplantation became safe and easier using rapamycin because of its anti-proliferative properties and low side effect (Wojarski et al. 2018). Researchers have also identified that rapamycin can regulate cholesterol biosynthesis and can also improve spatial learning ability of APP/S1 genetical mice (Wang et al. 2019). Their finding also suggests that rapamycin has potential energy to delay the progression of Alzheimer’s disease. The ability of rapamycin to stop breast cancer has been employed recently by inactivation of mTOR pathway and telomerase–telomere dynamics in human breast cancer cells (Gopalakrishnan et al. 2018). Rapamycin antibiotic production is usually favoured by suboptimal growth conditions (Dutta et al. 2017). A microorganism that stops dividing in the stationary phase produces antibiotic synthetase which catalyses surplus primary metabolites into secondary metabolites (Chater 1984; Riesenberg and Bergter 1979). In summary, cell growth is the fate of two simple biological phenomena, nutrient uptake from the environment and conversion of it into a metabolic end product. A careful and impeccable kinetic study is required to establish the relationship between cell growth, substrate utilization and product formation anomaly. Different kinetic model has been developed with the in-depth knowledge of mathematical equations to meet the requirement for the analysis of experimental outcome with simulated value.
In the current research work, we have incorporated different well-established unstructured kinetic models to define the complex mechanism of rapamycin production in bioreactor by the mutant strain of S. hygroscopicus NTG-30-27. The strain was previously engineered in our lab through random mutagenesis using N-methyl-N-nitro-N-nitrosoguanidine (NTG) as chemical agent (Dutta et al. 2017). Among different models tested for nonlinear regression and analysis of complex kinetic behaviour of the metabolic pathway for rapamycin synthesis, Andrew’s model was found satisfactory and agreed well with our experimental findings. However, nonlinear models are relatively challenging for parameter approximation than generalized linear regression models (Okpokwasili and Nweke 2006). As per the preliminary knowledge, antibiotic is mainly synthesized in stationary phase of growth. Keeping this information in mind, Luedeking–Piret model was incorporated in our study to establish the product formation phenomena. This model was first introduced during lactic acid fermentation by homo-fermentative bacteria Lactobacillus delbrueckii (Luedeking and Piret 1959). Any metabolite or product whether it is growth associated or not can be easily found out from this model. Optimum value of physicochemical factors (temp, agitation, pH) responsible for higher yield of rapamycin by NTG-30-27 has been already carried out and established (Dutta et al. 2017). For bioreactor operation of filamentous microorganism in submerged culture, one very crucial factor for scale-up is the rate of oxygen supply through proper aeration (Song et al. 2017; Wang and Zhang 2007). In this context, determination of volumetric mass transfer coefficient (KLa) is very important as it plays a vital role in scale-up and to determine aeration capacity of a reactor during aerobic fermentation (Puthli et al. 2005). Optimum oxygen concentration throughout the fermentation is essential for effective metabolite production which mainly governs by KLa of the reactor (Stanbury et al. 2013). However, the value of KLa depends on agitation rate, impeller design, aeration rate of the reactor; it is unaffected by dissolved oxygen concentration (DO, mg/l) of the broth (Stanbury et al. 2013). In our experiment, dissolved oxygen concentration was maintained above 30% of air saturation (as low solubility of oxygen in water) during fermentation run. After scale-up in the bioreactor, final rapamycin yield was around 59% higher than that was obtained in shake flask study with NTG-30-27 (Dutta et al. 2017). Further characterization and purification of rapamycin were carried out in liquid chromatography–mass spectroscopy (LC–ESI-MS-TOF) method.
Experimental
Chemicals
All chemicals used for the present research work were of analytical and HPLC grades and purchased from Sigma-Aldrich (presently known as Merck, USA), Himedia (India) and Tokyo Chemical Industries (TCI, India). HPLC-grade (> 95% pure) rapamycin standard (1 mg) was purchased from Cayman Chemical, USA. Deionized water used for HPLC analysis was prepared by Ultrapure Water System (Arium®, 611UF, Sartorius, Germany).
Microorganisms
All the kinetic studies were carried out using chemical mutant of Streptomyces hygroscopicus NTG-30-27 previously generated in our lab (Dutta et al. 2017). Candida albicans MTCC-227 was purchased in a lyophilized vial from microbial type culture collection (MTCC), Institute of Microbial Technology (IMTECH), Chandigarh, India. It was used as a test organism to check antibiotic concentration via disc diffusion method and potency check by turbidimetric assay. S. hygroscopicus NTG-30-27 was grown and maintained in a medium consisting of (g/l) glucose 4; yeast extract 4; malt extract 10; CaCO3 2. The pH of the medium was maintained at 7.2. C. albicans was grown on MYGP medium having the following composition (g/l): malt extract 3; yeast extract 3; peptone 5; glucose 10 (pH 7). Media composition described here is provided with the strain when procured from IMTECH, Chandigarh. The nutrients present in the media were used for growth and subsequent subculturing purpose of the strains (NTG-30-27, MTCC 4003 and MTCC 227).
Methods
Inoculum preparation
Inoculum (seed culture) was prepared by inoculating thawed S. hygroscopicus spore into 250-ml Erlenmeyer flask containing 50 ml growth medium with the help of a sterile inoculating loop under aseptic condition. Flasks were then incubated at 25 °C and 120 rpm for 7 days. S. hygroscopicus and C. albicans were maintained by bimonthly transfer to fresh medium and stored at 4 °C after incubation at 25 °C for 5 days.
Production medium and cultivation in bioreactor
After adequate growth of mycelium, vegetative cells of S. hygroscopicus NTG-30-27 were transferred (5% v/v) to production medium for generating a maximum yield of rapamycin. Kinetic study of rapamycin production including determination of KLa was carried out in 3-L stirred tank reactor (Lark hygiene, India) with 2.2 L working volume and Lark innovative-data scanner software for advanced online real-time monitoring of fermentation conditions. Optimized media components for rapamycin production in bioreactor were as follows (g/l): fructose 25, mannose 5, l-lys HCl 7.5, (NH4)2SO4 2.5, K2HPO4 2, NaCl 2.5, CaCO3 3 and Na2SO4 0.35 with supplementation of sterile trace element solution (1×). Physicochemical parameters for rapamycin production inside the reactor were pH 7.6, temperature 23 °C, agitation 300 rpm and air flow rate of 1 LPM (~ 0.8 vvm). Standard calibration of pH probe was carried out before autoclaving the reactor with suitable buffer solutions (pH 4, pH 7 and pH 9) and was set to be 7.6 whereas dissolve oxygen (DO) probe (Mettler Toledo®) was calibrated after autoclaving and set value was 100%. pH was maintained constant at set point 7.6 by supplying 2(N) NaOH and 1(N) HCl automatically inside the reactor through peristaltic pumps. “dead band” value for pH was set at 1. As reported, “P band” value is around 10% of dead band value, hence the pH of fermentation media was always maintained between 7.5 and 7.7 (Gerson et al. 1988). DO value was controlled via constant agitation by six-bladed turbine impellers and supply of compressed sterile air through multiple point sparger inside the vessel for 10 days at 1 LPM.
Kinetic study of rapamycin production
Monod’s kinetic model relates growth rate with the concentration of a single growth-limiting substrate over the parameters µmax and KS. It also provides the idea of yield coefficient (Yx/s) to the µ (Okpokwasili and Nweke 2006). Among the substrate inhibition models prescribed by the researchers, Andrew’s equation is widely used (Okpokwasili and Nweke 2006; Tan et al. 1996). This model was used for simulation study to fit the experimental data. Among several kinetic models studied, the Andrew’s model was best fitted and gave highest R2 value.
Product formation relationship can be established using Luedeking–Piret model. It is a globally accepted model for the determination of growth-associated, non-growth-associated as well as mixed growth-associated product (Ozergin-Ulgen and Mavituna 1993). The model says product formation rate (dp/dt) depends on both the rapid biomass concentration (X, g/l) and the growth rate (dx/dt) in a linear fashion (Lu et al. 2011a). The expression for mixed growth-associated product is
where \(\alpha\) and \(\beta\) are Luedeking–Piret model kinetic constants which vary with fermentation dynamics. Dividing both sides of Eq. (2) by ‘x’ yields
i.e. \(\frac{1}{x}\left( {\frac{{{\text{d}}p}}{{{\text{d}}t}}} \right) = \alpha \mu + \beta ;\) (3)by plotting experimental values of \(\frac{1}{x}\left( {\frac{{{\text{d}}p}}{{{\text{d}}t}}} \right)\) against µ, a straight line curve is being generated. \(\alpha\) is the slope of that curve and \(\beta\) will be the intercept (Luedeking and Piret 1959). The value of \(\beta\) can also be generated from stationary phase data using the following equation:
where \(x_{\text{m}}\) is maximum attainable biomass (when dx/dt = 0, at stationary phase) (Elibol and Mavituna 1999; Lu et al. 2011a).
Analytical procedure
Rapamycin extracted from fermentation broth was checked via “Agar disc diffusion method” for preliminary confirmation (Abdel-Fattah 2008; Dutta et al. 2014; Kojima et al. 1995). Further validation was carried out by High-performance liquid chromatography (HPLC) technique (Dutta et al. 2014; Dutta et al. 2017). Amount of total carbohydrate or residual sugar was estimated by phenol–sulfuric acid method (Nielsen 2015). During fermentation in shake flask, the microbial growth under the submerged conditions appeared as spherical pellets. Hence, after inoculation, 50 ml of sample was taken from shake flask in 50-ml centrifuge tube at every 24-h interval starting from 0th hr and centrifuged at 3500 rpm for 15 min at 4 °C (Cheng et al. 1995). After centrifugation, the supernatant obtained was taken in a separate centrifuge tube, and the cell pellet was washed twice with 3 ml of methanol by centrifuging at 200 rpm for 15 min. For determination of cell mass, before every sampling, 50-ml empty falcon tubes were kept in hot air oven (Khera Instrument, Model: KI-26) at 80 °C for 48 h to make constant weight and also to minimize the error. Luedeking–Piret model was used to describe the rapamycin production kinetics in bioreactor (Elibol and Mavituna 1999; Lu et al. 2011a; Luedeking and Piret 1959). Volumetric oxygen transfer coefficient (KLa) was calculated using “Dynamic gas in-gas out method” (Bandyopadhyay et al. 1967). Structural analysis of standard rapamycin, as well as extracted sample, was carried out by mass spectroscopy-ESI-TOF (Maxis impact, Bruker Daltonics, USA) method.
Purification and structural prediction of rapamycin by LC–MS study
Around 750 µl of fermented broth was taken in a microcentrifuge tube (2 ml). Mycelium separation was initially carried out by standard filtration technique. After that, equal volume (750 µl) of HPLC-grade methanol was added to it, since rapamycin is highly soluble in methanol (~ 25 mg/ml). The mixture was vortexed vigorously and centrifuged at 14,000 rpm for 20 min at 4 °C. Following centrifugation, methanol was separated by passing the clear supernatant through methanol-absorbing HPLC-grade PTFE filter (0.22 micron pore size). The final volume of the extract was around 550 µl (0.55 ml), as methanol and little amount of water were absorbed by PTFE filter.
Structural integrity and fold of purification of rapamycin was determined by mass spectroscopy-electrospray ionization (MS-ESI-TOF) method. Initial purification of rapamycin was carried out in HPLC and the purified sample was injected into mass spectrometer (LC–ESI-MS-QTOF, Maxis Impact, Bruker Daltonics). Around 200 µl of purified sample was injected through auto-injector (Kd Scientific, USA). Result was analysed by compass data analysis software whereas online mass peak data acquisition was carried out by “Otof control”. Smart formula of all the related masses can also be generated with the software. Standard rapamycin sample (1 mg, > 99.999% purified, Sigma-Aldrich) was injected into the instrument followed by sample extract of 5th and 6th day of fermentation. Total run time for both the culture was around 2 min. Data scan started at 80 m/z (where m is the molecular mass of analyte and z is the net charge) and continued up to 1300 m/z to determine the most appropriate parent ion for rapamycin (Earla et al. 2012). Dry heater temperature was set at 180 °C and ionization polarity was positive throughout the analysis.
Results and discussion
Kinetic study of rapamycin in bioreactor using potent mutant strain S. hygroscopicus NTG-30-27
The objective of the research work was to study the effect of some factors on the growth kinetics of mutant strain S. hygroscopicus NTG-30-27 in bioreactor. Major factors that influence the microbial growth are sugar concentration, temperature, pH and oxygen transfer rate. The effect of sugar concentration, temperature and pH were studied extensively in shake flask where the determination of oxygen transfer rate was carried out in laboratory-scale fermenter. Growth and production kinetic studies were carried out with parameters optimized previously in our lab (Dutta et al. 2017).
Cell growth kinetics
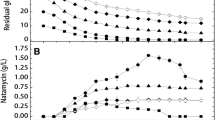
The specific growth rate (µ) was determined from the slope of the regression line of the plot of ln(Xt/Xo) vs t during the exponential phase noting that µ remains constant during the exponential growth (Jagannath and Ramachandran 2010). µ was calculated experimentally, and the value was found to be 0.04 h−1 as depicted in Fig. 1. It is observed that specific growth rate increases with residual substrate concentration up to a certain value beyond which it starts to decrease. Analysis of specific growth rate under different substrate concentration conditions suggested that it regulates the growth rate of microorganism. Various unstructured kinetic models were tested and compared in this study to find out the best possible model for the analysis of S. hygroscopicus growth pattern. Models used in this study were Monod, Andrews, Aiba, etc. Experimental data have been fitted to different unstructured models and were analysed using GraphPad Prism 6 software.
Monod and Andrew’s kinetic models
The specific growth rate during fermentation time interval has been fitted initially to Monod equation (Monod, 1949) to generate kinetic parameters, namely maximum specific growth rate (µmax) and saturation constant (Ks) and is shown in Fig. 2. \(\mu = \frac{{\mu_{\hbox{max} } S}}{{K_{\text{S}} + S}},\) where µmax is the maximum specific growth rate (hr−1), Ks is the saturation constant (g/l) in Monod equation, S is residual substrate concentration (g/l) and µ is specific growth rate (hr−1). The values of kinetic parameters such as µmax and Ks were found to be equal to 0.03096 h−1 and 0.5329 g/l, respectively. The correlation coefficient (R2) was found to be 0.7674. Lower correlation coefficient value of Monod model using experimental data might be due to either product or substrate inhibition. Very few substrate inhibition models are reported in the literature for rapamycin production; hence, effect of substrate inhibition is only used for simulation purpose (Dutta et al. 2014; Schindler and Zäuhner 1973). Andrew’s model was found satisfactory as the experimental data fitted well in the model equation (Eq. 1) with higher regression coefficient (R2 = 0.9981) as indicated in Fig. 3.
Value of inhibition constant (Ki) was found to be very low (Ki~ 0.009 g/l) and thus can be neglected. Lower Ki value (Ki~ 0.00534 M) was also reported in the literature for chlorothricin production by Bacillus stearothermophilus (Schindler and Zäuhner 1973).
Different biokinetic parameters, i.e. maximum specific growth rate (µmax), doubling time (td) and saturation constant (Ks) were found to be 0.03985 h−1, 17.39 h and 2.076 g/l, respectively. From the data obtained, it is quite interesting that the value of specific growth rate for Andrew’s model (0.039 h−1) agreed well with our experimental µ (0.04 h−1).
Similar kinetic study of rapamycin production was carried out with wild-type Streptomyces hygroscopicus MTCC 4003 in a 3-L fermenter (working volume 2.2 L) (Dutta et al. 2014). For wild-type strain, Andrew’s model was also found to fit the experimental data well since R2 value was obtained around 0.9849 (Dutta et al. 2014). Values of μmax, KS and Ki were found to be 0.0083 h−1, 2.835 g/l and 0.073 g/l, respectively. It was found that specific growth rate of wild-type organism was very low (µmax= 0.008 h−1) with doubling time of 86.625 h. It can be easily predicted that low cell activity during the end of fermentation time can be one possibility of slower growth and lower production of rapamycin by wild-type organism (Lu et al. 2011b). The average YX/S value for wild type was calculated to be 0.107 g g−1 whereas for mutant strain it was around 0.290 g g−1.
Rapamycin production kinetics
The fermenter was operated with a constant impeller speed of 300 rpm for 8 consecutive days. Dissolved oxygen (DO) plays a vital role in rapamycin production. The air flow rate was kept constant at 1 LPM throughout the fermentation process. Initially, DO was set at 100% at the time of inoculation, while during the lag phase microorganisms did not start to utilize the nutrient source present the media. However, when the fermentation entered log phase, dissolved oxygen concentration rapidly decreased as a result of increase in oxygen demand by the cells. Therefore, the dissolved oxygen level drops and finally reached a certain stationary value. Subsequently, the DO level was maintained above 30% of saturation level all the time. Final rapamycin yield was found to be 531.4 mg/l on its 5th day of fermentation. Around 1.68-fold (from 315 mg/l to 531 mg/l) higher yield was obtained in the reactor than shake flask study (Dutta et al. 2017). Higher production of rapamycin by mutant strain than wild type may be possible due to increased activity of enzymes responsible for antibiotic production upon UV irradiation or using chemical mutagen (Lu et al. 2011b). This can also be attributed to the fact that higher transcription and translation may also be responsible for enhanced macrolide yield (Lu et al. 2011b). Combined kinetic study of growth, substrate utilization as well as production of rapamycin by mutant strain of S. hygroscopicus NTG-30-27 is described in Fig. 4. It clearly shows that the exponential phase of growth is very crucial for rapamycin production (Sánchez and Brana 1996; Zhu et al. 2010). The synthesis of rapamycin was noticed after 48 h of incubation and reached a maximum at 120 h. Similar trends were also reported in the literature (Lee et al. 1997; Xu et al. 2005). Rapamycin production was observed to increase rapidly during the end of the exponential phase and early stationary phase (Xu et al. 2005).
Product formation kinetics
Luedeking–Piret model was used for the determination of growth-associated (\(\alpha\)) and non-growth-associated (\(\beta\)) product formation coefficients during rapamycin production. Globally, this model is already used for different antibiotic production, i.e. daptomycin from Streptomyces roseosporus and actinorhodin from Streptomyces coelicolor [180, 209]. Value of \(\alpha\) for both the cases was found to be “non-zero” as depicted in Fig. 5 which indicates that antibiotic production is growth associated. The result is in good accordance with rapamycin production by wild-type strain which reports that rapamycin synthesis started on exponential phase and continued up to late stationary phase (Dutta et al. 2014). Value of specific production rate (qp, mg/g-hr−1) was plotted against specific growth rate (µ) and the equation obtained is Y = 35.03X + 0.0512 with regression coefficient (R2) of 0.9942. \(\alpha\) was calculated from the slope of the graph whereas the intercept value indicated \(\beta\). Figure 5 clearly indicates that rapamycin production is half growth associated with alpha and beta values of 35.03 mgrap/gcell-hr and 0.0512 mgrap/gcell-hr−1, respectively. In case of pleuromutilin production, \(\alpha\) value was found to be 161.04 mg-product/g-cell/hr, which indicates that antibiotic production is growth associated (Benkortbi et al. 2007). The overall pattern of experimental values with simulated model indicates that Andrew’s and Luedeking–Piret models described the mechanism of cell growth, substrate inhibition and product formation phenomena satisfactorily. Table 1 represents all biokinetic parameters for mutant strain obtained from the present research work with their level of significance.
Determination of volumetric mass transfer coefficient (K L a)
KLa determination was carried out using dynamic gassing out method with NTG-30-27 mutant organism in Lark hygiene 3-L fermenter. We have used integral as well as differential methods for the calculation of volumetric mass transfer coefficient obtained from the fermentation run. Unlike bacterial growth, S. hygroscopicus growth rate is meagre as observed from our kinetic analysis. Hence, oxygen consumption rate was also very low just after the inoculation of active culture in the reactor. CL value was measured automatically using a chart recorder in every 30-min interval during “air-off” condition. A linear fall of dissolved oxygen concentration was observed up to 300 min, where the O2 tension was just above critical oxygen concentration (~ 30%) of the broth. The digital signal value of dissolved oxygen (DO) was converted to ppm by the following formula:
where C* is the saturation oxygen concentration in the broth (~ 8 mg/l), CL is the oxygen concentration at any point of time during de-oxygenation step and CL0 is the oxygen concentration at time ‘t’ = 0. At the beginning of experiment, CL0 = CL = C* = 8 mg/l.
Table 2 describes the change in dissolved oxygen concentration (mg/l) with time during “air-off” condition. CL values were plotted against time, and QO2X was calculated from the slope of the graph which was found to be 0.0079 g/l-s−1 as illustrated in Fig. 6.
Re-aeration was started after 300 min and continued for 2 h until the value reached to saturation state. Sufficient aeration and agitation was needed for KLa estimation. However, very high aeration can increase shear force on the cell which may result in reduced antibiotic titer (Yinliang et al. 1999). KLa was calculated by plotting CL against (dCL/dt + QO2X) at different time intervals. A linear relationship was obtained, i.e. Y = − 1.1002X + 7.9946 with higher regression coefficient (R2) of 0.97. Reciprocal of the slope is around 0.909 min. Hence, KLa value was found to be 54.31 h−1 at 300 rpm as depicted in Fig. 7.
To the best of our knowledge, a very few study was conducted worldwide for the determination of KLa during rapamycin production using S. hygroscopicus in bioreactor. It has been noticed that during biosurfactant production by Streptomyces sp. R1, researchers have obtained KLa around 50.94 h−1 in a 3-L fermenter which is quite supportive for this study (Zambry et al. 2017). Hence, it is quite apparent that agitation at 250–400 rpm is desirable for higher antibiotic production. Cruz et al. (1999) used a neural network technique to determine KLa for cephalosporin antibiotic production by Cephalosporium acremonium. They have estimated KLa to be around 69.92 h−1 in a 5-L fermenter (Cruz et al. 1999). Air flow rate was around 0.8–1.0 vvm throughout the whole fermentation run which was found to be significant for higher rapamycin yield. Oxygen transfer rate can be considered as one of the important parameters for maximal production of rapamycin in batch STR operation and can be controlled by employing both aeration and agitation for better product formation. Scale-up studies are performed for the measurement of KLa from shake flask to laboratory-scale bioreactor and from lab-scale bioreactor to pilot plant, and successively to industrial level. As dissolved oxygen is the rate-limiting factor because of its less solubility in the aqueous solutions, it affects the cell growth and concentration of products in the aerobic fermentation (Çalik et al. 2003).
Purification and characterization of rapamycin
To the best of our knowledge, effective purification and characterization of rapamycin is necessary for some specific applications (kidney transplantation, injecting into mice model, immunosuppressant, etc.).
After HPLC analysis, 531.4 mg/l of rapamycin concentration was obtained with mutant strain S. hygroscopicus NTG-30-27. However, rapamycin concentration in crude extract was found to be 430 mg/l. Hence, after extraction, amount of pure rapamycin present in 550 µl sample was around
However, before methanol extraction, amount of rapamycin present was
Hence, the % of purification is \(\left( {\frac{0.29227}{0.3225}} \right) \times 100\% = 90.62\% .\).
Characterization of rapamycin
LC–MS study was carried out for the determination of structural integrity and analysis of molecular mass for standard as well as extracted rapamycin. Both the samples showed a dominant peak at molecular mass of 936 D. After analysing molecular mass, it can be predicted that in both the cases (standard and sample), rapamycin was detected in the ESI-TOF as sodium adduct ion [M+ Na+] (Earla et al. 2012; Rosu et al. 2006; Vidal et al. 1998). Use of sodium acetate buffer as LC mobile phase is advantageous because it never clogs the peak tubes and aseptic seals in the column (Earla et al. 2012). Also, they are more stable in positive ionization mode. It can be a possible cause for sodium adduct peak in rapamycin chromatogram as depicted in Fig. 8. Subtracting 23 amu from 936 D gives our product of interest, i.e. rapamycin (C51H79NO13 ~ 914 D).
Conclusion
Rapamycin is produced by different strains of Streptomyces hygroscopicus around the world and its low yield limits the large-scale industrial production. Therefore, proper knowledge of nutritional requirements and kinetic behaviour of the organism needs to be studied thoroughly for the maximum output of this immunosuppressant drug. In the present research work, enhanced production of rapamycin followed by its kinetics, purification and characterization study by the mutated strain of Streptomyces hygroscopicus NTG-30-27 were investigated.
To generate higher rapamycin yield, an experiment was carried out in a 3-L fermenter with S. hygroscopicus NTG-30-27 mutant strain. Final rapamycin yield was found to be 531.4 mg/l on its 5th day of fermentation which is 6.7-fold higher than the wild-type strain S. hygroscopicus MTCC 4003. Kinetic model analysis was carried out to define various biokinetic parameters obtained during rapamycin production. An extensive investigation was carried out to describe the kinetics of cell growth, substrate utilization and product formation in a batch reactor system. Evaluation of biokinetic parameters was carried out using best fit unstructured model. Nonlinear regression analysis was taken into consideration for computational purpose. Andrew’s kinetic model showed comparatively better R2 value among all tested models. The values of maximum specific growth rate (µmax), doubling time (td), saturation constant (KS), inhibition constant (KI) and yield coefficient (YX/S) were found to be 0.03985 h−1, 17.16 h, 2.076 g/l, 0.009 g/l and 0.290 g g−1. Product formation pattern of rapamycin was analysed by Luedeking–Piret model. Non-zero constant values signify that rapamycin is a mixed growth-associated product. KLa value 54.53 h−1 suggests that sufficient aeration, agitation and mass transfer took place during the fermentation process in the bioreactor. Purification of extracted sample was carried out by High-performance liquid chromatography (HPLC) followed by mass spectroscopy (MS) analysis. Analysis of the chromatogram suggested that extracted rapamycin is sufficiently pure and no primary degradation product was found. Finally, around 90.62% of pure rapamycin was obtained from the study with a molecular mass of 936 D. Although very few literature studies are reported on S. hygroscopicus kinetic study, we have compared and validated our data with the available findings.
In the present research work, sufficient effort has been given to justify the research gaps on bioengineering aspects of rapamycin production. As a promising immunosuppressant and potent multifunctional drug, scale-up study in a large-scale fermenter using cheap raw material will be very helpful to produce it in a cost-effective manner.
Abbreviations
- dP/dt :
-
Production rate (mg/l-hr−1)
- K i :
-
Inhibition coefficient (g/l)
- K L a :
-
Volumetric mass transfer coefficient (hr−1)
- K S :
-
Saturation constant (g/l)
- M :
-
Cell maintenance coefficient
- P 0, P, P max :
-
Product concentration at 0th h of fermentation (mg/l), product concentration at any time of fermentation (mg/l), maximum product concentration at particular fermentation time (mg/l)
- R 2 :
-
Regression coefficient
- S 0, S :
-
Initial substrate concentration (g/l), limited substrate concentration (g/l)
- t :
-
Incubation time (hr)
- t d :
-
Doubling time of cell (hr)
- LPM:
-
Liter per minute
- mTOR:
-
Mechanistic target of rapamycin
- vvm:
-
Volume of air per volume of fermentation media per minute (m3)
- X 0, X, X max :
-
Initial cell mass concentration (g/l), cell mass concentration at any time of fermentation (g/l), maximum attainable call mass concentration (g/l)
- Y X/S :
-
Growth yield coefficient per unit substrate consumed (g/g)
- µ, µ max :
-
Specific growth rate (hr−1), maximum specific growth rate (hr−1)
- α :
-
Growth-associated product formation coefficient (mg/g)
- β :
-
Non-growth-associated product formation coefficient (mg/g-hr−1)
References
Abdel-Fattah YR (2008) Non-conventional method for evaluation and optimization of medium components for rapamycin production by Streptomyces hygroscopicus. Res J Microbiol 3:405–413
Bandyopadhyay B, Humphrey AE, Taguchi H (1967) Dynamic measurement of the volumetric oxygen transfer coefficient in fermentation systems. Biotechnol Bioeng 9:533–544
Barquilla A, Crespo JL, Navarro M (2008) Rapamycin inhibits trypanosome cell growth by preventing TOR complex 2 formation. Proc Natl Acad Sci 105:14579–14584
Benkortbi O, Hanini S, Bentahar F (2007) Batch kinetics and modelling of pleuromutilin production by Pleurotus mutilis. Biochem Eng J 36:14–18
Çalik P, Bilir E, Çalik G, Özdamar TH (2003) Bioreactor operation parameters as tools for metabolic regulations in fermentation processes: influence of pH conditions. Chem Eng Sci 58:759–766
Calne R, Lim S, Samaan A, Collier DSJ, Pollard S, White D, Thiru S (1989) Rapamycin for immunosuppression in organ allografting. Lancet 334:227
Chater KF (1984) Morphological and physiological differentiation in Streptomyces. Cold Spring Harbor Monogr Arch 16:89–115
Cheng YR, Hauck L, Demain AL (1995) Phosphate, ammonium, magnesium and iron nutrition of Streptomyces hygroscopicus with respect to rapamycin biosynthesis. J Ind Microbiol 14:424–427
Cohen DJ, Loertscher R, Rubin MF, Tilney NL, Carpenter CB, Strom TB (1984) Cyclosporine: a new immunosuppressive agent for organ transplantation. Ann Intern Med 101:667–682
Cruz AJG, Silva AS, Araujo MLGC, Giordano RC, Hokka CO (1999) Estimation of the volumetric oxygen transfer coefficient (K L a) from the gas balance and using a neural network technique. Br J Chem Eng 16:179–183
Douros J, Suffness M (1981) New antitumor substances of natural origin. Cancer Treat Rev 8:63–87
Dutta S, Basak B, Bhunia B, Chakraborty S, Dey A (2014) Kinetics of rapamycin production by Streptomyces hygroscopicus MTCC 4003. 3 Biotech 4:523–531. https://doi.org/10.1007/s13205-013-0189-2
Dutta S, Basak B, Bhunia B, Sinha A, Dey A (2017) Approaches towards the enhanced production of Rapamycin by Streptomyces hygroscopicus MTCC 4003 through mutagenesis and optimization of process parameters by Taguchi orthogonal array methodology. World J Microbiol Biotechnol 33:90
Earla R, Cholkar K, Gunda S, Earla RL, Mitra AK (2012) Bioanalytical method validation of rapamycin in ocular matrix by QTRAP LC–MS/MS: Application to rabbit anterior tissue distribution by topical administration of rapamycin nanomicellar formulation. J Chromatogr B 908:76–86
Elibol M, Mavituna F (1999) A kinetic model for actinorhodin production by Streptomyces coelicolor A3 (2). Process Biochem 34:625–631
Gerson DF, Kole MM, Ozum B, Oguztoreli MN (1988) Substrate concentration control in bioreactors. Biotechnol Genet Eng Rev 6:67–150
Gopalakrishnan K, Venkatesan S, Low ESH, Hande M (2018) Effects of rapamycin on the mechanistic target of rapamycin (mTOR) pathway and telomerase in breast cancer cells. Mutat Res 836:103–113
Harrison DE et al (2009) Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460:392–395
High KP (1994) The antimicrobial activities of cyclosporine, FK506, and rapamycin. Transplantation 57:1689–1700
Jagannath S, Ramachandran KB (2010) Influence of competing metabolic processes on the molecular weight of hyaluronic acid synthesized by Streptococcus zooepidemicus. Biochem Eng J 48:148–158
Khan SA, Salloum F, Das A, Xi L, Vetrovec GW, Kukreja RC (2006) Rapamycin confers preconditioning-like protection against ischemia–reperfusion injury in isolated mouse heart and cardiomyocytes. J Mol Cell Cardiol 41:256–264
Kojima I, Cheng YR, Mohan V, Demain AL (1995) Carbon source nutrition of rapamycin biosynthesis in Streptomyces hygroscopicus. J Ind Microbiol 14:436–439
Lee MS, Kojima I, Demain AL (1997) Effect of nitrogen source on biosynthesis of rapamycin by Streptomyces hygroscopicus. J Ind Microbiol Biotechnol 19:83–86
Lu W, Fan J, Wen J, Xia Z, Caiyin Q (2011) Kinetic analysis and modeling of daptomycin batch fermentation by Streptomyces roseosporus. Appl Biochem Biotechnol 163:453–462
Luedeking R, Piret EL (1959) A kinetic study of the lactic acid fermentation. Batch process at controlled pH. Biotechnol Bioeng 1:393–412
Nielsen S (2015) Food analysis laboratory manual, 2nd edn. Springer, US, pp 47–53
Okpokwasili GC, Nweke CO (2006) Microbial growth and substrate utilization kinetics. Afr J Biotechnol 5:305–317
Ozergin-Ulgen K, Mavituna F (1993) Actinorhodin production by Streptomyces coelicolor A3 (2): kinetic parameters related to growth, substrate uptake and production. Appl Microbiol Biotechnol 40:457–462
Puthli MS, Rathod VK, Pandit AB (2005) Gas–liquid mass transfer studies with triple impeller system on a laboratory scale bioreactor. Biochem Eng J 23:25–30
Riesenberg D, Bergter F (1979) Dependence of macromolecular composition and morphology of Streptomyces hygroscopicus on specific growth rate. Z allg Mikrobiol 19:415–430
Rosu F, Pirotte S, De Pauw E, Gabelica V (2006) Positive and negative ion mode ESI-MS and MS/MS for studying drug–DNA complexes. Int J Mass Spec 253:156–171
Sánchez L, Brana AF (1996) Cell density influences antibiotic biosynthesis in Streptomyces clavuligerus. Microbiology 142:1209–1220
Schindler PW, Zäuhner H (1973) Mode of action of the macrolide-type antibiotic, chlorothricin. FEBS J 39:591–600
Song X, Zhang Y, Xue J, Li C, Wang Z, Wang Y (2017) Enhancing nemadectin production by Streptomyces cyaneogriseus ssp. noncyanogenus through quantitative evaluation and optimization of dissolved oxygen and shear force. Bioresour technol 255:180–188
Stanbury PF, Whitaker A, Hall SJ (2013) Principles of fermentation technology. Elsevier, Oxford, UK
Steiner JP, Connolly MA, Valentine HL, Hamilton GS, Dawson TM, Hester L, Snyder SH (1997) Neurotrophic actions of nonimmunosuppressive analogues of immunosuppressive drugs FK506, rapamycin and cyclosporin A. Nat Med 3:421–428
Tan Y, Wang Z, Marshall KC (1996) Modeling substrate inhibition of microbial growth. Biotechnol Bioeng 52:602–608
Vezina C, Kudelski A, Sehgal S (1975) Rapamycin (AY-22, 989), a new antifungal antibiotic. J Antibiot 28:721–726
Vidal C, Kirchner G, Sewing KF (1998) Structural elucidation by electrospray mass spectrometry: an approach to the in vitro metabolism of the macrolide immunosuppressant SDZ RAD. J Am Soc Mass Spectrom 9:1267–1274
Wang YH, Zhang X (2007) Influence of agitation and aeration on growth and antibiotic production by Xenorhabdus nematophila. World J Microbiol Biotechnol 23:221–227
Wang X, Xia W, Li K, Zhang Y, Ge W, Ma C (2019) Rapamycin regulates cholesterol biosynthesis and cytoplasmic ribosomal proteins in hippocampus and temporal lobe of APP/PS1 mouse. J Neurol Sci. https://doi.org/10.1016/j.jns.2019.02.022
Wojarski J, Zeglen S, Ochman M, Karolak W (2018) Safety of Early Sirolimus Immunosuppression in Lung Transplantation. J Heart Lung Transplant 37:S455
Xu Z, Shen W, Chen X, Lin J, Cen P (2005) A high-throughput method for screening of rapamycin-producing strains of Streptomyces hygroscopicus by cultivation in 96-well microtiter plates. Biotechnol Lett 27:1135–1140
Yinliang C, Jeffrey K, Weimin H, Julia PC (1999) New process control strategy used in a rapamycin fermentation. Process Biochem 34:383–389
Zambry NS, Ayoib A, Noh NAM, Yahya ARM (2017) Production and partial characterization of biosurfactant produced by Streptomyces sp. R1. Bioprocess Biosyst Eng. https://doi.org/10.1007/s00449-017-1764-4
Zhu X, Zhang W, Chen X, Wu H, Duan Y, Xu Z (2010) Generation of high rapamycin producing strain via rational metabolic pathway based mutagenesis and further titer improvement with fed batch bioprocess optimization. Biotechnol Bioeng 107:506–515
Acknowledgements
This work was financially supported by the Council of Scientific and Industrial Research (CSIR), Ministry of Science and Technology, Govt. of India [Grant no.: 09/973(0012)/2014 EMR-I].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Dutta, S., Bhunia, B., Raju, A. et al. Enhanced rapamycin production through kinetic and purification studies by mutant strain of Streptomyces hygroscopicus NTG-30-27. Chem. Pap. 73, 2053–2063 (2019). https://doi.org/10.1007/s11696-019-00767-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11696-019-00767-0